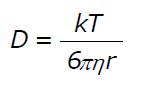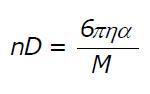Copyright
©The Author(s) 2018.
World J Hepatol. Feb 27, 2018; 10(2): 186-212
Published online Feb 27, 2018. doi: 10.4254/wjh.v10.i2.186
Published online Feb 27, 2018. doi: 10.4254/wjh.v10.i2.186
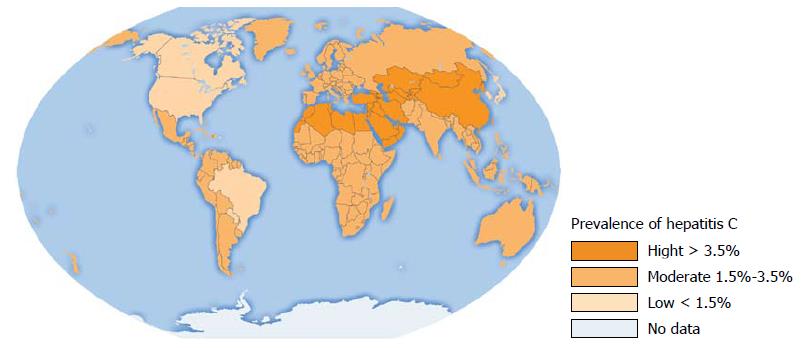
Figure 1 Hepatitis C virus infection in the World.
Analysis of seroprevalence[2].
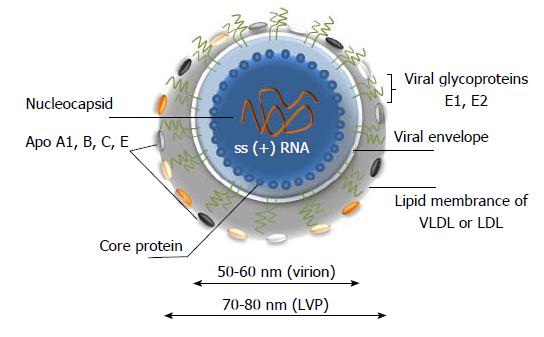
Figure 2 A model of hepatitis C virus lipoviral particle.
Lipid membrane formed by low density lipoproteins (LDL) and very low-density lipoproteins (VLDL) on the surface of the virion (given in grey). Viral core is given in blue and viral RNA is shown in orange. Heterodimers of glycoproteins E1 and E2 are partially embedded in the lipid bilayer and are forming 6 nm long spikes (projections) on the surface of the virion[14-19]. As a result of association with LDL and VLDL, the morphology of the virion is not icosahedral. Depending on the viral source, the shape and size of the particles might vary.
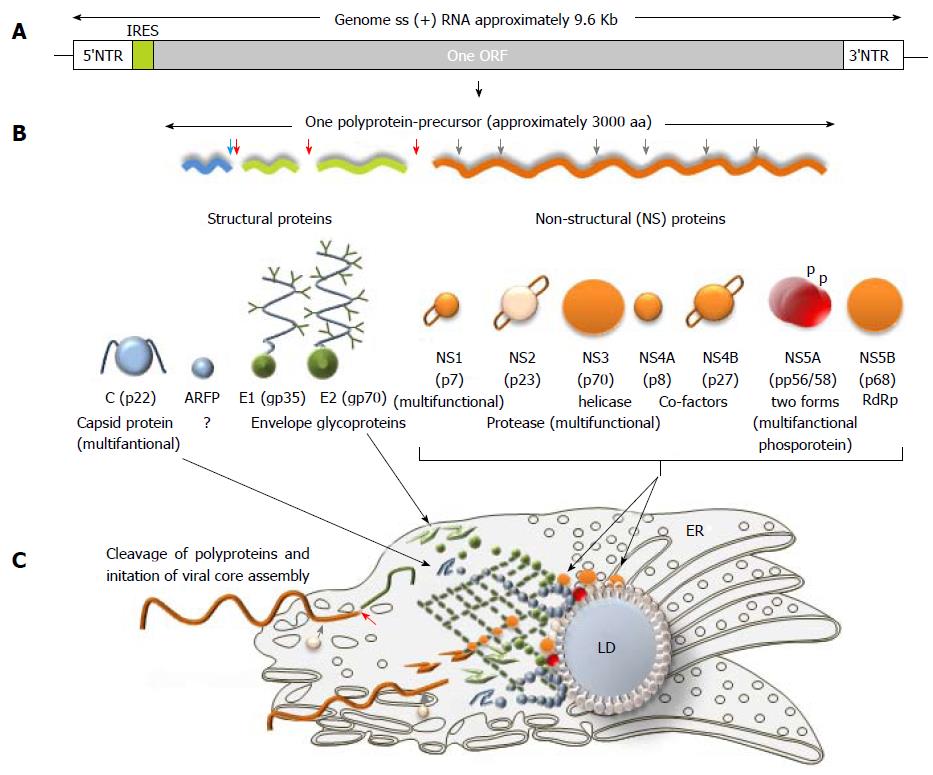
Figure 3 Hepatitis C virus genome, polyprotein precursor and the initial steps of core assembly in endoplasmic reticulum.
A: Being structurally identical, the genomes of seven hepatitis C virus (HCV) genotypes demonstrated approximately 30% of sequence diversity[140]. Two non-translated regions (5’- and 3’-NTR) are flanking a single open reading frame (ORF) shown in grey; B: Polyprotein precursor composed of about 3000 amino acids translated from a single ORF. Three structural proteins - a core protein (shown in blue) and two envelope proteins (shown in green) and seven non-structural (NS) proteins NS1, NS3, NS4A, NS4B, NS5A, and NS5B (shown in orange) that are involved in cleavage, assembly, transcription and some other functions are shown. Alternative reading frame protein (ARFP) that overlaps with the core protein sequence is given in blue. Serine protease NS2 is given in pale yellow. Transmembrane fragments in non-structural proteins are shown as staples; p- phosphoprotein; C: A model of initial steps of the virion core assembly in the endoplasmic reticulum (membranous web is given in dark green) on lipid droplet (LD). Polyprotein precursor is cleaved by cellular C-terminal signal peptidase (red arrow) and cellular signal peptidase (blue arrow) to release capsid protein. NS3-NS4A serine protease cleaves the remaining proteins, while NS2-NS3 (grey arrow) protease cleaves itself. Pre-assembled cores are transported to the LD, where the final steps of assembly take place[183].
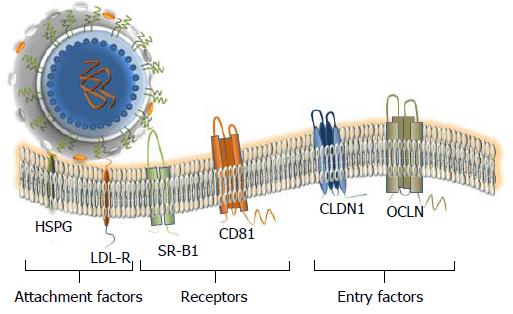
Figure 4 Attachment factors, receptor and entry factors utilized by hepatitis C virus.
At least six membrane proteins are essential for the virus attachment and entry. Heparan sulfate proteoglycan (HSPG) - a type of glycosaminoglycan (GAG) and low density lipoproteins (LDL)-receptor that is promoting the LDL endocytosis are considered as binding factors for hepatitis C virus (HCV). Scavenger Receptor class B member 1 (SR-B1) and CD81 that are ubiquitously expressed on the cell surface, are considered as true receptors. The receptor–viral cargo complex is then moving to the cell-cell contacts (not shown), where the interaction with the tight-junction proteins Claudin 1 (CLDN1) and Occludin (OCLN) takes place. Both proteins are required at the late stage of the entry. Additional interaction partners that are likely engaged in the HCV entry are described in the text. The docking of the LVP to the cellular membrane is shown.
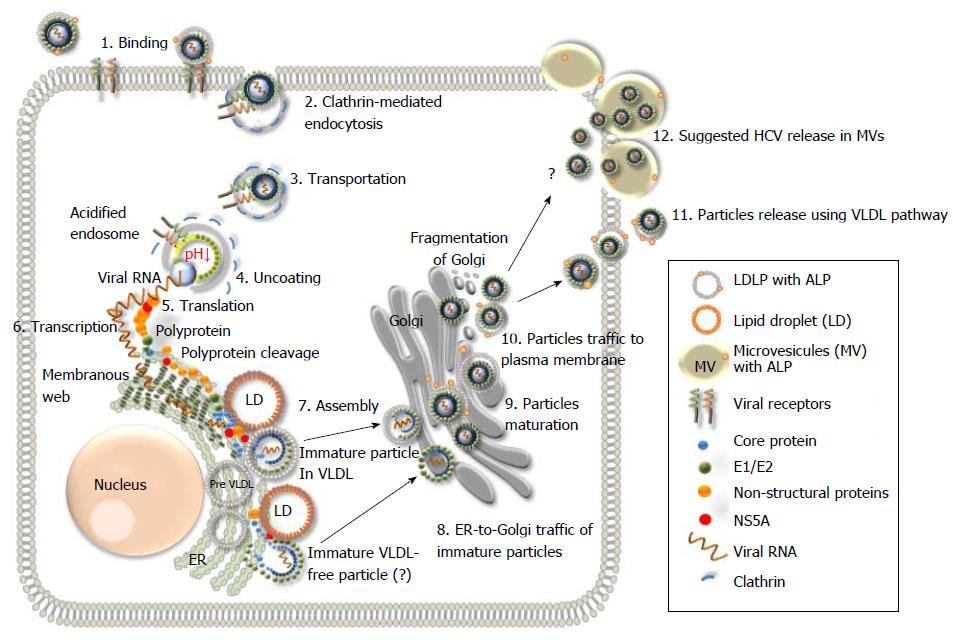
Figure 5 Schematic representation of the hepatitis C virus life cycle.
(1) The lipoviral particles (LVP) binds to entry factors and receptors on the surface of hepatocyte; (2) The virus enters into the cell by a clathrin-mediated endocytosis; (3) Transportation of the virus in endosome; (3) Acidification of endosome, un-coating of the virion and dissociation of the viral core; (4) Release of the viral RNA; (5 and 6) Translation and replication of the viral RNA in the ER in the convoluted membrane structure called the membranous web (shown in dark green); (7) Cleavage of the protein precursor by cellular and viral proteases, assembly of the core on the surface of the lipid droplet (LD) and recruitment of the newly synthesized viral RNA to the viral core during the formation. The mechanism of the viral RNA recruitment to the site of the assembly is not known. It is also unclear whether the core maturation is finalized at this stage; (8) The viral nucleocapsid egresses into the lumen side of the ER, likely interacted with very low-density lipoproteins (VLDL) and translocated to Golgi. It is not clear if some VLDL-free particles can also be produced (?). Details of traffic machinery are not well defined; (9) Final maturation of viruses in Golgi and virus induced partial fragmentation of Golgi; (10) The hepatitis C virus (HCV) complex with VLDL is directed to the plasma membrane using the VLDL secretory pathway; (11) Release of particles from the cell (adapted from Ref.[16,19,135]); and (12) The HCV release in MV cannot be excluded as another pathway for the virus release during active replication.
Math 1 Math(A1).
Math 2 Math(A1).
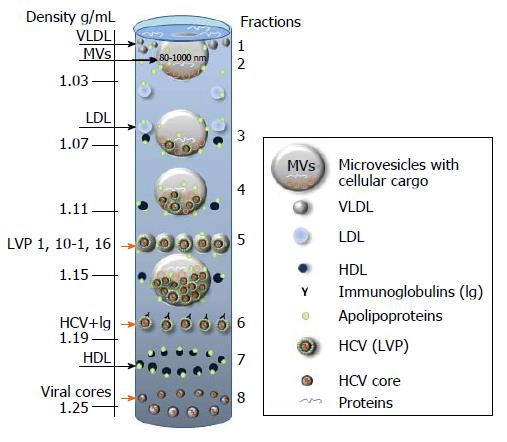
Figure 6 Putative distribution of lipoproteins (LDL, VLDL, HDL), lipoviral particles and viral cores after density gradient centrifugation of plasma from infected individual.
The picture is based on the known buoyant densities of analyzed elements[15,170,184-187]. The density values (g/mL) are given on the left. The initial material that has been used for the virus isolation, contributes greatly to the associated lipid content of the virions and, as a consequence, it influences the buoyant density[177,183-187]. Because of an irregular protein to cholesterol amounts, high density lipoproteins (HDL) of 5-15 nm in diameter can be detected in the different fractions (1.06-1.21 g/mL). The family of HDL is given as dark blue spheres. The buoyant density of microvesicles (MVs) may vary from 1.02 g/mL to 1.16 g/mL, depending on MVs diameter and cargo (miRNA, tRNA, fragments of mRNA and proteins). If hepatitis C virus (HCV) is released in MVs with similar diffusion parameters (size and mass), the buoyant density of vesicle should be viral cargo-dependent. Fractions 1: Single cellular or viral proteins “floating” on the surface, very low-density lipoproteins (VLDL, diameter 50-75 nm, density 0.95-1.006 g/mL); Fraction 2: Low density lipoproteins (LDL, diameter 18-25 nm, density 1.019-1.06 g/mL) and virus-free MVs with a low cellular cargo content; Fraction 3: MVs that carry few viral particles, possible overlap with LDL and HDL; Fraction 4: MVs with a higher viral load, possible overlaps with HDL and with densely loaded virus-free MVs; Fraction 5: Lipoviral particles (LVP, diameter 60-80 nm, density 1.10-1.16 g/mL) and MVs with a significant viral and/or mixed load. Overlaps with HDL, virus-loaded MVs and “dense” virus-free MVs are possible; Fraction 6: Lipid-free virions with attached immunoglobulins, possible overlap with HDL; Fractions 7: HDL with a high protein load; Fraction 8: Non-enveloped viral cores that might overlap with dense HDL. Small grey spheres - VLDL. Light blue spheres - LDL. Orange arrows indicated fractions of the gradient that are most likely to contain viruses and viral cores.
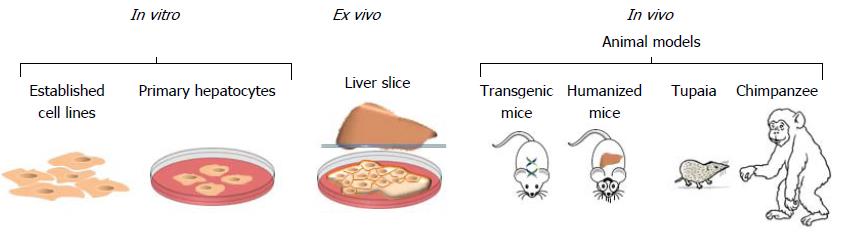
Figure 7 In vitro, ex vivo and in vivo models to study hepatitis C virus.
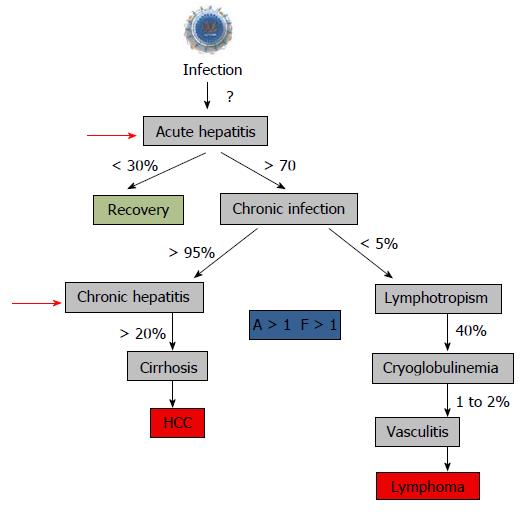
Figure 8 Natural history of hepatitis C virus infection and start to treat.
The recommendation to treat chronic hepatitis is usually a “significant” fibrosis as defined by a Child–Pugh score (A to C) and a fibrosis grade (F) greater than 1 by the Metavir scoring system, with usually a significant necrotic-inflammation as defined by an activity stage greater than 1 by the Metavir scoring system[288,289]. The Child–Pugh score employs five clinical measures of liver disease: Total bilirubin, Serum albumin, Prothrombin time, Ascites, Hepatic encephalopathy. The letter F refers to the scars of the liver caused by the aggression. It is classified from F0 to F4: F1, F2 are minimal to moderate fibrosis, F3 corresponds to a pre-cirrhotic stage and F4 corresponds to cirrhosis. Red arrows indicated the time to start treatment[290].
- Citation: Morozov VA, Lagaye S. Hepatitis C virus: Morphogenesis, infection and therapy. World J Hepatol 2018; 10(2): 186-212
- URL: https://www.wjgnet.com/1948-5182/full/v10/i2/186.htm
- DOI: https://dx.doi.org/10.4254/wjh.v10.i2.186