Copyright
©The Author(s) 2022.
World J Hepatol. Jul 27, 2022; 14(7): 1382-1397
Published online Jul 27, 2022. doi: 10.4254/wjh.v14.i7.1382
Published online Jul 27, 2022. doi: 10.4254/wjh.v14.i7.1382
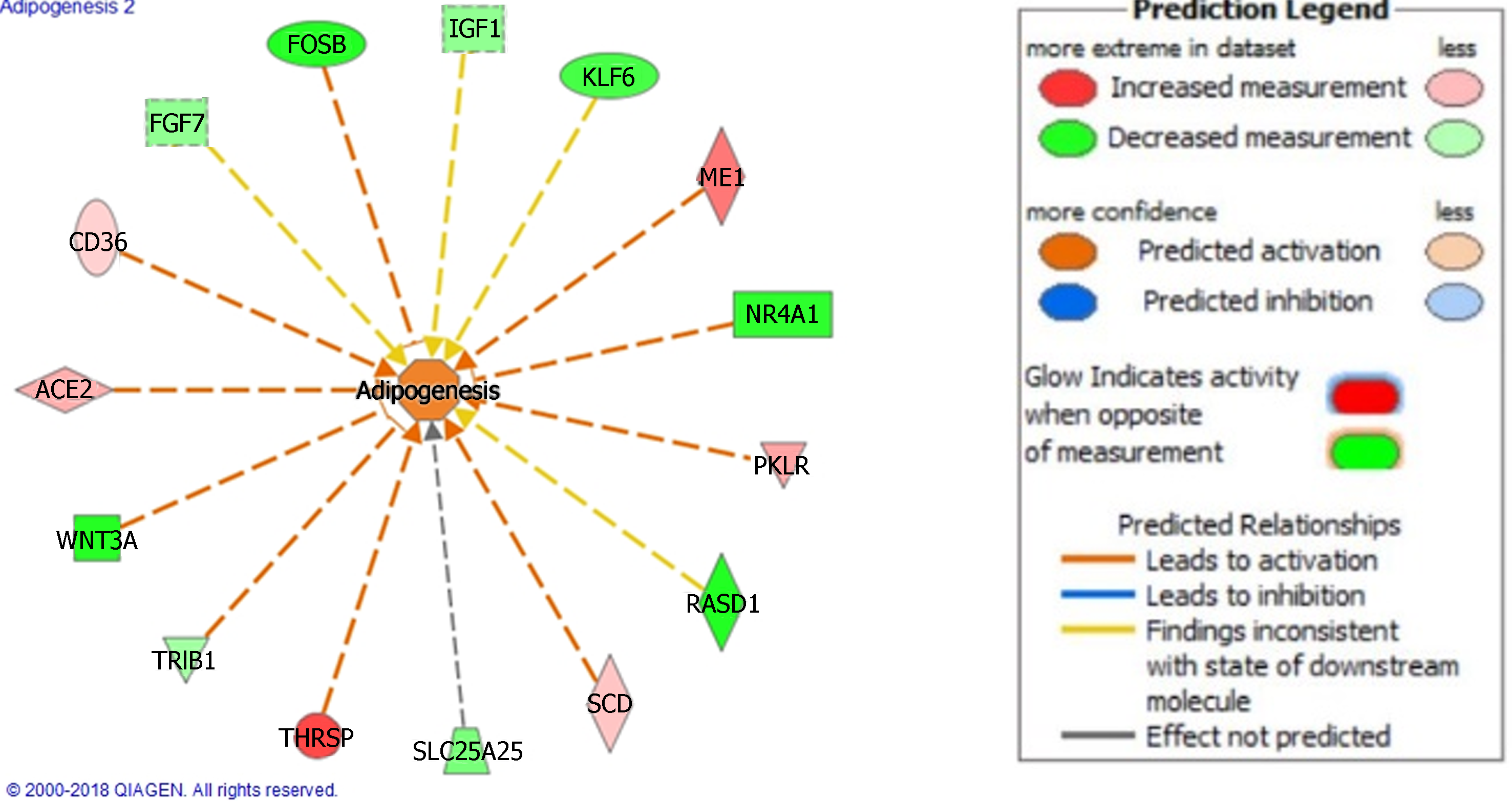
Figure 1 We used Ingenuity Pathway Analysis, gene function feature to define pathologic processes in our non-alcoholic steatohepatitis analysis.
This Figure highlights the adipogenic changes in hepatocytes from non-alcoholic steatohepatitis patients. Prediction legend illustrates relations of molecules and Figure generated using Ingenuity Pathway Analysis.
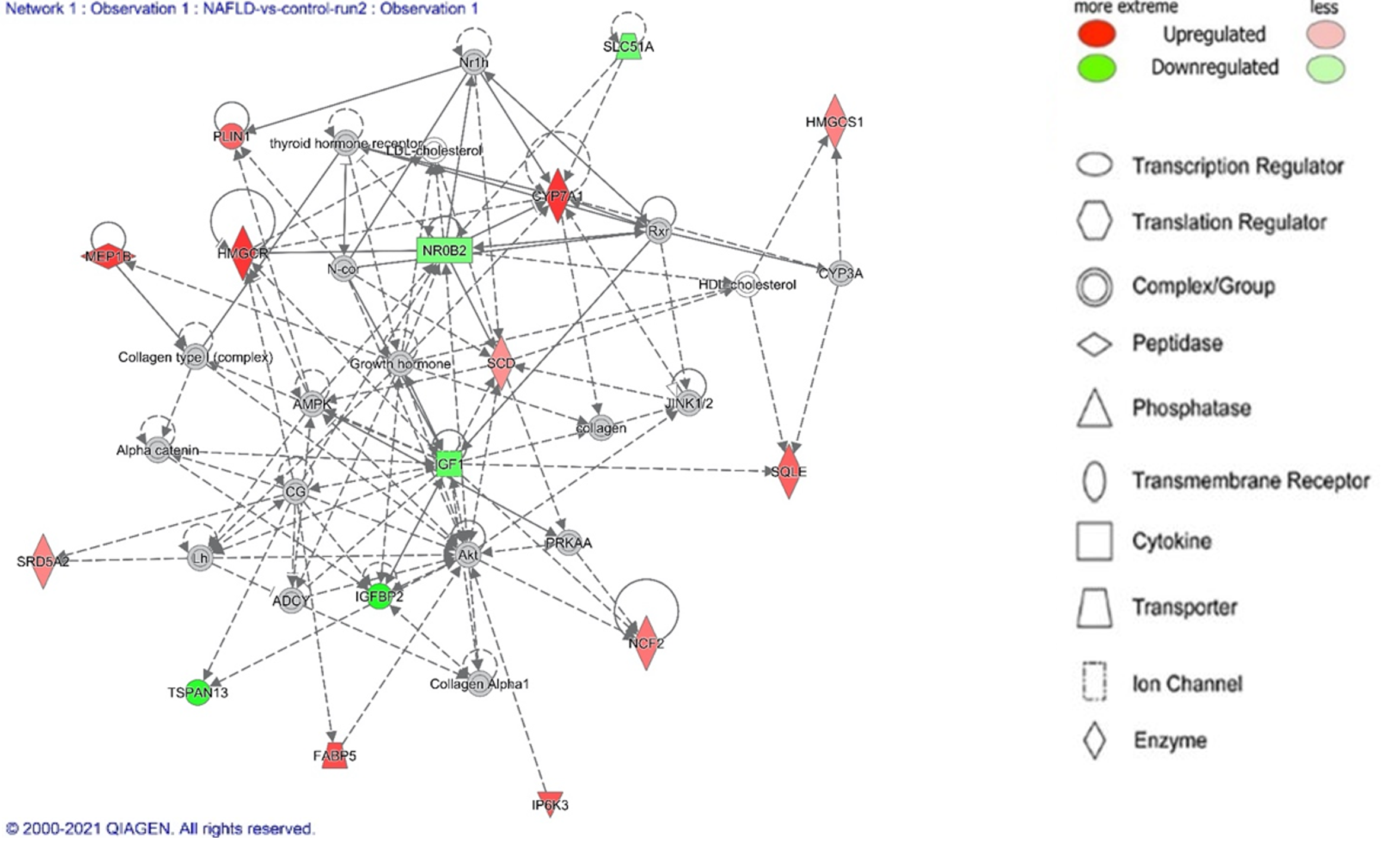
Figure 2 Top network (Lipid metabolism, small molecule biochemistry, vitamin and mineral metabolism) identified by Ingenuity Pathway Analysis Network analysis of non-alcoholic fatty liver disease.
Legend illustrates class of the gene. Red indicates upregulation and green downregulation, with shade depicting magnitude of change. Solid and dashed lines depict direct and indirect, respectively, relationship between genes. Figure generated using Ingenuity Pathway Analysis.
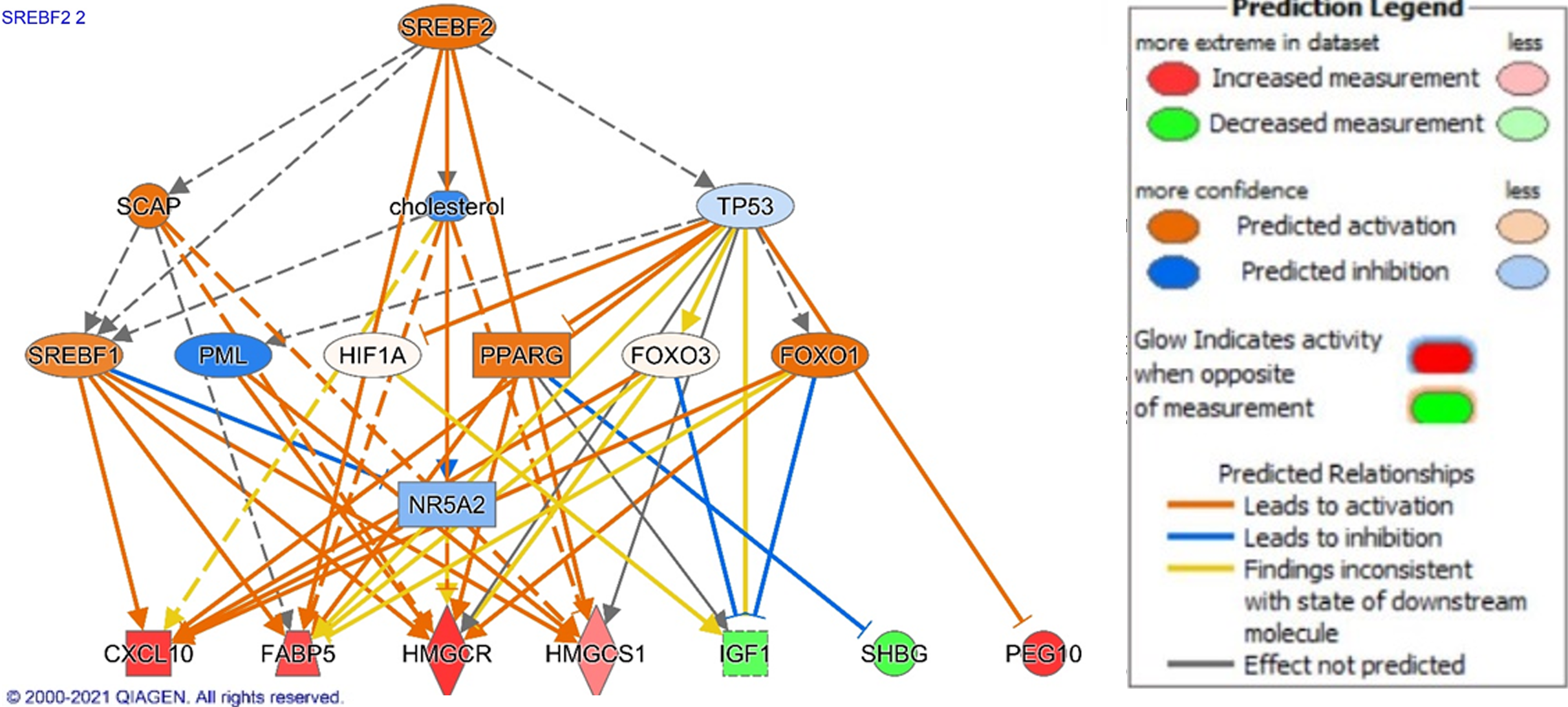
Figure 3 Ingenuity Pathway Analysis of SREBF1 signaling in non-alcoholic fatty liver disease.
Genes are implicated in several potential disease processes including the inflammation, metabolism, and transport. Legend illustrates relationship between genes. See Figure 2 legend for identification of shapes. Figure generated using Ingenuity Pathway Analysis.
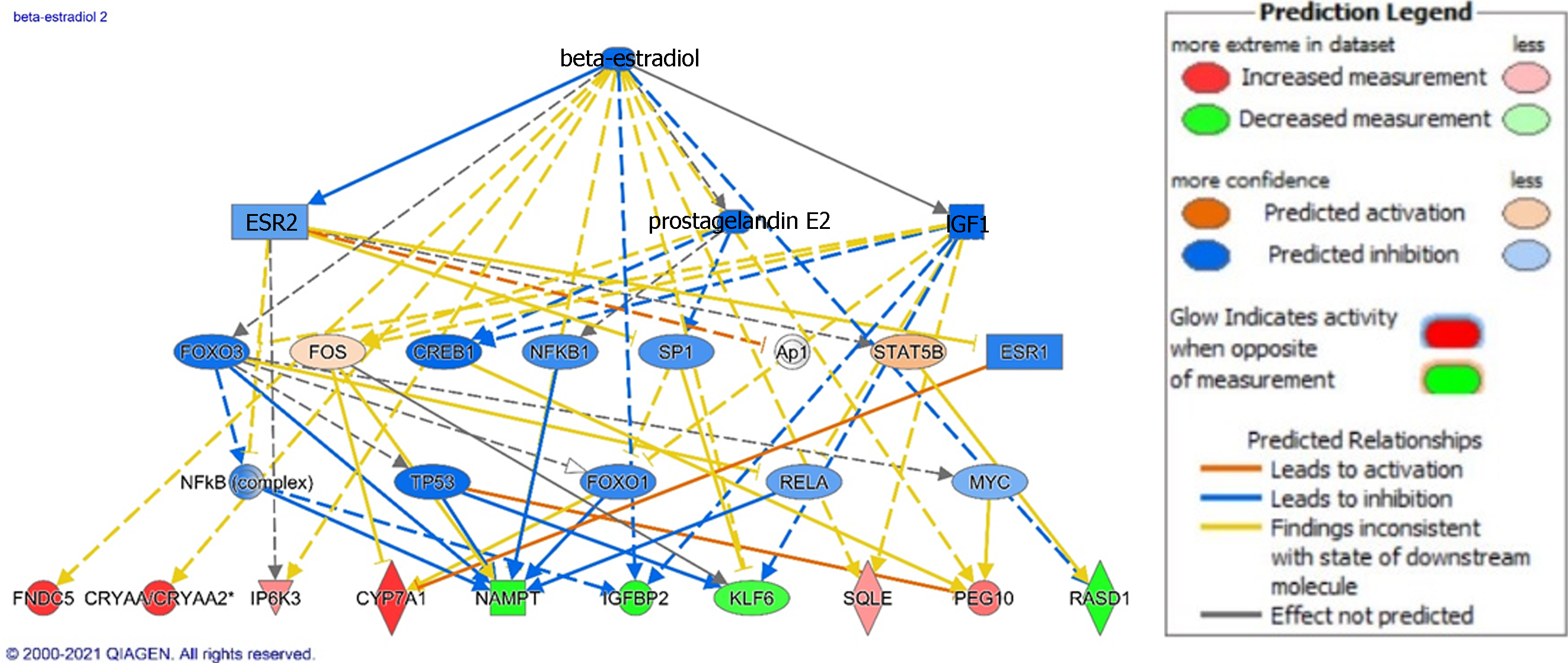
Figure 4 Ingenuity Pathway Analysis of beta-estradiol signaling in non-alcoholic steatohepatitis.
Genes are implicated in several potential disease processes including the metabolism, cancer development, bile acid synthesis, and cell survival. Legend illustrates relationship between genes. See Figure 2 legend for identification of shapes. Figure generated using Ingenuity Pathway Analysis.
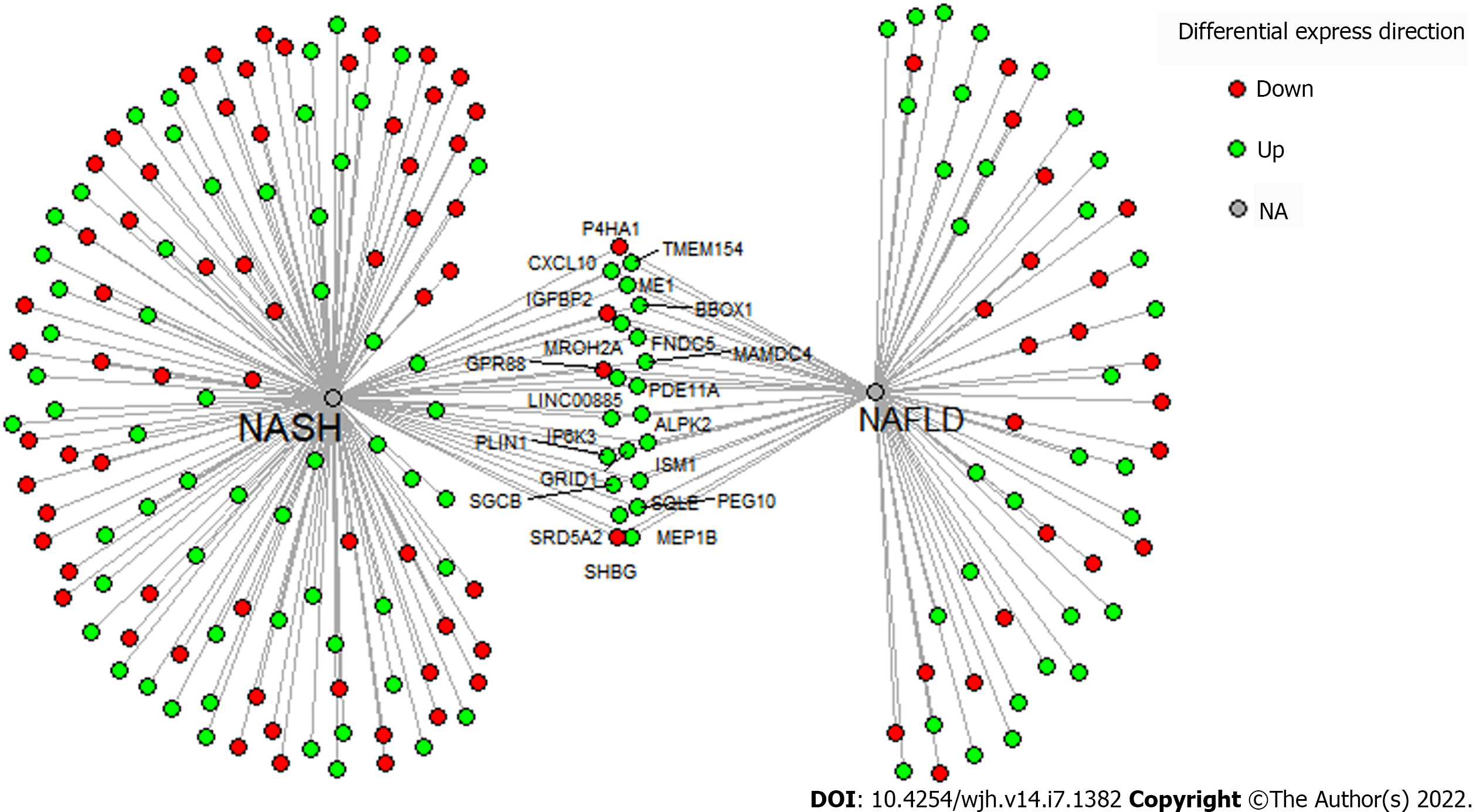
Figure 5 Pathologic gene patterns shared in the non-alcoholic fatty liver disease and non-alcoholic steatohepatitis meta-analyeses are highlighted above.
This dataset was inputted in clue.io to identify potential drug targets. We found riciribine (an AKT inhibitor) and ZSTK-474 (a PI3K inhibitor) as drugs that best targeted the gene expression above.
- Citation: Aljabban J, Rohr M, Syed S, Khorfan K, Borkowski V, Aljabban H, Segal M, Mukhtar M, Mohammed M, Panahiazar M, Hadley D, Spengler R, Spengler E. Transcriptome changes in stages of non-alcoholic fatty liver disease. World J Hepatol 2022; 14(7): 1382-1397
- URL: https://www.wjgnet.com/1948-5182/full/v14/i7/1382.htm
- DOI: https://dx.doi.org/10.4254/wjh.v14.i7.1382