Copyright
©The Author(s) 2024.
World J Hepatol. Nov 27, 2024; 16(11): 1306-1320
Published online Nov 27, 2024. doi: 10.4254/wjh.v16.i11.1306
Published online Nov 27, 2024. doi: 10.4254/wjh.v16.i11.1306
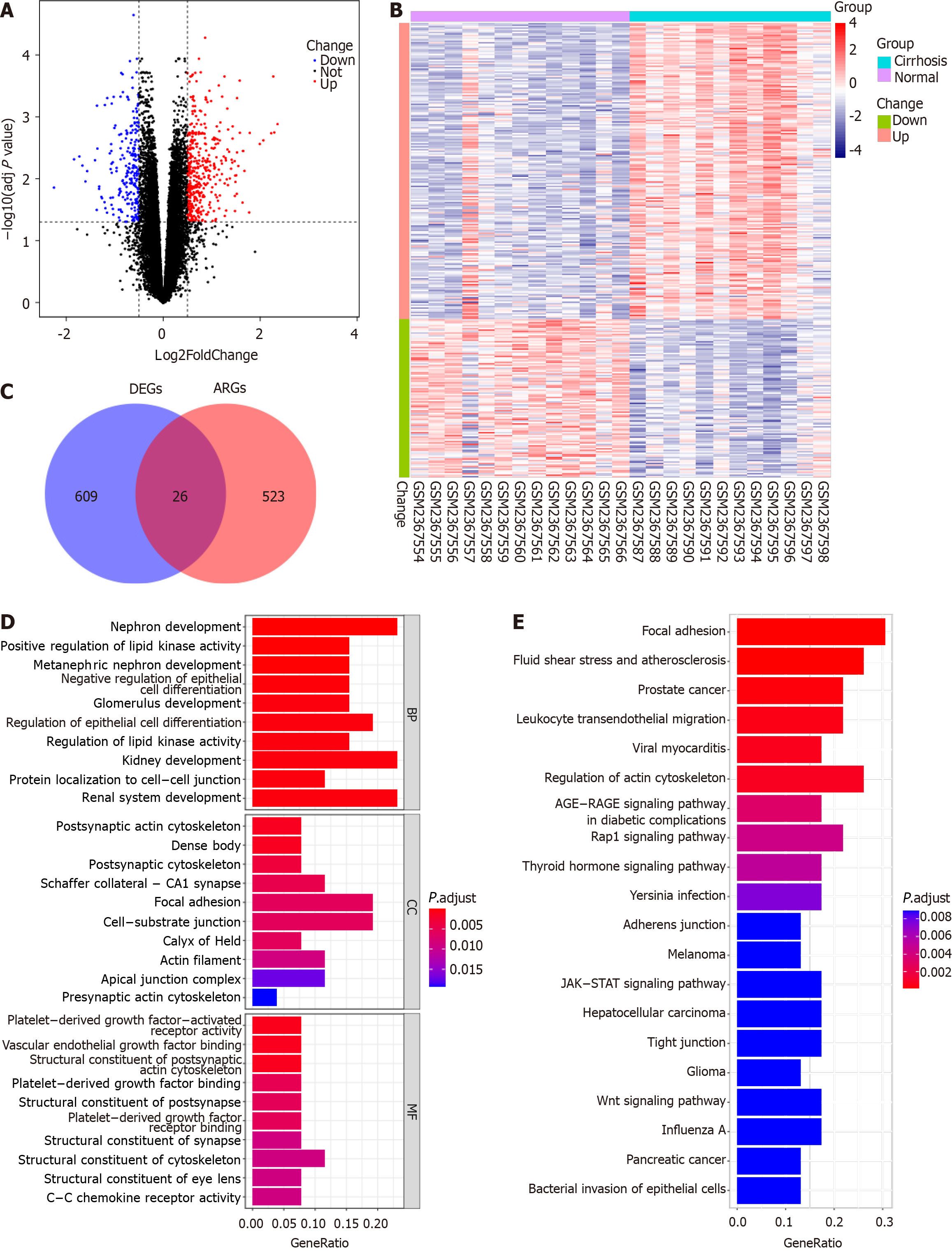
Figure 1 Identification and functional enrichment analysis of differentially expressed anoikis-related genes.
A: Volcano maps of differentially expressed genes (DEGs); B: Heat map of DEGs; C: Venn diagram of differentially expressed anoikis-related genes (DEARGs); D: Gene ontology analysis of DEARGs based on three domains, namely, cellular component, molecular function, and biological process; E: Kyoto Encyclopedia of Genes and Genome pathway analysis. DEGs: Differentially expressed gene; ARG: Anoikis-related gene.
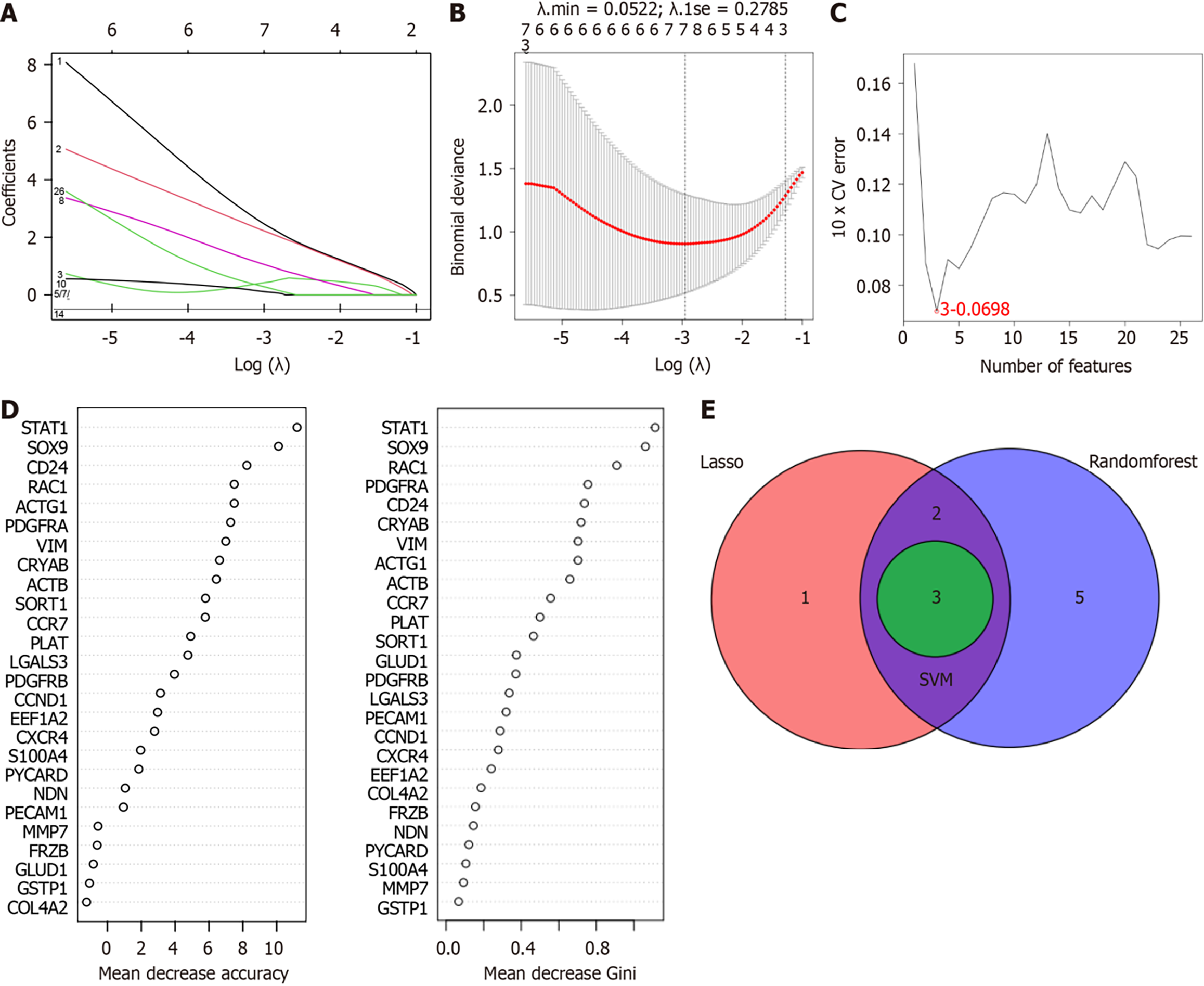
Figure 2 Identification of biomarkers through machine learning algorithms.
A: Least absolute shrinkage and selection operator coefficients profiles; B: Cross-validation for tuning parameter selection. Variables with non-zero coefficients were excluded; C: When n was 3, the classifier had the minimum error in the support vector machine recursive feature elimination model; D: Interpretation chart of two important evaluation indices; E: Venn diagram showing three biomarkers for cirrhosis.
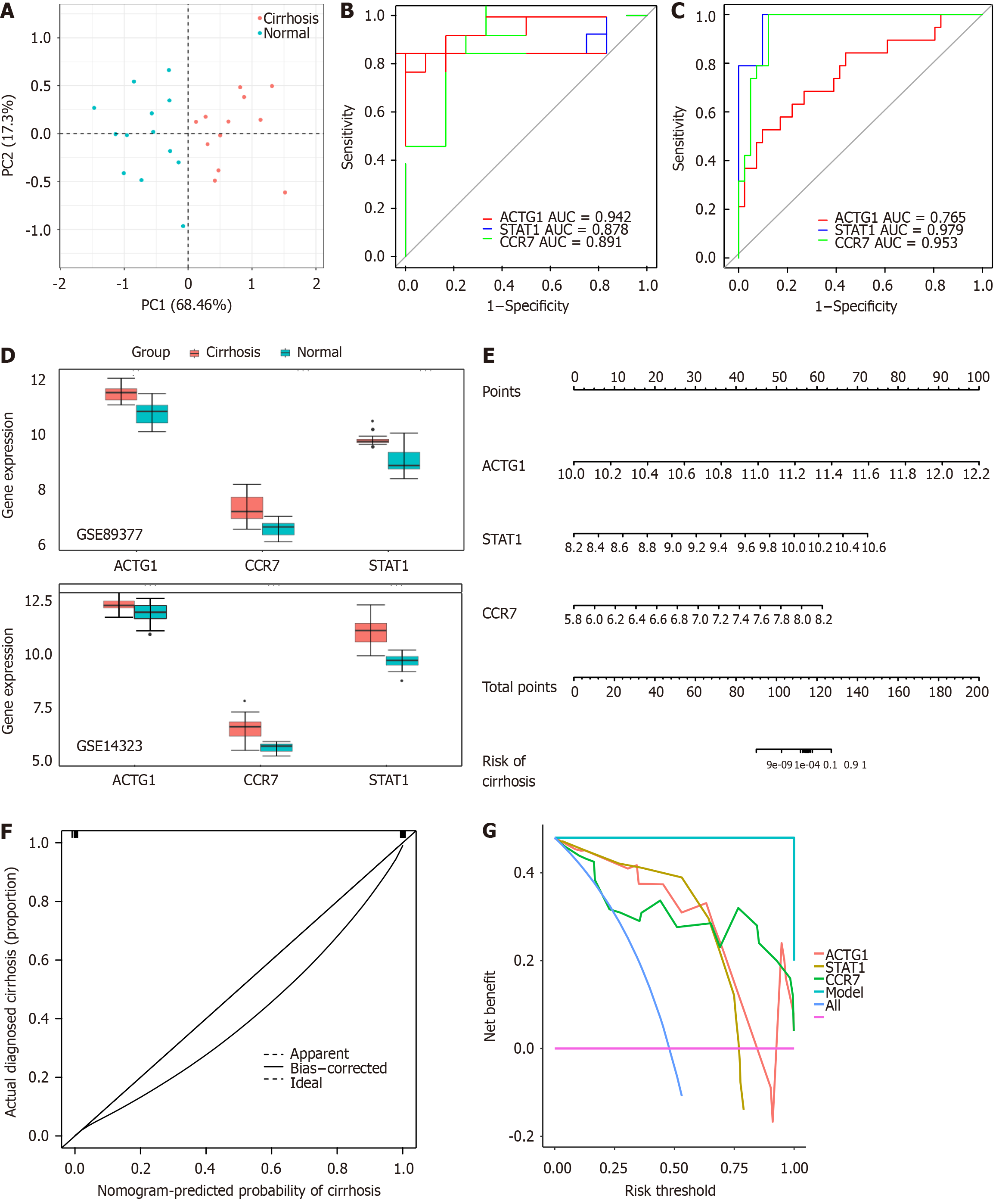
Figure 3 Assessment and validation of the diagnostic efficacy of biomarkers.
A: Principal component analysis was performed to analyze whether the three biomarkers could distinguish between control and cirrhotic samples; B: Receiver operating characteristic (ROC) curves of the biomarkers in the GSE89377 dataset; C: ROC curves of the biomarkers in the GSE14323 dataset; D: Expression of the biomarkers in the GSE89377 and GSE14323 datasets; E: A nomogram based on the biomarkers; F: Calibration curve of the nomogram; G: Decision curve analysis of the nomogram. AUC: Area under curve.
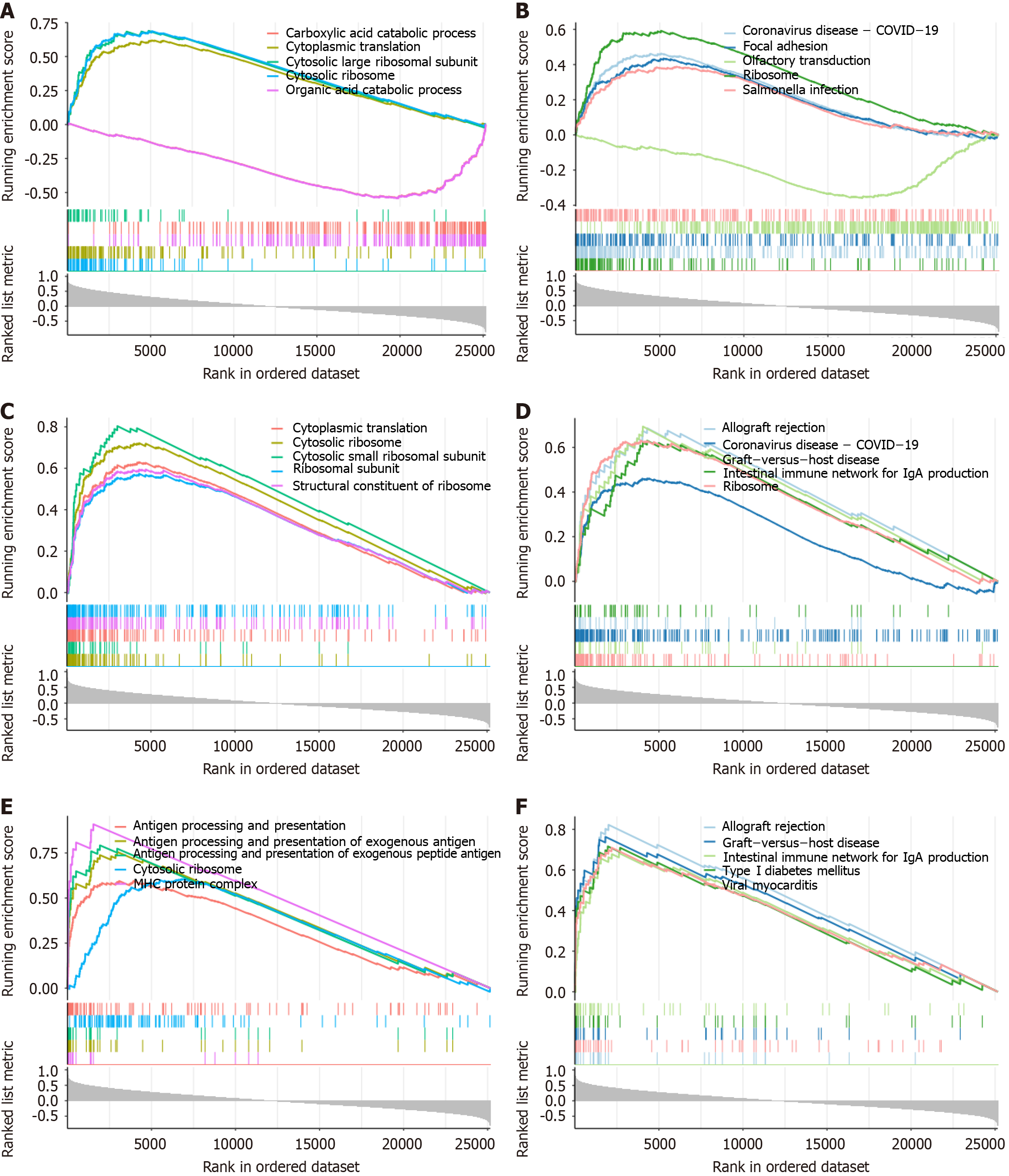
Figure 4 Gene set enrichment analysis.
A: Gene ontology (GO) enrichment analysis of ACTG1; B: Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of ACTG1; C: GO enrichment analysis of CCR7; D: KEGG enrichment analysis of CCR7; E: GO enrichment analysis of STAT1; F: KEGG enrichment analysis of STAT1. COVID-19: Coronavirus disease 2019.
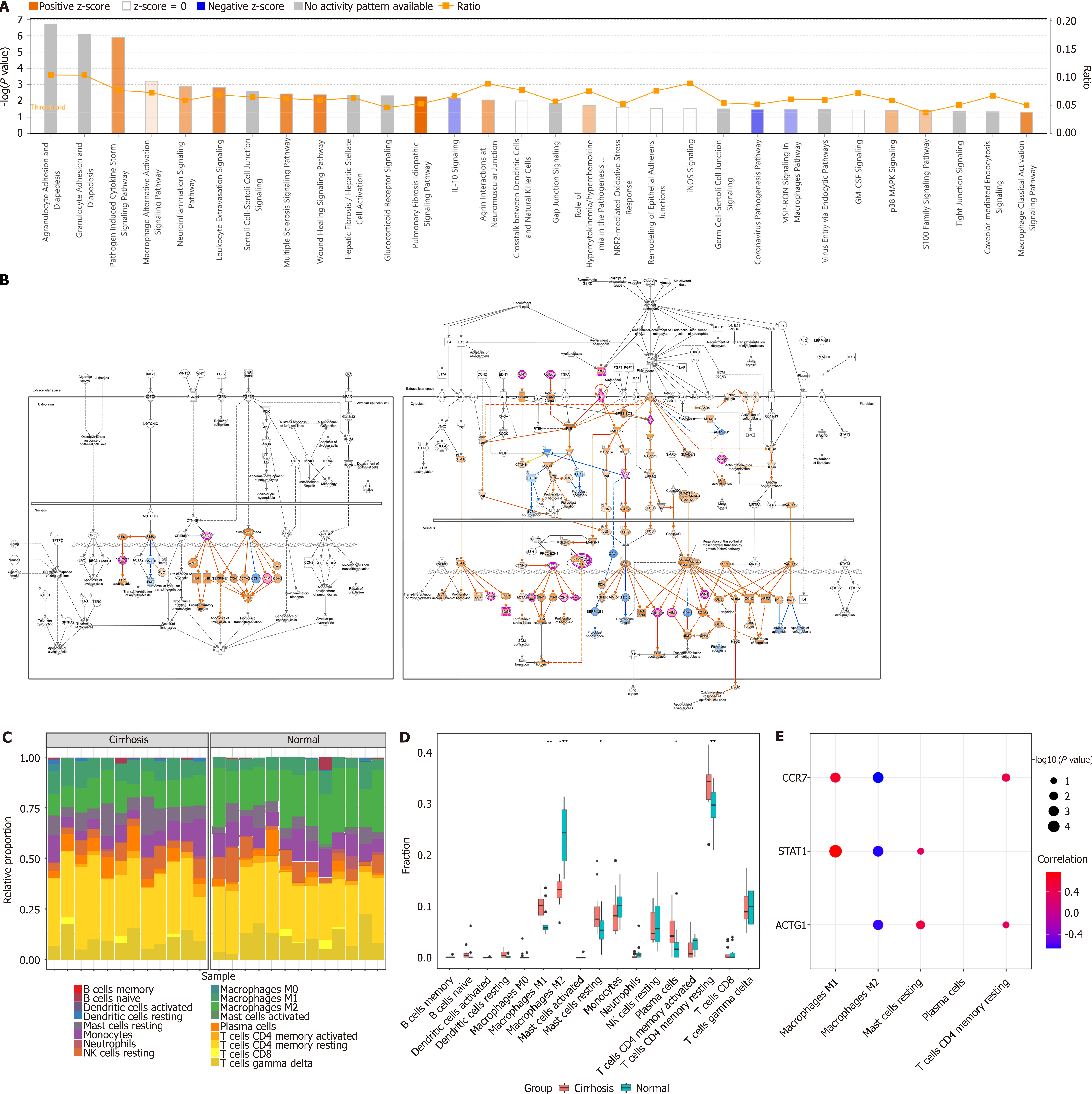
Figure 5 Analysis of classical signaling pathways and immune infiltration.
A: Classical pathways involving the three biomarkers; B: Idiopathic pulmonary fibrosis-related signaling pathway; C: Proportional stacking plot of immune cell distribution in each sample; D: Boxplot of immune cell distribution between the cirrhosis and control groups; E: Correlation between the biomarkers and differential immune cells.
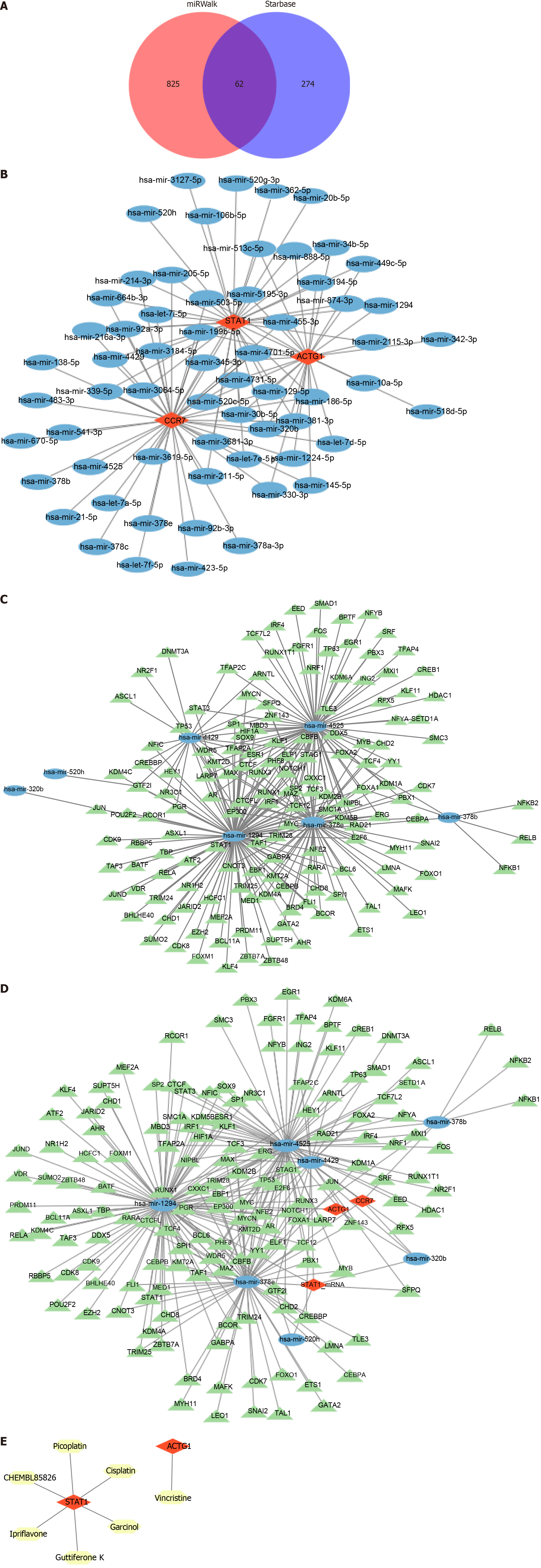
Figure 6 Analysis of classical signaling pathways and immune infiltration.
A: Classical pathways involving the three biomarkers; B: Idiopathic pulmonary fibrosis-related signaling pathway; C: Proportional stacking plot of immune cell distribution in each sample; D: Boxplot of immune cell distribution between the cirrhosis and control groups; E: Correlation between the biomarkers and differential immune cells.
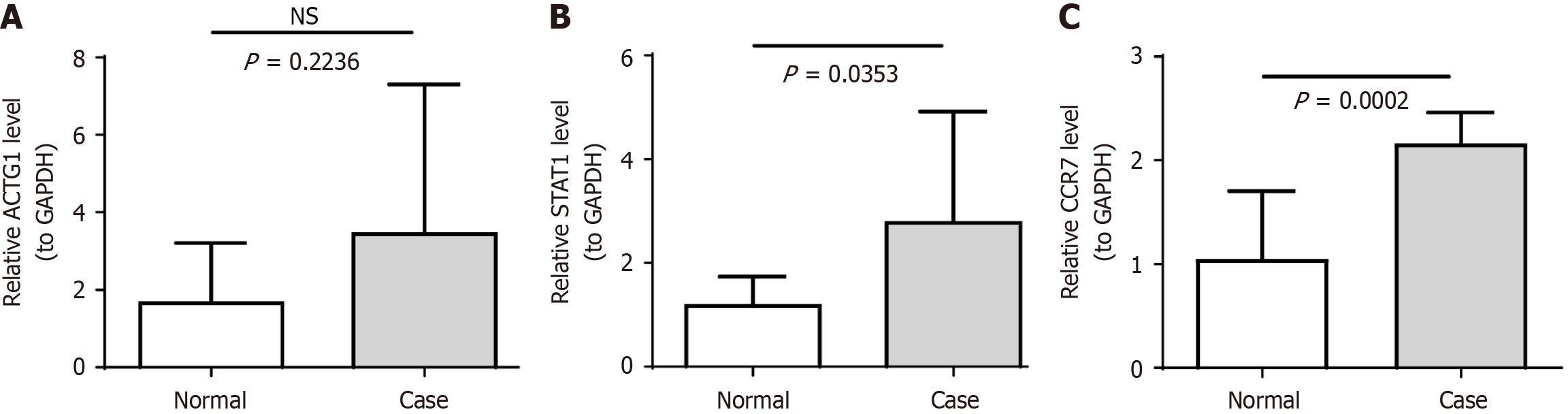
Figure 7 Expression of biomarkers in clinical samples.
A: ACTG1; B: STAT1; C: CCR7; NS: Not significant.
- Citation: Luo JY, Zheng S, Yang J, Ma C, Ma XY, Wang XX, Fu XN, Mao XZ. Development and validation of biomarkers related to anoikis in liver cirrhosis based on bioinformatics analysis. World J Hepatol 2024; 16(11): 1306-1320
- URL: https://www.wjgnet.com/1948-5182/full/v16/i11/1306.htm
- DOI: https://dx.doi.org/10.4254/wjh.v16.i11.1306