Copyright
©The Author(s) 2015.
World J Hepatol. Apr 28, 2015; 7(6): 859-873
Published online Apr 28, 2015. doi: 10.4254/wjh.v7.i6.859
Published online Apr 28, 2015. doi: 10.4254/wjh.v7.i6.859
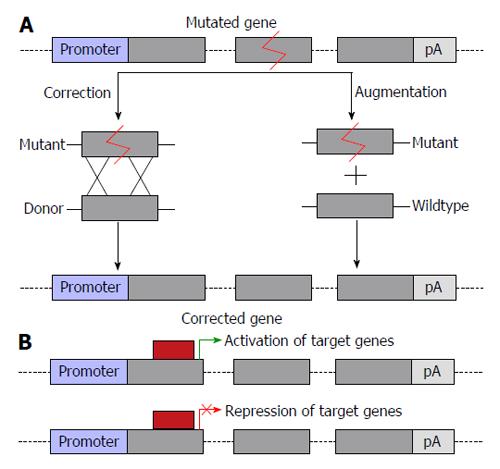
Figure 1 Potential outcomes of gene therapy.
A: Correction of mutations or augmentation of gene function by introduction of a second copy of the gene of interest into a safe harbor locus; B: Changes to transcriptional regulation of a gene of interest, allowing for correction of gene function either by transcriptional activation or suppression. The grey boxes represent exons and linking lines represent introns.
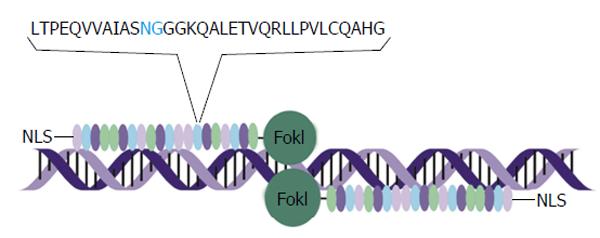
Figure 2 Schematic representation of transcription activator-like effector nuclease interaction with DNA.
A pair of transcription activator-like effector (TALE) nucleases interacts with its target through a tandem arrangement of DNA-binding modules, represented by oval structures in the schematic. Each DNA-binding module comprises 34 amino acids, with a repeat variable diresidue (RVD), in blue, at amino acids 12 and 13. The RVD specifies the DNA base to be targeted. There is a nuclear localisation signal on the N-terminal and a FokI effector bound to the C-terminal. As well as FokI nuclease domains, TALEs may also be bound to transcriptional regulators such as Krüppel-associated box or VP16/64. NLS: Nuclear localisation signal.
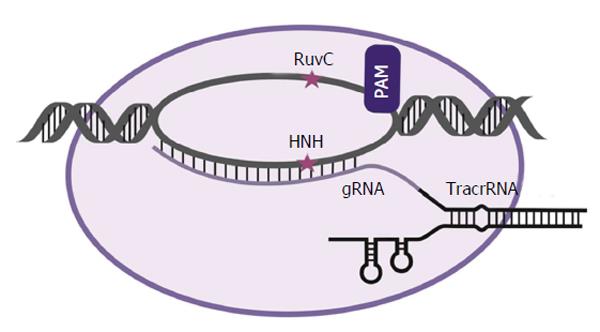
Figure 3 Schematic representation of the clustered regularly interspaced palindromic repeats/clustered regularly interspaced palindromic repeats associated 9 complex and its interaction with DNA.
With the CRISPR/Cas system, a 20 nucleotide guide RNA is processed and bound to the Cas9 protein. When adjacent to a proto-spacer–associated motif element, the guide recognises and binds a complementary sequence in the DNA to direct target cleavage by the Cas9 protein. A catalytically inactive Cas9 protein may also be coupled to other effectors such as Krüppel-associated box or VP16/64 domains. CRISPR/Cas: Clustered regularly interspaced palindromic repeats/CRISPR associated. CRISPR: Clustered regularly interspaced palindromic repeats; Cas: CRISPR associated; tracrRNA: Trans-activating CRISPR RNA; gRNA: RNA guide.
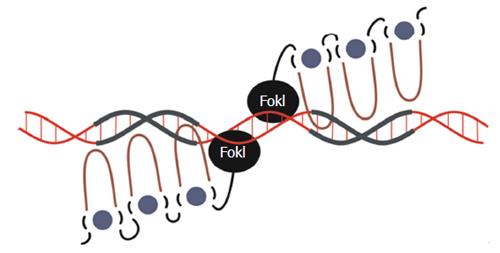
Figure 4 Schematic representation of zinc finger nuclease interactions with DNA.
Each zinc finger interacts with 3-4 bp in the major groove of the DNA double helix. The Hys2Cys2 side chains (depicted as the loop) bind to the DNA, which is facilitated by the interaction between the zinc finger protein (ZFP) and a Zinc ion (depicted as green circle) which stabilizes the protein structure. As well as FokI nuclease domains, ZFPs may also be bound to other effectors such as Krüppel-associated box or VP16/64.
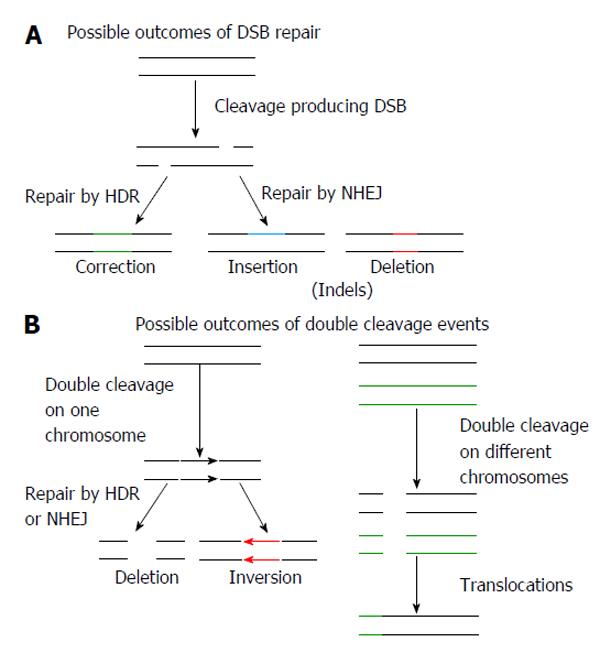
Figure 5 Potential effects of using genome editing endonucleases.
A: Nuclease induced double strand breaks (DSB) may be repaired by homology directed repair (HDR), resulting in corrections that may be to a single base pair or thousands of base pairs. Repair by non-homologous end joining (NHEJ) may result in non-specific insertions and deletions (indels); B: The introduction of two nucleases targeting the same chromosome may result in large deletions or inversions of the DNA. Chromosomal translocations may occur when two DSBs are introduced on different chromosomes.
- Citation: Nicholson SA, Moyo B, Arbuthnot PB. Progress and prospects of engineered sequence-specific DNA modulating technologies for the management of liver diseases. World J Hepatol 2015; 7(6): 859-873
- URL: https://www.wjgnet.com/1948-5182/full/v7/i6/859.htm
- DOI: https://dx.doi.org/10.4254/wjh.v7.i6.859