Copyright
©The Author(s) 2018.
World J Hepatol. Oct 27, 2018; 10(10): 645-661
Published online Oct 27, 2018. doi: 10.4254/wjh.v10.i10.645
Published online Oct 27, 2018. doi: 10.4254/wjh.v10.i10.645
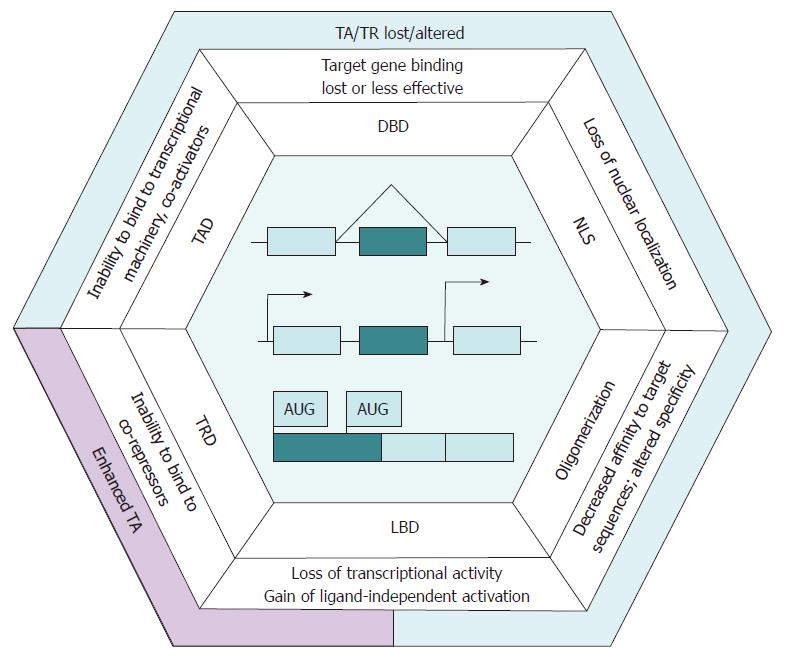
Figure 1 Alternative transcription factor isoforms possess regulatory properties distinct from those of canonical variants.
Schematic representation of the common isoform formation mechanisms - alternative splicing, alternative promoter usage and alternative initiation of translation - is shown in the central part of the hexagon. Light boxes correspond to constitutive exons, and dark ones designate the exons harboring domains and regions indicated in the adjacent frame. TF isoform properties divergent from those of the canonical variants and resulting from functional domain loss are indicated at the corresponding hexagon edges on the outer side of the frame. The brace marks exon exclusion - a particular case of alternative splicing. The arrows represent transcription start sites. AUG: Start codon; TR: Transcription repression; TRD: Transcription repression domain; TF: Transcription factor; TAD: Trans-activation domain; LBD: Ligand-binding domains; DBD: DNA-binding; NLS: Nuclear localization signal.
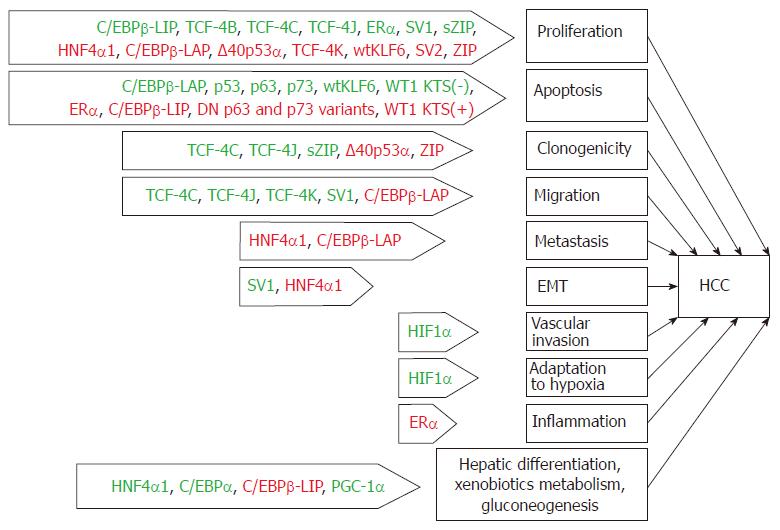
Figure 2 The key processes implicated in hepatocarcinogenesis that are regulated by transcription factors or their specific isoforms.
TFs with stimulatory effects on these properties are depicted in green color, and TFs acting as repressors are colored in red. Sharp arrows indicate the stimulation of HCC development, and inhibition is indicated by blunt arrows. TF: Transcription factor; HCC: Hepatocellular carcinoma; EMT: Epithelial-mesenchymal transition.
- Citation: Krivtsova O, Makarova A, Lazarevich N. Aberrant expression of alternative isoforms of transcription factors in hepatocellular carcinoma. World J Hepatol 2018; 10(10): 645-661
- URL: https://www.wjgnet.com/1948-5182/full/v10/i10/645.htm
- DOI: https://dx.doi.org/10.4254/wjh.v10.i10.645