Copyright
©The Author(s) 2015.
World J Hepatol. Jun 8, 2015; 7(10): 1377-1389
Published online Jun 8, 2015. doi: 10.4254/wjh.v7.i10.1377
Published online Jun 8, 2015. doi: 10.4254/wjh.v7.i10.1377
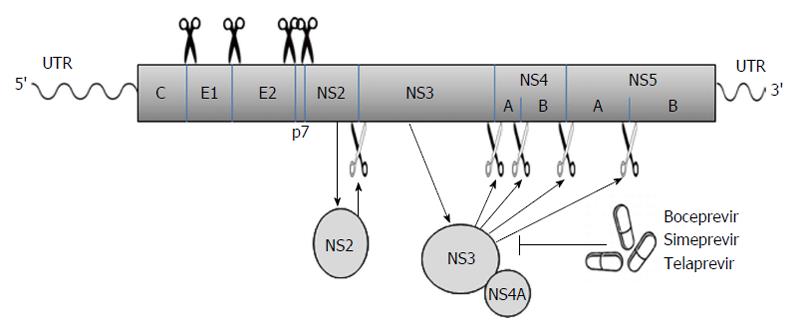
Figure 1 The hepatitis C virus poly-protein is processed co- and posttranslationally by host and viral proteases into at least 10 different proteins, which are arranged in the order of NH2-C-E1-E2-p7-NS2-NS3-NS4A-NS4B-NS5A-NS5B-COOH.
Host signal peptidase is required for the cleavages at C-E1, E1-E2, E2-p7, and p7-NS2 junctions. NS2 cleaves the site between NS2 and NS3; NS3/4A serine protease cleaves the sites at NS3-NS4A, NS4A-NS4B, NS4B-NS5A, and NS5A-NS5B junctions. Several new DAAs (e.g., boceprevir, simeprevir, and telaprevir) specifically designed to inhibit the NS3/4A protease are now becoming available. The wavy lines mark the un-translated regions (UTR) of hepatitis C virus genomic RNA while the rectangle represents the poly-protein derived from the long open reading frame.
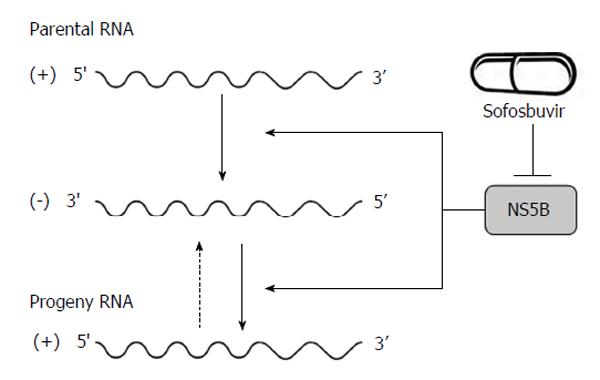
Figure 2 Hepatitis C virus NS5B acts as RNA-dependent RNA polymerase and plays an important role in the synthesis of new RNA genomes.
As the central component of the hepatitis C virus replication complex, NS5B has emerged as a major target for antiviral treatment. New direct acting antivirals, specifically designed to inhibit the NS5B are now becoming available (e.g., sofosbuvir).
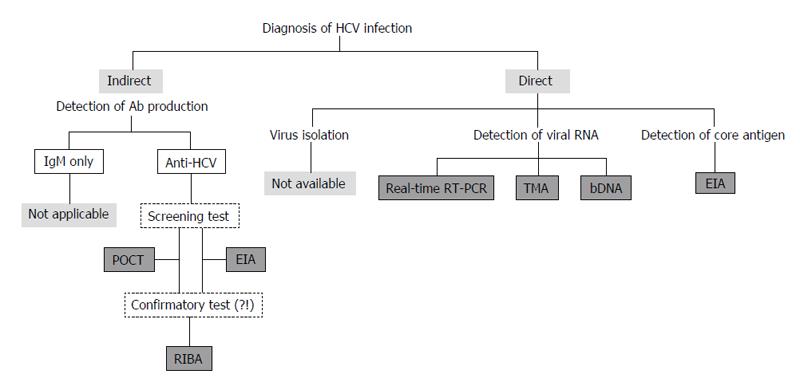
Figure 3 Assays to detect anti-hepatitis C virus antibody, viral core antigen, and viral genomic RNA are used to diagnose HCV infection in clinical practice.
HCV: Hepatitis C virus; POCT: Point-of-care test; EIA: Enzyme immuno-assay; RIBA: Recombinant immunoblot assays; RT-PCR: Reverse transcription-polymerase chain reaction; TMA: Transcription-mediated amplification; bDNA: Branched DNA.
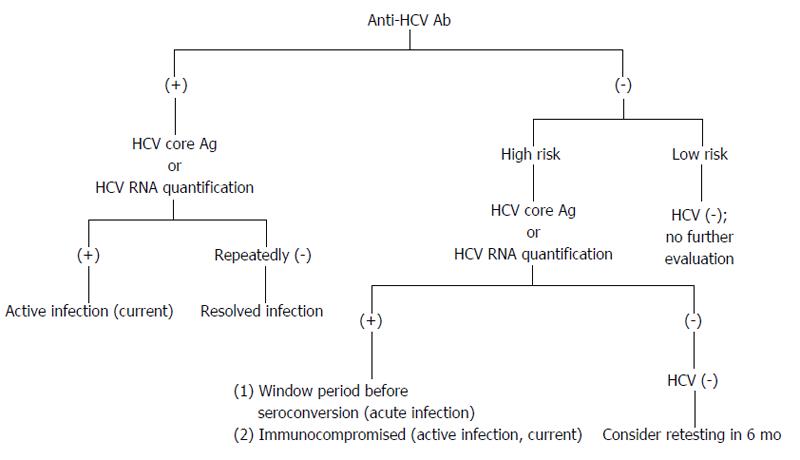
Figure 4 Possible diagnostic results for hepatitis C virus infection.
Individuals are in high risk, e.g., persons who have been exposed to HCV; persons with elevated alanine aminotransferase; persons who are immunocompromised. HCV: Hepatitis C virus.
- Citation: Li HC, Lo SY. Hepatitis C virus: Virology, diagnosis and treatment. World J Hepatol 2015; 7(10): 1377-1389
- URL: https://www.wjgnet.com/1948-5182/full/v7/i10/1377.htm
- DOI: https://dx.doi.org/10.4254/wjh.v7.i10.1377