Copyright
©2012 Baishideng Publishing Group Co.
World J Hepatol. Dec 27, 2012; 4(12): 342-355
Published online Dec 27, 2012. doi: 10.4254/wjh.v4.i12.342
Published online Dec 27, 2012. doi: 10.4254/wjh.v4.i12.342
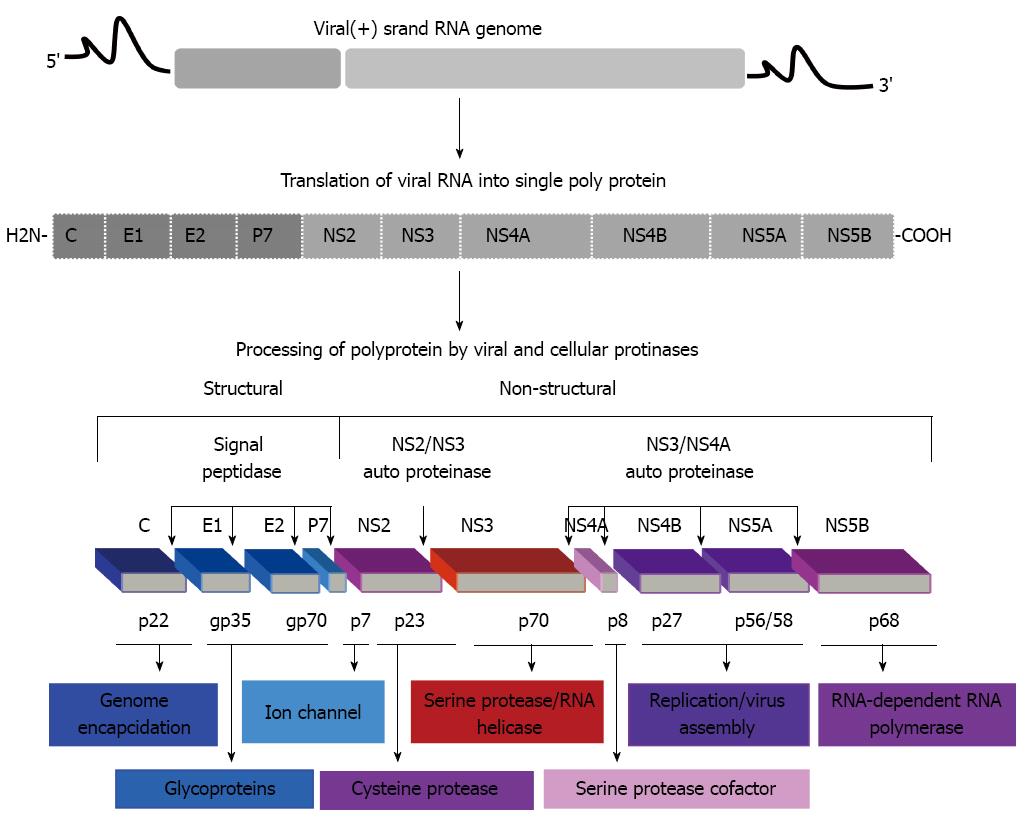
Figure 1 Hepatitis C virus genome including 5’ and 3’ noncoding regions, and the long open reading frame encoding for polyprotein precursor of 3010 amino acids.
This polyprotein precursor can be cleaved functionally by co- and post-translationally processes mediated by cellular and viral proteases into ten different products, including structural and non-structural proteins. The structural proteins core (C), envelop 1 (E1) and E2 are located in the N-terminal third, whereas, the non-structural/replicative proteins (NS2, NS3, NS4A, NS4B, NS5A, NS5B) are located in the remainder of the polyprotein. Putative functions of the cleavage products are shown.
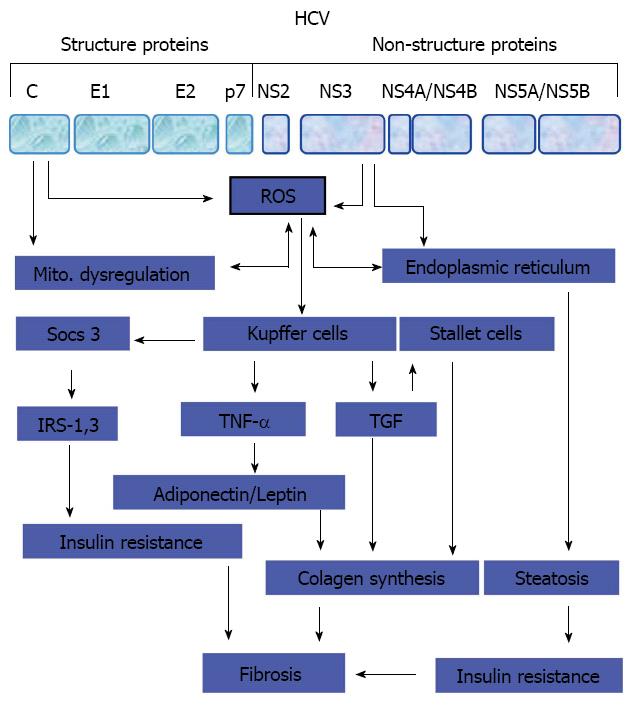
Figure 2 Pathogenesis of liver fibrosis in chronic hepatitis C infected patients.
Potential mechanisms that thought be involved in the regulation of hepatitis C virus -associated hepatic fibrosis. HCV: Hepatitis C virus; ROS: Reactive oxygen species; IRS: Insulin receptor substrate; TNF: Tumor necrosis factor; TGF: Transforming growth factor.
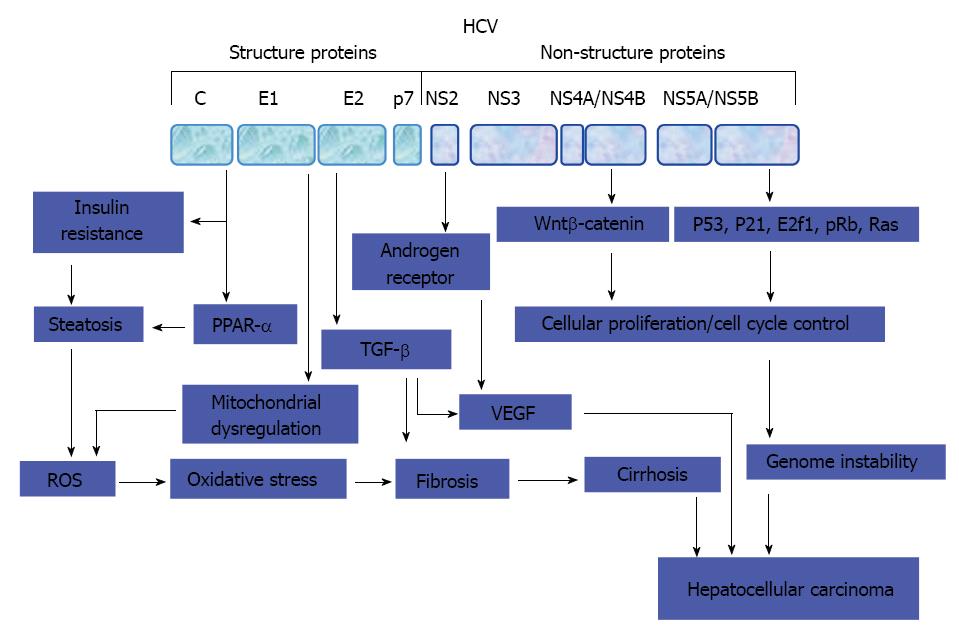
Figure 3 Molecular mechanisms of hepatitis C virus-mediated hepatocarcinogenesis.
Key steps that thought to be involved in the development of hepatitis C virus-associated hepatocellular carcinoma. HCV: Hepatitis C virus; ROS: Reactive oxygen species; IRS: Insulin receptor substrate; TGF: Transforming growth factor; VEGF: Vascular endothelial growth factor; PPAR: Peroxisome proliferator-activated receptor.
- Citation: Selimovic D, El-Khattouti A, Ghozlan H, Haikel Y, Abdelkader O, Hassan M. Hepatitis C virus-related hepatocellular carcinoma: An insight into molecular mechanisms and therapeutic strategies. World J Hepatol 2012; 4(12): 342-355
- URL: https://www.wjgnet.com/1948-5182/full/v4/i12/342.htm
- DOI: https://dx.doi.org/10.4254/wjh.v4.i12.342