修回日期: 2021-03-24
接受日期: 2021-06-02
在线出版日期: 2021-07-28
N6-甲基腺嘌呤(N6-methyladenosine, m6A)是高等真核生物信使RNA (messenger RNA, mRNA)上最多的化学修饰之一, 与mRNA加工、核输出、翻译以及降解密切相关. m6A修饰主要通过甲基转移酶和去甲基化酶以动态可逆的方式进行调控. m6A在表观遗传水平上通过调控原癌基因、抑癌基因的表达而影响肿瘤生长. 越来越多的证据表明, m6A与消化系统恶性肿瘤的发生和进展有关, 有望成为肿瘤诊断的分子标志物或药物治疗的潜在靶点. 本文就m6A在消化系统恶性肿瘤中的作用进行述评.
核心提要: 本文就N6-甲基腺嘌呤(N6-methyladenosine, m6A)甲基化修饰及其在消化系恶性肿瘤中的作用进行述评. m6A通过一系列复杂的机制参与多种肿瘤的发生、进展及化疗耐药的调控, 有望成为肿瘤防治的新型靶标.
引文著录: 梁锐煌, 朱南星, 侯钦, 吴灵飞. m6A甲基化在消化系统恶性肿瘤发生与进展中的作用. 世界华人消化杂志 2021; 29(14): 747-757
Revised: March 24, 2021
Accepted: June 2, 2021
Published online: July 28, 2021
N6-methyladenosine (m6A) is the most common modification in higher eukaryotic messenger RNA (mRNA), which is closely related to the mRNA processing, nuclear output, translation, and degradation. M6A modification is regulated by methyltransferase and demethylase dynamically and reversibly. M6A plays an essential role in tumors progression by regulating epigenetic modification of tumor suppressor genes and oncogenes. In recent years, more and more studies have shown that m6A is related to the occurrence and development of digestive system malignant tumors and may serve as a novel potential biomarker for the diagnosis and prognosis of digestive cancer. This article reviews the latest progress in the research of m6A in digestive system malignant tumors.
- Citation: Liang RH, Zhu NX, Hou Q, Wu LF. Role of m6A methylation in occurrence and progression of digestive system malignancies. Shijie Huaren Xiaohua Zazhi 2021; 29(14): 747-757
- URL: https://www.wjgnet.com/1009-3079/full/v29/i14/747.htm
- DOI: https://dx.doi.org/10.11569/wcjd.v29.i14.747
近年来, 随着人们饮食结构和生活方式的改变, 消化系统肿瘤的发病率呈逐年上升趋势. 消化道作为人类恶性肿瘤的好发部位之一, 肿瘤发生的机制十分复杂, 多条通路参与其中, 其中表观遗传学相关机制受到广泛关注. 表观遗传修饰是指在不改变DNA核苷酸序列的前提下引起基因表达的可遗传性改变, 其内容不但涉及DNA甲基化、组蛋白修饰, 还包括染色质重构等. 早期的研究主要集中于DNA和蛋白质水平, 随着基因测序和生物信息学技术的进步, RNA 水平的化学修饰逐渐成为生命科学领域的研究热点. 其中, N6-甲基腺嘌呤(N6-methyladenosine, m6A)尤其引人注目. 目前已证实, m6A受多种因子的调节, 在细胞增殖、侵袭、转移等一系列恶性生物学行为中发挥重要作用. 本文就m6A相关蛋白的功能及其与消化系肿瘤的研究进展作一述评.
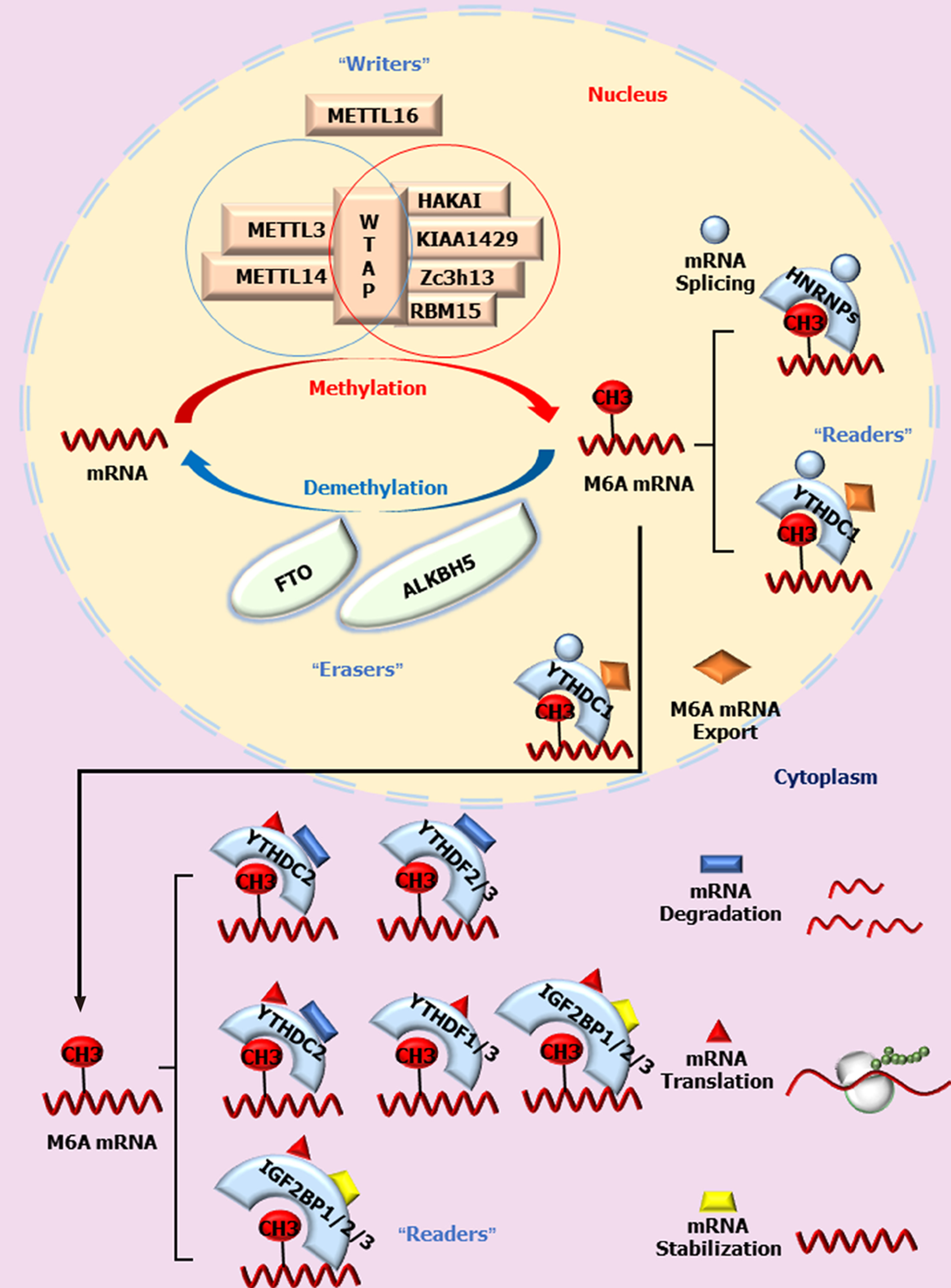
RNA 修饰是一个新兴的领域, 常见的RNA甲基化包括5-甲基胞嘧啶(m5C)、N1-甲基腺苷(m1A)[1,2]和m6A[3]等. 目前, 在生物体内已发现有163种化学修饰[4], 分布在各种RNA上, 其中信使RNA(mRNA)的修饰尤其重要, 因为它具有将DNA的遗传信息转录到蛋白质的功能. m6A甲基化修饰最早在酵母tRNA中发现, tRNA发生m6A修饰后有助于维持其三叶草结构的稳定性, 提高翻译效率[5]. 1974年密歇根州立大学Desrosiers首次明确m6A甲基化是真核生物mRNA与长链非编码RNA (long noncoding RNA, lncRNA)之间最常见的内部修饰. m6A甲基转移酶复合物利用S-腺苷甲硫氨酸(S-adenosyl methionine, SAM)为甲基供体, 将腺嘌呤第6位氮原子上的氢甲基化而形成m6A. 这是mRNA转录后最丰富和保守的转录后表观遗传修饰.据统计, m6A甲基化大约占mRNA化学修饰的50%, 在生命起源及生物生长发育过程中起重要作用[6,7]. m6A虽然发现较早, 但限于当时的实验技术和条件, 较长时间并未取得进展. 由于mRNA半衰期较短, 其修饰的作用时间非常有限而在过去常被认为是静态的. 随着免疫共沉淀等生物技术的进步, m6A甲基化位点被识别, m6A修饰的作用重新引起了人们的关注. 2011年, Jia等[8]发现高度保守的脂肪质量与肥胖相关蛋白(fat mass and obesity-associated protein, FTO)参与了m6A的去甲基化调控. m6A这一可逆的动态过程极大拓展了人们对其生理功能的认识. 目前基于anti-m6A抗体的meRIP技术、miCLIP技术以及新近报道的不依赖抗体的MAZTER-seq和DART-seq技术的研究结果表明, 哺乳动物RNA中约0.1%-0.4%的腺苷存在m6A甲基化修饰[6]. 这种修饰并非随机分布在成熟转录本中, 而是在特定的位点如终止密码子周围、3ʹUTR和内部长外显子内富集, 具有一致的motif, 即RRACH序列(R: A/G, H: A/C/U)[7,9,10]. 随着m6A甲基转移酶(Writers), 去甲基化酶(Erasers)和甲基识别蛋白(Readers)的不断发现, m6A与mRNA之间的关系被不断揭示. m6A可逆、动态的修饰不仅参与了mRNA转运出核、翻译及降解等生理过程, 而且与肿瘤生长、转移及化疗耐药密切相关[11-13]. 下面对新近取得的进展进行述评.
第一类蛋白质是m6A甲基转移酶. 它们编码的基因被称为m6A"writers". Knuckles等[14]在小鼠胚胎干细胞中过表达METTL3, 通过TAP-LC-MS技术筛选鉴定与METTL3相互作用的蛋白. 将在高盐条件下洗脱获得的复合物命名为m6A-METTL复合物(m6A methyltransferase complex, MAC). 将在低盐条件下洗脱获得的复合物命名为m6A-METTL相关蛋白复合物(m6A methyltransferase associated complex, MACOM). 其中, MAC主要包括甲基转移酶样蛋白3 (methyltransferase-like 3, METTL3)和METTL14蛋白, 两者以1:1比例形成稳定的复合物并共定位于细胞核内[15]. METTL3作为主要的甲基化催化中心, 催化m6A合成, 它与METTL14、Wilm's肿瘤1相关蛋白(Wilm's tumor 1-associating protein, WATP)形成复合物. METTL14本身并无催化活性[16], 但可提高METTL3催化酶的活性[17]. WTAP对甲基化修饰亦无直接催化作用, 它通过招募METTL3-METTL14二聚体定位于核散斑中, 参与RNA的选择性剪切并保持MAC酶活性. MACOM主要是由接头蛋白WTAP、VIRMA/KIAA1429、RBM15、Zc3h13和CBLL1/HAKAI相互结合而形成复合物, 与MAC共同参与m6A甲基化修饰. WTAP作为接头蛋白招募MAC定位到核散斑中[18]. 接头蛋白KIAA1429、RBM15、HAKAI、Zc3h13等能够与WTAP结合, 决定MAC的正确定位[19,20]. 此外, METTL16被鉴定为单独存在的具有催化活性的m6A甲基转移酶[21].它与METTL3同源, 可结合U6 snRNA, 导致U6第43位腺嘌呤m6A甲基化, 影响U6 剪切因子对mRNA前体的剪接[21]. Pendleton等[22]证实METTL16通过诱导甲硫氨酸腺苷转移酶2A (MAT2A)合成参与snRNA甲基化调控. MAT2A编码大部分细胞的SAM, 我们既往的研究表明, SAM与腺苷的代谢过程密切相关[23]. 细胞内甲硫氨酸水平下降可导致MAT2A表达增加. METTL16通过调控MAT2A mRNA腺苷的甲基化水平而影响MAT2A生成, 进而调控snRNA的活性及其生物学功能.
第二类蛋白质是m6A去甲基化酶, 其编码基因称为m6A"Erasers", 它的功能是将RNA分子中已进行m6A修饰的甲基去除. 目前已证实参与m6A去甲基化修饰过程的有FTO和AlkB同源蛋白5 (AlkB homolog 5, ALKBH5). FTO定位于核散斑及细胞质, 对RNA上的m6A具有较强的去甲基活性. FTO与核斑点的部分共域化实验结果表明mRNA中的m6A是FTO的作用底物[8]. Su等[24]分析了FTO催化m6A和N6, 2'-O-二甲基腺苷(m6Am)修饰的亲和力, 结果证实FTO与其底物的结合与FTO蛋白的定位有关. 在细胞核中FTO只催化m6A去甲基化, 而在细胞质中FTO既可以介导m6A又可以催化m6Am的去甲基化. 有趣的是, Mauer等[25]研究表明FTO对m6Am的去甲基化酶活性大约是m6A的100倍, 显示FTO的作用底物更可能是m6Am, 这与Jia等人的研究结果有所不同[8]. 目前多数学者认为, FTO在细胞核中对m6A亲和力较高, 而在细胞质中对m6Am亲和力更强[26,27]. ALKBH5主要定位在核散斑, 影响mRNA的出核转运[28]. FTO与ALKBH5对m6A修饰的去甲基化作用是相互独立的. ALKBH5可直接将m6A氧化为普通的腺苷, 而FTO则是分级代谢, 依次将m6A氧化为N6-羟甲基腺苷(hm6A)和N6-甲酰腺苷(f6A), 最后水解为腺嘌呤. 此外, 二者对底物的选择性和序列的亲和力也存在差别, 即m6A的"Erasers"可能因底物的"构象标记"不同而作用相异. 如FTO在小鼠大脑中表达水平高, 而ALKBH5在小鼠睾丸中表达较高. 这种在不同组织间的表达差异及其对底物的选择性差别显示了去甲基化酶生理作用的复杂性.
第三类与m6A甲基化位点结合并读取其信息的蛋白质被称为m6A"Readers", 即m6A识别蛋白.它们能够选择性识别靶RNA上的m6A并参与RNA的多种代谢过程. Readers包括YTHs家族蛋白、HNRNPs超家族蛋白和IGFBPs家族蛋白三组成员. 人具有5个含有YTH结构域的蛋白: 如定位于细胞质中的YTHDF1、YTHDF2、YTHDF3、YTHDC2和定位于细胞核的YTHDC1. YTHDF蛋白虽然结构相似, 但功能不完全相同. YTHDF2在细胞质中促进mRNA的降解[29-31]. YTHDF1结合m6A后促进mRNA的翻译[32]. YTHDF2和YTHDF1的不同功能可能在维持mRNA降解和翻译的平衡中发挥重要作用. YTHDF3与YTHDF1协同促进蛋白合成, 并通过YTHDF2介导mRNA降解[33]. YTHDC1促进外显子的剪切以及控制其靶标的出核过程[34]. YTHDC2能够与RNA解旋酶相互作用, 调节mRNA翻译延伸[35]. 总之, YTHDF2、YTHDF3、YTHDC2可以促进mRNA降解; YTHDF1、YTHDF3、YTHDC2可以促进靶mRNA的翻译; YTHDC1则影响mRNA的剪接及其核输出过程. HNRNPs超家族蛋白包括三个成员: HNRNPA2B1、HNRNPC和HNRNPG. 这3个蛋白均在核内发挥作用. 其中HNRNPC和HNRNPG影响mRNA定位和可变剪切[36]. HNRNPA2B1促进miRNA初级转录本的剪接加工[37]. IGF2BPs包括IGF2BP1-3. 这类蛋白通过常见的RNA结合域识别具有m6A的RNA. IGF2BPs识别结合m6A修饰位点后, 增加mRNA的稳定性, 避免其降解并促进其翻译[38]. 此外, 真核起始因子eIF3能够直接和mRNA 5ʹUTR的m6A修饰位点结合, 招募43S核糖体复合物并促进蛋白翻译[39]. 有趣的是, 细胞质中METTL3也可作为识别蛋白并促进某些特定类型细胞mRNA翻译[40].
总之, m6A甲基化是一个动态、可逆的过程, MAC、MACOM及METTL16均能催化m6A形成, 而FTO和ALKBH5可使其去甲基化. 已甲基化的RNA被相应的Readers识别后才进入下一步代谢过程以保证其有效发挥生物学功能, 包括维持RNA稳定、促进mRNA翻译、或加速其剪切和降解等. 具体见图1.
m6A既是mRNA加工和代谢的重要分子标志, 又在细胞分化进程中具有改变细胞状态的功能[41], 如与细胞异型增生有关的mRNA可变剪切常受m6A调控[42], 表明m6A与肿瘤发生有关. 研究证实m6A相关的蛋白在乳腺癌[43]、肺癌[40]、急性髓细胞白血病[44]、宫颈癌[45]存在过表达或者低表达, 并通过多种机制促进相应肿瘤进展. 下面总结m6A在消化系肿瘤中的作用.
Wu等[46]发现经过香烟烟雾冷凝物染毒后, 食管鳞状细胞癌(esophageal squamous cell carcinoma, ESCC)中ALKBH5表达升高. ALKBH5高表达介导LINC00278的去甲基化, 抑制LINC00278-sORF1翻译YY1BM. 而YY1BM可阻断YY1与雄激素受体(AR)相互作用, YY1BM表达降低可促进AR信号通路介导的eEF2K上调, 使肿瘤细胞适应缺乏营养的环境而减少凋亡.这项研究从m6A去甲基化角度探索了吸烟促进ESCC进展的分子机制. Liu等[47]研究发现FTO在ESCC组织中的表达亦高于邻近的正常组织, 并且与患者不良预后有关. FTO高表达正向调控癌基因MMP13, MMP13上调促进了ESCC转移. Yang等[48]研究证实与癌旁正常组织相比, ESCC组织中YTHDC2的表达下调, 低表达YTHDC2有助于ESCC扩散, 表明m6A去甲基化酶及个别识别蛋白参与了ESCC的发生及发展过程, 但仍缺乏全面挖掘. m6A甲基化酶是否参与食管癌的发病尚不清楚.
Ge等[49]收集100例胃癌(gastric carcinoma, GC)患者、30例良性胃病患者和75例健康对照的外周血液, 通过比色法检查发现GC组患者外周血RNA中m6A水平明显高于胃病和健康对照组. 随着GC的进展m6A水平升高, 手术后则下降. 受试者工作特性(receiver operating characteristic, ROC)曲线评估显示, GC组m6A的曲线下面积明显大于癌胚抗原(CEA)和糖类抗原199(CA19-9)的面积, 表明外周血总RNA的m6A检测对GC诊断及预后有一定参考价值, 其作用优于常规的癌胚抗原, 显示出m6A作为GC诊断标记物有较好的临床应用前景, 但其特异性和敏感性仍需扩大病例数进行临床大样本、多中心验证.
Yue等[50]发现METTL3在GC细胞中表达上调, METTL3表达水平升高提示预后不良. 进一步研究发现, ZMYM1是METTL3的m6A修饰靶标. METTL3通过对ZMYM1进行m6A修饰而增强ZMYM1 mRNA的稳定性. 另外, ZMYM1通过形成CtBP/LSD1/CoREST复合物与上皮细胞钙黏蛋白(E-cadherin)基因启动子结合并抑制其表达, 从而促进上皮细胞-间充质转化(epithelial mesenchymal transition, EMT)和癌细胞转移. Liu等[51]同样发现METTL3在GC细胞中表达上调, 其表达水平随着肿瘤分期和分级的进展而逐渐升高. 敲低METTL3显著抑制了GC细胞的总体RNA的m6A水平, 也显著降低了与GFI-1和α-平滑肌肌动蛋白的表达, 从而抑制GC细胞的迁移.
METTL14与METTL3虽同属于m6A"Writers", 但Zhang等[52]研究发现敲降METTL14激活了Wnt和PI3K-Akt信号传导通路, 促进GC细胞的增殖和侵袭, 反而显示出METTL14的抑癌作用. METTL3与METTL14在GCm6A甲基化过程中的不同结果显示出"Writers" 调控机制的复杂性. Xu等[53]证实与癌旁非肿瘤组织相比, GC组织中FTO在蛋白水平和mRNA水平的表达均上调. FTO表达水平与细胞低分化、淋巴结转移、患者的不良预后密切相关. 进一步研究发现, FTO的表达与GC的TNM分期正相关, FTO表达下调可显著抑制GC细胞的增殖、迁移和侵袭. 然而, Li等[54]发现, FTO蛋白在印戒细胞癌和GC组织中表达水平显著下调. FTO在GC中的作用有待进一步研究.
Wu等[55]发现lncRNA RP11 (RP11-138 J23.1)在结直肠癌(colorectal carcinoma, CRC)组织中高表达, 其表达水平随着CRC分期的上升而增加. RP11有正向调节CRC细胞的迁移、EMT、促进CRC细胞肝转移的功能. 作者发现在HCT-15和HCT-8细胞中, 过表达的METTL3可提高RP11表达水平, 增加RP11和hnRNPA2B1之间的结合, 促进RP11/hnRNPA2B1/mRNA复合物的形成. Zeb1作为EMT转录因子之一, 其上调促进了细胞迁移. 研究发现, 两种泛素E3连接酶(Siah1和Fbxo45)通过泛素-蛋白酶体途径诱导Zeb1降解, 而RP11/hnRNPA2B1/mRNA复合物可促进Siah1和Fbxo45的降解, 使Zeb1水平增高, 从而有助于CRC细胞EMT及迁移, 表明METTL3具有促癌作用. Peng等[56]发现METTL3在CRC细胞表达上调, 其高表达与CRC转移呈正相关. METTL3通过甲基化pri-miR-1246使其成熟为miR-1246. miR-1246可抑制抑癌基因SPRED2的表达, 降低其在Raf/MEK/ERK通路中发挥的抑癌作用, 从而促进CRC细胞迁移、侵袭, 并诱导EMT.
多项研究证实, METTL3通过稳定mRNA的方式, 促进CRC进展. Li等[57]通过分析TCGA数据库, 发现METTL3在多数恶性肿瘤中表达水平增加. 高表达METTL3的CRC患者总生存期和无病生存期缩短. 他们发现Y染色体性别决定区-box 2 (SOX2)是METTL3的下游基因, METTL3通过维持SOX2在CRC中的表达以促进CRC细胞的干性. 进一步研究发现, METTL3提高了SOX2转录本的甲基化水平, 阻止其被降解[58,59]. Zhang等[60]通过qRT-PCR和IHC发现在化疗耐药的结肠癌组织中CBX8明显过表达. CBX8将KMT2b招募到LGR5启动子上, 使其维持H3K4me3状态以促进LGR5的表达, 增加肿瘤细胞的干性, 并降低其化疗敏感性. 进一步研究发现, METTL3的甲基化修饰和IGF2BP1的结合有助于维持CBX8 mRNA的稳定性. 另外, Zhu等[61]发现METTL3以m6A依赖的方式稳定CCNE1的mRNA, 从而促进CRC细胞的增殖. 与上述研究结果不同的是, Deng等[62]发现METTL3的下调导致p-p38和p-ERK的激活, 促进结肠癌细胞的增殖、迁移和侵袭. Chen等[63]发现同样作为"Writer"的METTL14在CRC组织和细胞系中表达下调, 并与患者总生存期密切相关. METTL14基因的敲低显著降低总RNA中的m6A水平, 促进CRC细胞生长. 相反, METTL14过表达则增加总RNA中m6A水平, 抑制CRC细胞生长和转移. 上述研究表明, 在CRC中METTL14发挥抑癌作用, 这与GC的结果相一致[52].
Wang等[64]研究证实与IGF2BP2稳定性有关的lincRNA LINRIS在CRC组织中上调. LINRIS通过自噬-溶酶体途径阻断IGF2BP2的K139泛素化, 抑制IGF2BP2降解, 促进IGF2BP2的下游效应, 尤其是MYC介导的糖酵解过程, 使CRC耐药性增强[65]. YTHDF已被多项研究证明与CRC的发生和进展有关. Bai等[66]证实YTHDF1在CRC中高表达, 且与肿瘤TNM分期、远处转移和肿瘤组织学分型有关. 研究表明, 癌基因c-Myc可与YTHDF1的5ʹUTR结合, 促进YTHDF1的表达. 而敲除YTHDF1则可抑制Wnt/β-catenin信号通路, 表明YTHDF1通过调控Wnt/β-catenin通路促进肿瘤进展[67]. Ni等[68]报道, lncRNA GAS5促进内源性YAP从细胞核到胞浆的移位和磷酸化而加速YAP降解, 进而在体内外抑制CRC进展, 而YTHDF3通过促进lncRNA GAS5降解而阻遏YAP降解, 促进CRC的增殖及转移. 上述研究表明"Readers"在CRC中发挥促癌作用. 然而, 是否存在起抑癌作用的m6A识别蛋白以及METTL3在CRC中究竟扮演何种角色尚有待进一步研究.
Chen等[69]发现, METTL3在肝癌(hepatocellular carcinoma, HCC)中表达上调, 降低了SOCS2 mRNA稳定性, 从而下调抑癌基因SOCS2的表达, 促进HHC病变进展并缩短患者的生存期. Zuo等[70]报道METTL3表达上调可通过METTL3 -LINC00958-HDGF轴促进HCC的发展. Cui等[71]发现, 在肝母细胞瘤组织中METTL3与miR186的表达显著负相关, METTL3表达上调通过Wnt/β-catenin途径促进肝母细胞瘤进展. Ma等[72]证明METTL14可通过与抑癌因子pri-miR-126 DGCR8结合, 抑制HCC细胞转移, METTL14低表达则增强HCC的侵袭和转移能力. Chen等[73]发现WTAP在HCC中高表达, 通过Hur-EtS1 -P21/P27通路促进HCC的发生和发展. Lan等[74]证实KIAA1429在HCC组织中表达上调. GATA3是KIAA1429介导的m6A修饰的下游靶点. KIAA1429在HCC中高表达促进了GATA3 pre-mRNA的3ʹUTR发生m6A修饰, 导致RNA结合蛋白HuR的分离和GATA3 pre-mRNA的降解, 从而加速肿瘤的增殖和转移. 此外, KIAA1429通过上调ID2 mRNA上m6A修饰而抑制ID2的表达, 促进HCC细胞的迁移和侵袭[75].
Zhao等[76]报道, YTHDF1在HCC患者中表达明显上调并与病理分期呈正相关. 作者通过对YTHDF1共表达基因的GO和KEGG通路分析, 发现YTHDF1在调节HCC细胞周期和代谢方面起着重要作用. 有趣的是, Zhou等[77]发现, METTL3与YTHDF1共过表达的患者预后差. KEGG分析表明, 两者介导HCC不良预后的机制与DNA复制、RNA的剪切和降解有关, METTL3和YTHDF1联合检测具有判断HCC恶性程度和评估预后的价值.
Yang等[78]发现YTHDF2在HCC中表达上调, 并受到miR145负性调节. Zhang等[79]的研究表明, 敲除YTHDF2后, Hep3B和Huh7两种HCC细胞株的干细胞特性受损, 而YTHDF2过表达会增加肿瘤干细胞(CSC)的表型. 在HCC中YTHDF2过表达通过调控OCT4 mRNA的m6A甲基化使OCT4表达上调, 促进CSC生长, 维持CSC表型, 从而促进HCC转移. 与上述结果相反, Hou等[80]发现在YTHDF2缺陷的HCC中, STAT3的磷酸化以及IL-11和SERPINE2的表达上调, 而缺氧可以增强这种作用. 缺氧条件下诱导的YTHDF2下调加剧了HCC组织中的炎症反应, 促进了HCC组织中的细胞增殖及血管异常化, 促进HCC细胞生长和病灶转移, 表明YTHDF2发挥了抑癌作用. 有趣的是, Zhong等[81]的研究表明同样在缺氧环境下, 下调YTHDF2诱导ERK磷酸化, 激活ERK/MAPK通路, 从而促进癌细胞的增殖. YTHDF2在HCC组织中直接结合EGFR 3'UTR的m6A修饰位点, 促进EGFR mRNA的降解, 进而抑制ERK/MAPK信号通路发挥抑癌作用. 多项研究表明IGF2BPs 与HCC细胞的增殖、转移、侵袭能力和集落形成能力呈正相关[82,83], 但一些相互矛盾的结果显示m6A相关蛋白在HCC中的作用机制仍需进一步研究.
Li等[84]发现低m6A水平的HCC中FTO表达明显升高. 丙酮酸激酶M2 (pyruvate kinase M2, PKM2)在肿瘤细胞供能、EMT及侵袭转移方面有重要作用, 是FTO的靶蛋白之一. 敲除FTO后, HCC组织增殖与克隆能力减弱, 同时可减少PKM2 mRNA表达及其蛋白生成, 而过表达PKM2可逆转敲除FTO的表型改变, 表明FTO诱导的PKM2 mRNA的去甲基化具有促癌作用. 有趣的是, Rong[85]等发现FTO在肝内胆管细胞癌(ICC)细胞中的表达下调, 且FTO的表达与CA19-9的表达呈负相关, Kaplan-Meier生存分析显示, FTO低表达预示ICC预后不良, 这表明FTO可能有抑制ICC进程的作用. 体外实验发现FTO还可抑制ICC细胞生长和迁移. 另外, 裸鼠体内过表达FTO抑制了ICC肿瘤的生长. 这可能与FTO提高了TEAD2mRNA的稳定性有关. 结合上述研究结果, METTL14在GC、CRC、HCC三种肿瘤中均起抑癌作用[52,63,72], 而METTL3主要发挥促癌作用[50,57,69]. FTO在多种肿瘤中出现异常表达, 但其作用及机制均有待进一步明确.
Xia等[86]发现与正常细胞相比, 胰腺癌(pancreatic carcinoma, PC)组织中METTL3和mRNA水平显著升高, 敲除METTL3后, RNA m6A修饰减少, 抑制了肿瘤细胞增殖、侵袭和迁移, 表明METTL3具有促癌作用. 此外, Taketo等[87]发现m6A修饰与PC化疗药物的敏感度有关, METTL3缺失的细胞对抗癌药如吉西他滨、5-氟尿嘧啶、顺铂和辐射显示出更高的敏感性. 进一步研究表明, 包括MAPK级联反应、泛素化过程、RNA剪接等多条关键途径可能均是METTL3的作用靶点. 这些信号通路的改变直接导致了肿瘤进展和化疗耐药, 表明METTL3参与了PC放疗及化疗耐药的调控. 此外, 他们还发现METTL3的下调加速了PC细胞凋亡. m6A还可以通过影响miRNA或lncRNA调节PC的发生. Zhang等[88]发现胰管上皮细胞中, 香烟烟雾冷凝物促使有致癌作用的miR-25成熟依赖于METTL3过表达介导的m6A甲基化. METTL3在PC中的促癌作用, 与其在GC、CRC及HCC中所起的作用一致[50,57,69].
Tang等[89]发现ALKBH5在基于吉西他滨治疗患者的异种移植瘤模型中下调, 其过表达增加PC对化疗的敏感性. 在体外和体内, 沉默ALKBH5显著增加PC增殖、迁移和侵袭, 而过表达ALKBH5导致完全相反的效果. 进一步研究发现ALKBH5通过降低Wnt抑制因子1的m6A水平及阻碍Wnt信号通路来抑制PC的发生. He等[90]的研究发现, ALKBH5过表达后, KCNK15-AS表达随之增加, 而PC细胞的EMT受到了抑制, 提示KCNK 15-AS1对PC细胞迁移和侵袭具有抑制作用, 也间接证实ALKBH5可抑制EMT进程. Cho等[91]证实ALKBH5是PC患者的独立预后因素. ALKBH5表达与患者预后显著正相关. Chen等[92]发现与正常组织相比, PC组织中YTHDF2的表达在基因和蛋白质水平上均上调. 高表达的YTHDF2在PC组织中显示出双重作用: 一方面促进癌细胞增殖; 另一方面YTHDF2通过YAP信号调控EMT, 抑制PC细胞的转移和侵袭. 此外, Tang[93]等研究亦表明, FTO在PC中高表达. 敲除FTO后可抑制肿瘤的形成和进展, 增加PC细胞凋亡, 表明FTO具有促癌作用.
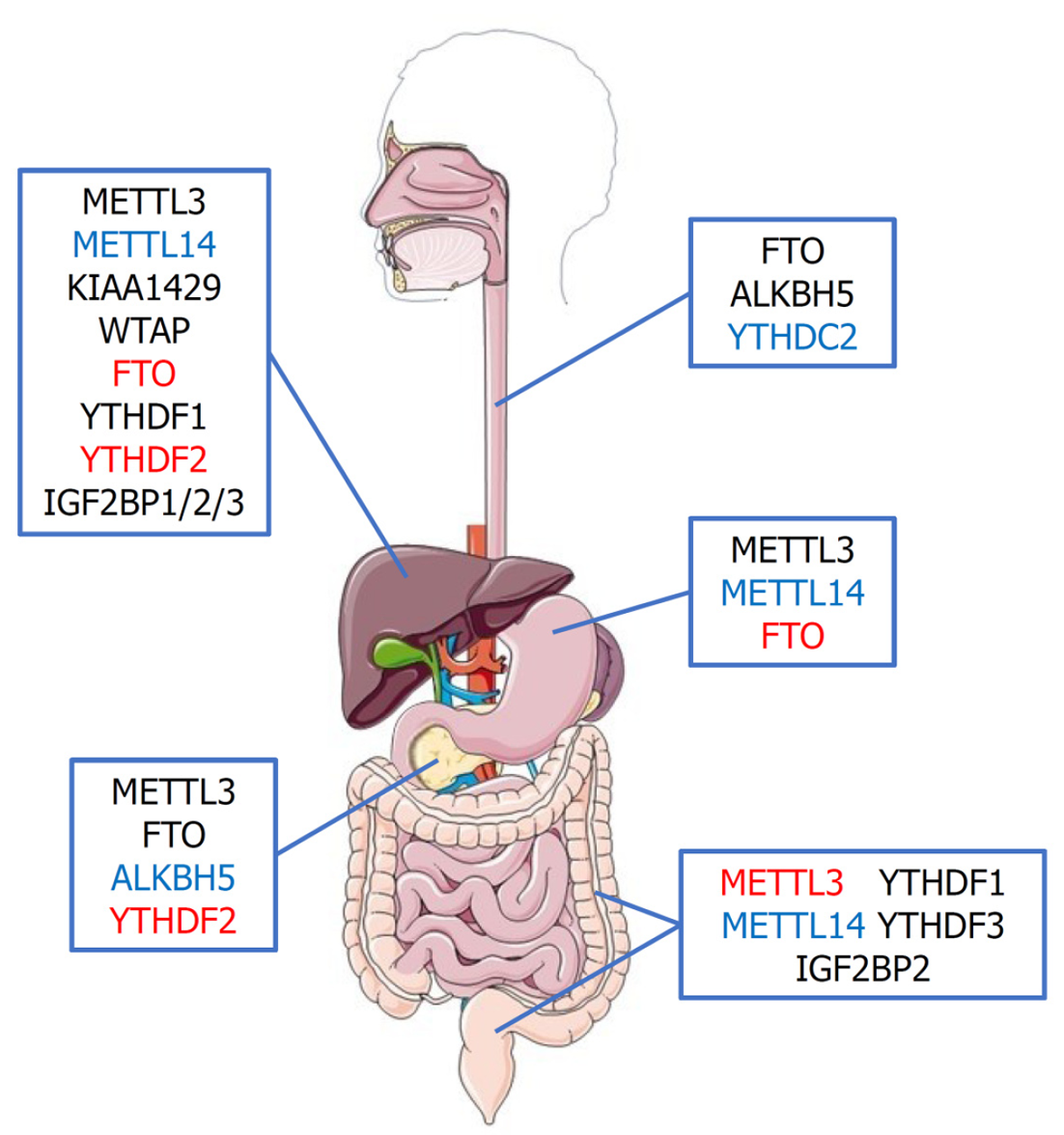
RNA m6A甲基化修饰相关蛋白在消化系统食管、胃、肝脏、胰腺及结直肠恶性肿瘤发病中的作用如图2所示.
m6A甲基化修饰的发现丰富了表观遗传学的内容. m6A甲基转移酶、去甲基化酶和识别蛋白各司其职, 共同调节靶基因上的m6A含量, 进而影响肿瘤的发生. 以m6A修饰为核心的靶向治疗已经成为临床研究的新热点. 其中包括: FTO抑制剂、METTL3-14/WTAP激活剂及其联合用药. 目前m6A相关蛋白抑制剂或激活剂存在活性低、特异性差、在细胞内确切作用机制不清等问题, 其开发尚处于起步阶段. 许多问题仍未解决: (1)未知的m6A修饰相关蛋白有待进一步鉴定; (2)新的m6A修饰机制有待进一步阐明; (3)鉴于甲基化酶和去甲基化酶在m6A修饰中起相反的作用, 如果去甲基化酶在某种癌症中发挥癌基因的作用, 是否甲基转移酶一定扮演抑癌基因的角色? (4)有些m6A修饰蛋白具有双重身份, 既可促癌又能抑癌, 这种"双刃刀"作用的触发机制又是什么? (5)"Readers"是如何选择识别并结合靶RNA? 不同"Reader"之间是相互竞争还是具有协同作用? 哪些m6A甲基化蛋白具有早癌筛查和/或肿瘤预后判断的临床价值? 相信随着m6A甲基化研究的不断深入, 终将揭开这些谜团, 最终为肿瘤患者带来福音.
学科分类: 胃肠病学和肝病学
手稿来源地: 广东省
同行评议报告学术质量分类
A级 (优秀): 0
B级 (非常好): B
C级 (良好): C, C, C, C
D级 (一般): 0
E级 (差): 0
科学编辑: 刘继红 制作编辑:张砚梁
| 1. | Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, Dai Q, Di Segni A, Salmon-Divon M, Clark WC, Zheng G, Pan T, Solomon O, Eyal E, Hershkovitz V, Han D, Doré LC, Amariglio N, Rechavi G, He C. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016;530:441-446. [PubMed] [DOI] |
| 2. | Li X, Xiong X, Wang K, Wang L, Shu X, Ma S, Yi C. Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat Chem Biol. 2016;12:311-316. [PubMed] [DOI] |
| 3. | Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, Sorek R, Rechavi G. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201-206. [PubMed] [DOI] |
| 4. | Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, de Crécy-Lagard V, Ross R, Limbach PA, Kotter A, Helm M, Bujnicki JM. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46:D303-D307. [PubMed] [DOI] |
| 5. | HALL RH. A general procedure for the isolation of "minor" nucleosides from ribonucleic acid hydrolysates. Biochemistry. 1965;4:661-670. [PubMed] [DOI] |
| 6. | Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci USA. 1974;71:3971-3975. [PubMed] [DOI] |
| 7. | Wei CM, Gershowitz A, Moss B. Methylated nucleotides block 5' terminus of HeLa cell messenger RNA. Cell. 1975;4:379-386. [PubMed] [DOI] |
| 8. | Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, He C. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885-887. [PubMed] [DOI] |
| 9. | Ke S, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A, Haripal B, Zucker-Scharff I, Moore MJ, Park CY, Vågbø CB, Kusśnierczyk A, Klungland A, Darnell JE, Darnell RB. A majority of m6A residues are in the last exons, allowing the potential for 3' UTR regulation. Genes Dev. 2015;29:2037-2053. [PubMed] [DOI] |
| 10. | Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. 2012;149:1635-1646. [PubMed] [DOI] |
| 11. | Liu ZX, Li LM, Sun HL, Liu SM. Link Between m6A Modification and Cancers. Front Bioeng Biotechnol. 2018;6:89. [PubMed] [DOI] |
| 12. | Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, Jaffrey SR. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369-373. [PubMed] [DOI] |
| 13. | Alarcón CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519:482-485. [PubMed] [DOI] |
| 14. | Knuckles P, Lence T, Haussmann IU, Jacob D, Kreim N, Carl SH, Masiello I, Hares T, Villaseñor R, Hess D, Andrade-Navarro MA, Biggiogera M, Helm M, Soller M, Bühler M, Roignant JY. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m6A machinery component Wtap/Fl(2)d. Genes Dev. 2018;32:415-429. [PubMed] [DOI] |
| 15. | Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, Dai Q, Chen W, He C. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93-95. [PubMed] [DOI] |
| 16. | Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, Gong Z, Wang Q, Huang J, Tang C, Zou T, Yin P. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016;534:575-578. [PubMed] [DOI] |
| 17. | Wang P, Doxtader KA, Nam Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol Cell. 2016;63:306-317. [PubMed] [DOI] |
| 18. | Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, Zhao X, Li A, Yang Y, Dahal U, Lou XM, Liu X, Huang J, Yuan WP, Zhu XF, Cheng T, Zhao YL, Wang X, Rendtlew Danielsen JM, Liu F, Yang YG. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177-189. [PubMed] [DOI] |
| 19. | Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, Cheng T, Gao M, Shu X, Ma H, Wang F, Wang X, Shen B, Wang Y, Feng X, He C, Liu J. VIRMA mediates preferential m6A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4:10. [PubMed] [DOI] |
| 20. | Horiuchi K, Kawamura T, Iwanari H, Ohashi R, Naito M, Kodama T, Hamakubo T. Identification of Wilms' tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J Biol Chem. 2013;288:33292-33302. [PubMed] [DOI] |
| 21. | Warda AS, Kretschmer J, Hackert P, Lenz C, Urlaub H, Höbartner C, Sloan KE, Bohnsack MT. Human METTL16 is a N6-methyladenosine (m6A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017;18:2004-2014. [PubMed] [DOI] |
| 22. | Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP, Conrad NK. The U6 snRNA m6A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell. 2017;169:824-835.e14. [PubMed] [DOI] |
| 23. | Wu LF, Wei BL, Guo YT, Ye YQ, Li GP, Pu ZJ, Feng JL. Apoptosis induced by adenosine involves endoplasmic reticulum stress in EC109 cells. Int J Mol Med. 2012;30:797-804. [PubMed] [DOI] |
| 24. | Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, Deng X, Wang Y, Weng X, Hu C, Yu M, Skibbe J, Dai Q, Zou D, Wu T, Yu K, Weng H, Huang H, Ferchen K, Qin X, Zhang B, Qi J, Sasaki AT, Plas DR, Bradner JE, Wei M, Marcucci G, Jiang X, Mulloy JC, Jin J, He C, Chen J. R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m6A/MYC/CEBPA Signaling. Cell. 2018;172:90-105.e23. [PubMed] [DOI] |
| 25. | Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, Linder B, Pickering BF, Vasseur JJ, Chen Q, Gross SS, Elemento O, Debart F, Kiledjian M, Jaffrey SR. Reversible methylation of m6Am in the 5' cap controls mRNA stability. Nature. 2017;541:371-375. [PubMed] [DOI] |
| 26. | Wu W, Feng J, Jiang D, Zhou X, Jiang Q, Cai M, Wang X, Shan T, Wang Y. AMPK regulates lipid accumulation in skeletal muscle cells through FTO-dependent demethylation of N6-methyladenosine. Sci Rep. 2017;7:41606. [PubMed] [DOI] |
| 27. | Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC, Shi H, Cui X, Su R, Klungland A, Jia G, Chen J, He C. Differential m6A, m6Am, and m1A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol Cell. 2018;71:973-985.e5. [PubMed] [DOI] |
| 28. | Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, Lu Z, Bosmans RP, Dai Q, Hao YJ, Yang X, Zhao WM, Tong WM, Wang XJ, Bogdan F, Furu K, Fu Y, Jia G, Zhao X, Liu J, Krokan HE, Klungland A, Yang YG, He C. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18-29. [PubMed] [DOI] |
| 29. | Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, Ren B, Pan T, He C. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117-120. [PubMed] [DOI] |
| 30. | Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, Ma J, Wu L. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626. [PubMed] [DOI] |
| 31. | Park OH, Ha H, Lee Y, Boo SH, Kwon DH, Song HK, Kim YK. Endoribonucleolytic Cleavage of m6A-Containing RNAs by RNase P/MRP Complex. Mol Cell. 2019;74:494-507.e8. [PubMed] [DOI] |
| 32. | Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161:1388-1399. [PubMed] [DOI] |
| 33. | Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, Liu C, He C. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017;27:315-328. [PubMed] [DOI] |
| 34. | Xu C, Wang X, Liu K, Roundtree IA, Tempel W, Li Y, Lu Z, He C, Min J. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol. 2014;10:927-929. [PubMed] [DOI] |
| 35. | Tanabe A, Tanikawa K, Tsunetomi M, Takai K, Ikeda H, Konno J, Torigoe T, Maeda H, Kutomi G, Okita K, Mori M, Sahara H. RNA helicase YTHDC2 promotes cancer metastasis via the enhancement of the efficiency by which HIF-1α mRNA is translated. Cancer Lett. 2016;376:34-42. [PubMed] [DOI] |
| 36. | Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, Calderaro J, Bioulac-Sage P, Letexier M, Degos F, Clément B, Balabaud C, Chevet E, Laurent A, Couchy G, Letouzé E, Calvo F, Zucman-Rossi J. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44:694-698. [PubMed] [DOI] |
| 37. | Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell. 2015;162:1299-1308. [PubMed] [DOI] |
| 38. | Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, Zhao BS, Mesquita A, Liu C, Yuan CL, Hu YC, Hüttelmaier S, Skibbe JR, Su R, Deng X, Dong L, Sun M, Li C, Nachtergaele S, Wang Y, Hu C, Ferchen K, Greis KD, Jiang X, Wei M, Qu L, Guan JL, He C, Yang J, Chen J. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285-295. [PubMed] [DOI] |
| 39. | Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR. 5' UTR m(6)A Promotes Cap-Independent Translation. Cell. 2015;163:999-1010. [PubMed] [DOI] |
| 40. | Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol Cell. 2016;62:335-345. [PubMed] [DOI] |
| 41. | Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2017;18:31-42. [PubMed] [DOI] |
| 42. | Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, Sun G, Lu Z, Huang Y, Yang CG, Riggs AD, He C, Shi Y. m6A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep. 2017;18:2622-2634. [PubMed] [DOI] |
| 43. | Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I, He X, Semenza GL. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proc Natl Acad Sci USA. 2016;113:E2047-E2056. [PubMed] [DOI] |
| 44. | Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, Chen Y, Sulman EP, Xie K, Bögler O, Majumder S, He C, Huang S. m6A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell. 2017;31:591-606.e6. [PubMed] [DOI] |
| 45. | Zhou S, Bai ZL, Xia D, Zhao ZJ, Zhao R, Wang YY, Zhe H. FTO regulates the chemo-radiotherapy resistance of cervical squamous cell carcinoma (CSCC) by targeting β-catenin through mRNA demethylation. Mol Carcinog. 2018;57:590-597. [PubMed] [DOI] |
| 46. | Wu S, Zhang L, Deng J, Guo B, Li F, Wang Y, Wu R, Zhang S, Lu J, Zhou Y. A Novel Micropeptide Encoded by Y-Linked LINC00278 Links Cigarette Smoking and AR Signaling in Male Esophageal Squamous Cell Carcinoma. Cancer Res. 2020;80:2790-2803. [PubMed] [DOI] |
| 47. | Liu S, Huang M, Chen Z, Chen J, Chao Q, Yin X, Quan M. FTO promotes cell proliferation and migration in esophageal squamous cell carcinoma through up-regulation of MMP13. Exp Cell Res. 2020;389:111894. [PubMed] [DOI] |
| 48. | Yang N, Ying P, Tian J, Wang X, Mei S, Zou D, Peng X, Gong Y, Yang Y, Zhu Y, Ke J, Zhong R, Chang J, Miao X. Genetic variants in m6A modification genes are associated with esophageal squamous-cell carcinoma in the Chinese population. Carcinogenesis. 2020;41:761-768. [PubMed] [DOI] |
| 49. | Ge L, Zhang N, Chen Z, Song J, Wu Y, Li Z, Chen F, Wu J, Li D, Li J, Wang C, Wang H, Wang J. Level of N6-Methyladenosine in Peripheral Blood RNA: A Novel Predictive Biomarker for Gastric Cancer. Clin Chem. 2020;66:342-351. [PubMed] [DOI] |
| 50. | Yue B, Song C, Yang L, Cui R, Cheng X, Zhang Z, Zhao G. METTL3-mediated N6-methyladenosine modification is critical for epithelial-mesenchymal transition and metastasis of gastric cancer. Mol Cancer. 2019;18:142. [PubMed] [DOI] |
| 51. | Liu T, Yang S, Sui J, Xu SY, Cheng YP, Shen B, Zhang Y, Zhang XM, Yin LH, Pu YP, Liang GY. Dysregulated N6-methyladenosine methylation writer METTL3 contributes to the proliferation and migration of gastric cancer. J Cell Physiol. 2020;235:548-562. [PubMed] [DOI] |
| 52. | Zhang C, Zhang M, Ge S, Huang W, Lin X, Gao J, Gong J, Shen L. Reduced m6A modification predicts malignant phenotypes and augmented Wnt/PI3K-Akt signaling in gastric cancer. Cancer Med. 2019;8:4766-4781. [PubMed] [DOI] |
| 53. | Xu D, Shao W, Jiang Y, Wang X, Liu Y, Liu X. FTO expression is associated with the occurrence of gastric cancer and prognosis. Oncol Rep. 2017;38:2285-2292. [PubMed] [DOI] |
| 54. | Li Y, Zheng D, Wang F, Xu Y, Yu H, Zhang H. Expression of Demethylase Genes, FTO and ALKBH1, Is Associated with Prognosis of Gastric Cancer. Dig Dis Sci. 2019;64:1503-1513. [PubMed] [DOI] |
| 55. | Wu Y, Yang X, Chen Z, Tian L, Jiang G, Chen F, Li J, An P, Lu L, Luo N, Du J, Shan H, Liu H, Wang H. m6A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol Cancer. 2019;18:87. [PubMed] [DOI] |
| 56. | Peng W, Li J, Chen R, Gu Q, Yang P, Qian W, Ji D, Wang Q, Zhang Z, Tang J, Sun Y. Upregulated METTL3 promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK signaling pathway. J Exp Clin Cancer Res. 2019;38:393. [PubMed] [DOI] |
| 57. | Li T, Hu PS, Zuo Z, Lin JF, Li X, Wu QN, Chen ZH, Zeng ZL, Wang F, Zheng J, Chen D, Li B, Kang TB, Xie D, Lin D, Ju HQ, Xu RH. METTL3 facilitates tumor progression via an m6A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer. 2019;18:112. [PubMed] [DOI] |
| 58. | Boumahdi S, Driessens G, Lapouge G, Rorive S, Nassar D, Le Mercier M, Delatte B, Caauwe A, Lenglez S, Nkusi E, Brohée S, Salmon I, Dubois C, del Marmol V, Fuks F, Beck B, Blanpain C. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature. 2014;511:246-250. [PubMed] [DOI] |
| 59. | Ku SY, Rosario S, Wang Y, Mu P, Seshadri M, Goodrich ZW, Goodrich MM, Labbé DP, Gomez EC, Wang J, Long HW, Xu B, Brown M, Loda M, Sawyers CL, Ellis L, Goodrich DW. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science. 2017;355:78-83. [PubMed] [DOI] |
| 60. | Zhang Y, Kang M, Zhang B, Meng F, Song J, Kaneko H, Shimamoto F, Tang B. m6A modification-mediated CBX8 induction regulates stemness and chemosensitivity of colon cancer via upregulation of LGR5. Mol Cancer. 2019;18:185. [PubMed] [DOI] |
| 61. | Zhu W, Si Y, Xu J, Lin Y, Wang JZ, Cao M, Sun S, Ding Q, Zhu L, Wei JF. Methyltransferase like 3 promotes colorectal cancer proliferation by stabilizing CCNE1 mRNA in an m6A-dependent manner. J Cell Mol Med. 2020;24:3521-3533. [PubMed] [DOI] |
| 62. | Deng R, Cheng Y, Ye S, Zhang J, Huang R, Li P, Liu H, Deng Q, Wu X, Lan P, Deng Y. m6A methyltransferase METTL3 suppresses colorectal cancer proliferation and migration through p38/ERK pathways. Onco Targets Ther. 2019;12:4391-4402. [PubMed] [DOI] |
| 63. | Chen X, Xu M, Xu X, Zeng K, Liu X, Sun L, Pan B, He B, Pan Y, Sun H, Xia X, Wang S. METTL14 Suppresses CRC Progression via Regulating N6-Methyladenosine-Dependent Primary miR-375 Processing. Mol Ther. 2020;28:599-612. [PubMed] [DOI] |
| 64. | Wang Y, Lu JH, Wu QN, Jin Y, Wang DS, Chen YX, Liu J, Luo XJ, Meng Q, Pu HY, Wang YN, Hu PS, Liu ZX, Zeng ZL, Zhao Q, Deng R, Zhu XF, Ju HQ, Xu RH. LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Mol Cancer. 2019;18:174. [PubMed] [DOI] |
| 65. | Smith B, Schafer XL, Ambeskovic A, Spencer CM, Land H, Munger J. Addiction to Coupling of the Warburg Effect with Glutamine Catabolism in Cancer Cells. Cell Rep. 2016;17:821-836. [PubMed] [DOI] |
| 66. | Bai Y, Yang C, Wu R, Huang L, Song S, Li W, Yan P, Lin C, Li D, Zhang Y. YTHDF1 Regulates Tumorigenicity and Cancer Stem Cell-Like Activity in Human Colorectal Carcinoma. Front Oncol. 2019;9:332. [PubMed] [DOI] |
| 67. | Nishizawa Y, Konno M, Asai A, Koseki J, Kawamoto K, Miyoshi N, Takahashi H, Nishida N, Haraguchi N, Sakai D, Kudo T, Hata T, Matsuda C, Mizushima T, Satoh T, Doki Y, Mori M, Ishii H. Oncogene c-Myc promotes epitranscriptome m6A reader YTHDF1 expression in colorectal cancer. Oncotarget. 2018;9:7476-7486. [PubMed] [DOI] |
| 68. | Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou A, Liu J, Che L, Li J. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m6A reader YTHDF3. Mol Cancer. 2019;18:143. [PubMed] [DOI] |
| 69. | Chen M, Wei L, Law CT, Tsang FH, Shen J, Cheng CL, Tsang LH, Ho DW, Chiu DK, Lee JM, Wong CC, Ng IO, Wong CM. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67:2254-2270. [PubMed] [DOI] |
| 70. | Zuo X, Chen Z, Gao W, Zhang Y, Wang J, Wang J, Cao M, Cai J, Wu J, Wang X. M6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J Hematol Oncol. 2020;13:5. [PubMed] [DOI] |
| 71. | Cui X, Wang Z, Li J, Zhu J, Ren Z, Zhang D, Zhao W, Fan Y, Zhang D, Sun R. Cross talk between RNA N6-methyladenosine methyltransferase-like 3 and miR-186 regulates hepatoblastoma progression through Wnt/β-catenin signalling pathway. Cell Prolif. 2020;53:e12768. [PubMed] [DOI] |
| 72. | Ma JZ, Yang F, Zhou CC, Liu F, Yuan JH, Wang F, Wang TT, Xu QG, Zhou WP, Sun SH. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N6 -methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017;65:529-543. [PubMed] [DOI] |
| 73. | Chen Y, Peng C, Chen J, Chen D, Yang B, He B, Hu W, Zhang Y, Liu H, Dai L, Xie H, Zhou L, Wu J, Zheng S. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol Cancer. 2019;18:127. [PubMed] [DOI] |
| 74. | Lan T, Li H, Zhang D, Xu L, Liu H, Hao X, Yan X, Liao H, Chen X, Xie K, Li J, Liao M, Huang J, Yuan K, Zeng Y, Wu H. KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol Cancer. 2019;18:186. [PubMed] [DOI] |
| 75. | Cheng X, Li M, Rao X, Zhang W, Li X, Wang L, Huang G. KIAA1429 regulates the migration and invasion of hepatocellular carcinoma by altering m6A modification of ID2 mRNA. Onco Targets Ther. 2019;12:3421-3428. [PubMed] [DOI] |
| 76. | Zhao X, Chen Y, Mao Q, Jiang X, Jiang W, Chen J, Xu W, Zhong L, Sun X. Overexpression of YTHDF1 is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Biomark. 2018;21:859-868. [PubMed] [DOI] |
| 77. | Zhou Y, Yin Z, Hou B, Yu M, Chen R, Jin H, Jian Z. Expression profiles and prognostic significance of RNA N6-methyladenosine-related genes in patients with hepatocellular carcinoma: evidence from independent datasets. Cancer Manag Res. 2019;11:3921-3931. [PubMed] [DOI] |
| 78. | Yang Z, Li J, Feng G, Gao S, Wang Y, Zhang S, Liu Y, Ye L, Li Y, Zhang X. MicroRNA-145 Modulates N6-Methyladenosine Levels by Targeting the 3′-Untranslated mRNA Region of the N6-Methyladenosine Binding YTH Domain Family 2 Protein. J Biol Chem. 2017;292:3614-3623. [PubMed] [DOI] |
| 79. | Zhang C, Huang S, Zhuang H, Ruan S, Zhou Z, Huang K, Ji F, Ma Z, Hou B, He X. YTHDF2 promotes the liver cancer stem cell phenotype and cancer metastasis by regulating OCT4 expression via m6A RNA methylation. Oncogene. 2020;39:4507-4518. [PubMed] [DOI] |
| 80. | Hou J, Zhang H, Liu J, Zhao Z, Wang J, Lu Z, Hu B, Zhou J, Zhao Z, Feng M, Zhang H, Shen B, Huang X, Sun B, Smyth MJ, He C, Xia Q. YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol Cancer. 2019;18:163. [PubMed] [DOI] |
| 81. | Zhong L, Liao D, Zhang M, Zeng C, Li X, Zhang R, Ma H, Kang T. YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett. 2019;442:252-261. [PubMed] [DOI] |
| 82. | Aguilo F, Zhang F, Sancho A, Fidalgo M, Di Cecilia S, Vashisht A, Lee D, Chen C, Rengasamy M, Andino B, Jahouh F, Roman A, Krig SR, Wang R, Zhang W, Wohlschlegel JA, Wang J, Walsh MJ. Coordination of m(6)A mRNA Methylation and Gene Transcription by ZFP217 Regulates Pluripotency and Reprogramming. Cell Stem Cell. 2015;17:689-704. [PubMed] [DOI] |
| 83. | Fawzy IO, Hamza MT, Hosny KA, Esmat G, El Tayebi HM, Abdelaziz AI. miR-1275: A single microRNA that targets the three IGF2-mRNA-binding proteins hindering tumor growth in hepatocellular carcinoma. FEBS Lett. 2015;589:2257-2265. [PubMed] [DOI] |
| 84. | Li J, Zhu L, Shi Y, Liu J, Lin L, Chen X. m6A demethylase FTO promotes hepatocellular carcinoma tumorigenesis via mediating PKM2 demethylation. Am J Transl Res. 2019;11:6084-6092. [PubMed] |
| 85. | Rong ZX, Li Z, He JJ, Liu LY, Ren XX, Gao J, Mu Y, Guan YD, Duan YM, Zhang XP, Zhang DX, Li N, Deng YZ, Sun LQ. Downregulation of Fat Mass and Obesity Associated (FTO) Promotes the Progression of Intrahepatic Cholangiocarcinoma. Front Oncol. 2019;9:369. [PubMed] [DOI] |
| 86. | Xia T, Wu X, Cao M, Zhang P, Shi G, Zhang J, Lu Z, Wu P, Cai B, Miao Y, Jiang K. The RNA m6A methyltransferase METTL3 promotes pancreatic cancer cell proliferation and invasion. Pathol Res Pract. 2019;215:152666. [PubMed] [DOI] |
| 87. | Taketo K, Konno M, Asai A, Koseki J, Toratani M, Satoh T, Doki Y, Mori M, Ishii H, Ogawa K. The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. Int J Oncol. 2018;52:621-629. [PubMed] [DOI] |
| 88. | Zhang J, Bai R, Li M, Ye H, Wu C, Wang C, Li S, Tan L, Mai D, Li G, Pan L, Zheng Y, Su J, Ye Y, Fu Z, Zheng S, Zuo Z, Liu Z, Zhao Q, Che X, Xie D, Jia W, Zeng MS, Tan W, Chen R, Xu RH, Zheng J, Lin D. Excessive miR-25-3p maturation via N6-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat Commun. 2019;10:1858. [PubMed] [DOI] |
| 89. | Tang B, Yang Y, Kang M, Wang Y, Wang Y, Bi Y, He S, Shimamoto F. m6A demethylase ALKBH5 inhibits pancreatic cancer tumorigenesis by decreasing WIF-1 RNA methylation and mediating Wnt signaling. Mol Cancer. 2020;19:3. [PubMed] [DOI] |
| 90. | He Y, Hu H, Wang Y, Yuan H, Lu Z, Wu P, Liu D, Tian L, Yin J, Jiang K, Miao Y. ALKBH5 Inhibits Pancreatic Cancer Motility by Decreasing Long Non-Coding RNA KCNK15-AS1 Methylation. Cell Physiol Biochem. 2018;48:838-846. [PubMed] [DOI] |
| 91. | Cho SH, Ha M, Cho YH, Ryu JH, Yang K, Lee KH, Han ME, Oh SO, Kim YH. ALKBH5 gene is a novel biomarker that predicts the prognosis of pancreatic cancer: A retrospective multicohort study. Ann Hepatobiliary Pancreat Surg. 2018;22:305-309. [PubMed] [DOI] |
| 92. | Chen J, Sun Y, Xu X, Wang D, He J, Zhou H, Lu Y, Zeng J, Du F, Gong A, Xu M. YTH domain family 2 orchestrates epithelial-mesenchymal transition/proliferation dichotomy in pancreatic cancer cells. Cell Cycle. 2017;16:2259-2271. [PubMed] [DOI] |
| 93. | Tang X, Liu S, Chen D, Zhao Z, Zhou J. The role of the fat mass and obesity-associated protein in the proliferation of pancreatic cancer cells. Oncol Lett. 2019;17:2473-2478. [PubMed] [DOI] |