修回日期: 2016-06-17
接受日期: 2016-06-27
在线出版日期: 2016-12-28
基因组编辑技术是位点特异性基因靶向操作的重要工具, 研究者可通过该技术实现对感兴趣的目标基因或DNA片段进行"编辑", 实现对靶DNA的定点敲除、敲入等基因组改造. 人们已经利用该技术实现包括从低等微生物到人的基因组改造的目标. 该技术的应用将在消化系肿瘤早期分期分型、精准治疗和预后判断等方面发挥重要的作用. 本文综述了目前较为成熟的以核酸酶为工具的包括类转录激活因子效应物核酸酶、锌指核酸酶以及成簇的规律间隔的短回文重复序列及相关基因在内的多种基因组编辑技术的原理及其在消化系肿瘤研究中的应用最新进展.
核心提要: 作为近年来发展和成熟起来的一系列的基因组定点改造技术, 基因组编辑技术已经在包括人类在内的多种生物体内获得成功. 本文综述了目前几种主要的基因组技术的基本原理及其在消化系肿瘤诊疗等方面的应用, 将为消化系肿瘤早期分期分型、精准治疗和预后判断等方面临床应用提供强有力的工具.
引文著录: 吕玉红, 李晓倩, 岳昌武, 王苗. 基因组编辑技术最新进展及其在消化系肿瘤研究中的应用. 世界华人消化杂志 2016; 24(36): 4772-4780
Revised: June 17, 2016
Accepted: June 27, 2016
Published online: December 28, 2016
Genome editing is a site-directed modification technology for gene targeting and a powerful tool to edit the target DNA by site-specific DNA knockout or knockin. Genome editing has achieved a considerable success from lower microbes to human in the past years and may play a very important role in tumor staging, precision medicine as well as prognosis evaluation in gastrointestinal cancers. This review discusses the mechanisms of different genome-editing strategies and describes each of the common nuclease-based platforms, including transcription activator-like effector nucleases, zinc finger nucleases and the CRISPR/Cas9 system. We also summarize the progress made in applying genome editing to the research of gastrointestinal cancers.
- Citation: Lv YH, Li XQ, Yue CW, Wang M. Application of genome editing technologies in gastrointestinal cancers. Shijie Huaren Xiaohua Zazhi 2016; 24(36): 4772-4780
- URL: https://www.wjgnet.com/1009-3079/full/v24/i36/4772.htm
- DOI: https://dx.doi.org/10.11569/wcjd.v24.i36.4772
消化系肿瘤是一大类严重威胁人类生命健康的重大疾病, 其发病率逐年升高且表现出低龄化趋势[1]. 目前, 全球范围而言, 恶性肿瘤已占所有死亡病因的1/4, 高居全部疾病死亡率的第2位[2], 中国消化系肿瘤发病率及病死率均高于全球平均水平, 其中胃癌、食管癌、结直肠癌和肝癌4种肿瘤在我国恶性肿瘤中居前列, 其死亡总数占恶性肿瘤前10位死亡总数的构成比达40.75%[3,4]. 世界卫生组织预计2020年全世界癌症发病率将比现在增加50%, 其中消化系肿瘤占重要部分[5,6]. 鉴于消化系肿瘤对人类健康的重要影响, 利用医学生命科学的最近研究进展, 发展新型的肿瘤诊疗手段十分必要[7-10].
随着各类组学技术、高通量生物大分子相互作用技术以及生物信息学分析等前沿技术巨大进步和成熟, 使得肿瘤全基因组研究成为可能[11-16]. 近年来飞速发展的基因组编辑技术为肿瘤发生、发展重要基因的靶向生物治疗、靶基因敲除或激活、影响细胞生长和增殖基因及参与细胞生长增殖调控等相关研究提供了强有力的工具, 已成为肿瘤防治研究新热点[16-20].
新一代基因组编辑技术主要包括锌指核酸酶(zinc finger nucleases, ZFNs)技术[21]、类转录激活因子效应物核酸酶(transcription activator-like effector nucleases, TALENs)技术[22]和成簇的规律间隔的短回文重复序列(clustered regularly interspaced short palindromic repeats, CRISPR)及相关基因(CRISPR-associated gene, Cas)编辑技术[23,24]. 本文对新一代基因组编辑技术的机制及在消化系肿瘤研究和治疗中的应用进展等方面作一述评.
自从40多年前人们提出可以利用基因靶向治疗的手段治疗癌症等多基因遗传疾病以来, 如何提高外源基因进入靶细胞以及外源DNA分子取代靶DNA的效率是困惑众多研究者的难题[25,26]. 直到今天, 维持治疗基因在靶细胞内稳定的复制和表达、降低或消除治疗基因对靶基因区周围的基因的表达影响、超长插入DNA片段的有效递送载体的选择、病毒基因组等非功能片段的去除等是基因治疗不得不面对的难题[27]. 新一代基因组编辑技术的出现和成熟为人们对基因组进行操作进而对肿瘤等基因遗传病进行精准治疗提供技术保证.
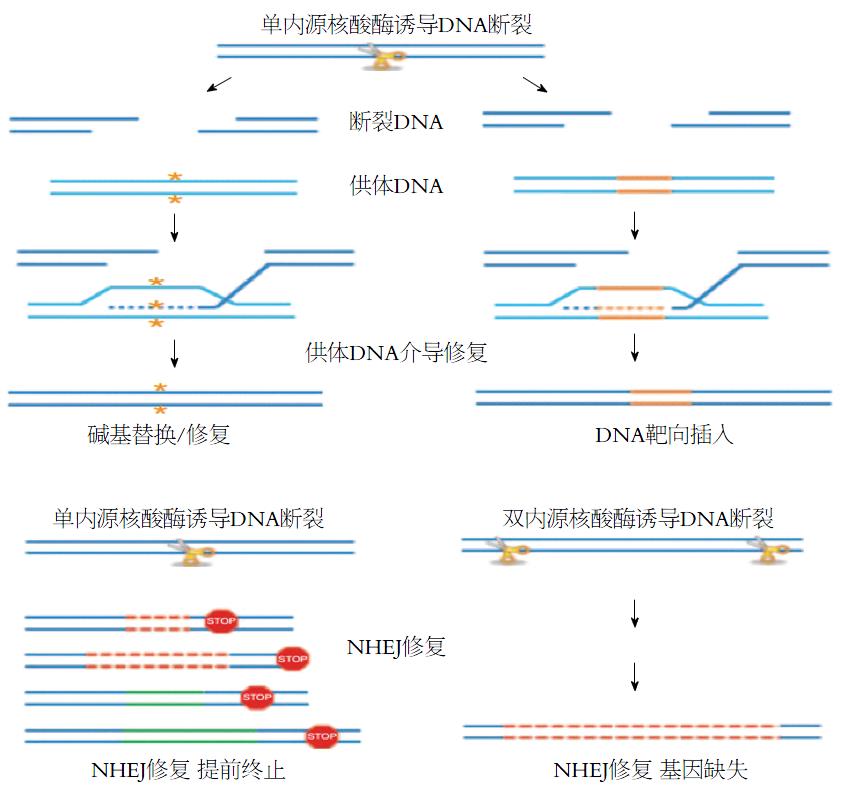
靶双链DNA断裂(DNA double strand breaks, DSBs)后细胞内源性修复机制的激活是基因组编辑技术的生物学基础, 肿瘤等疾病发生时, 细胞内DSBs通常是通过同源介导修复(homology-directed repair, HDR)[28]或非同源末端接合修复(nonhomologous end-joining, NHEJ)[29]. 只有当细胞核内存在与损伤DNA同源的DNA片段时, 细胞内可能发生HDR修复. 当同源DNA片段与受损DNA序列完全一致是, DNA损伤完全修复. 当同源序列与原受损DNA不一致时, 则在原DNA处引入相应突变(图1)[30]. 当细胞核内没有相应的同源DNA片段时, 细胞将利用NHEJ方式修复损伤, 其导致的结果是基因提前终止或发生大面积的DNA序列替换或缺失(图1)[31]. 利用上述原理, 人们可以有目的的将靶DNA序列替换成治疗基因DNA序列, 或者将相应的致病序列缺失突变, 以达到基因治疗的目的[32]. 基因组编辑的核心内容是依赖于DSBs修复机制, 将治疗基因精准的导入到相应的靶位点区进行靶DNA的替换、删除等. 人们已经发展了4种可诱导体内内源性DSBs修复机制进而进行基因组精准编辑的技术流程, 分别是ZFNs、TALENs以及CRISPR/Cas.
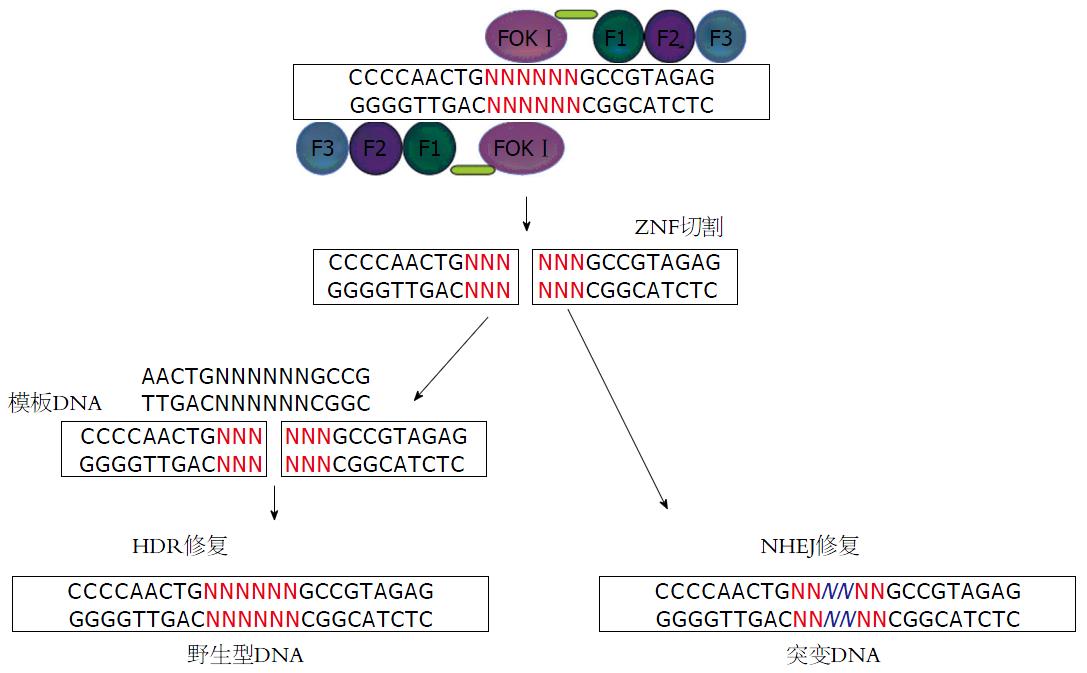
ZFNs又名锌指蛋白核酸酶, 他是一类人工构建的限制性内切酶, 由锌指DNA结合域F1\F2\F3与限制性内切酶的DNA切割域FokⅠ融合而成(图2), 其中由FokⅠ核酸内切酶来源的结构域被认为是真核细胞中含量最为丰富的核酸内切酶, 理论上可以切割所有的DNA序列[33,34]. ZFNs技术是一种新兴的基因高效靶向修饰和调控技术, 研究者可以通过加工改造ZFNs的锌指DNA结合域, 靶向定位结合目的DNA靶位点区, 从而使得ZFNs可以结合复杂基因组中的目的序列, 并由DNA切割域进行特异性切割. 此外, 通过将ZFNs技术和胞内DNA修复机制结合起来, 研究者还可以自如地在生物体内对基因组进行精确编辑[34].
目前已有很多关于ZFNs组装方法的研究, 并且ZFNs识别三联体DNA的方式, 已建立了相关文库, 随着对ZFNs的进一步研究, 科学家对FokⅠ进行了改造, 使其只能形成异源二聚体, 通过排除非预期的靶点进一步的提高特异性, 除此之外, 一些关于改造过具有更高可切割活性的FokⅠ核酸内切酶结构域也陆续被应用, 使得ZFNs获得更高的特异性[35-39].
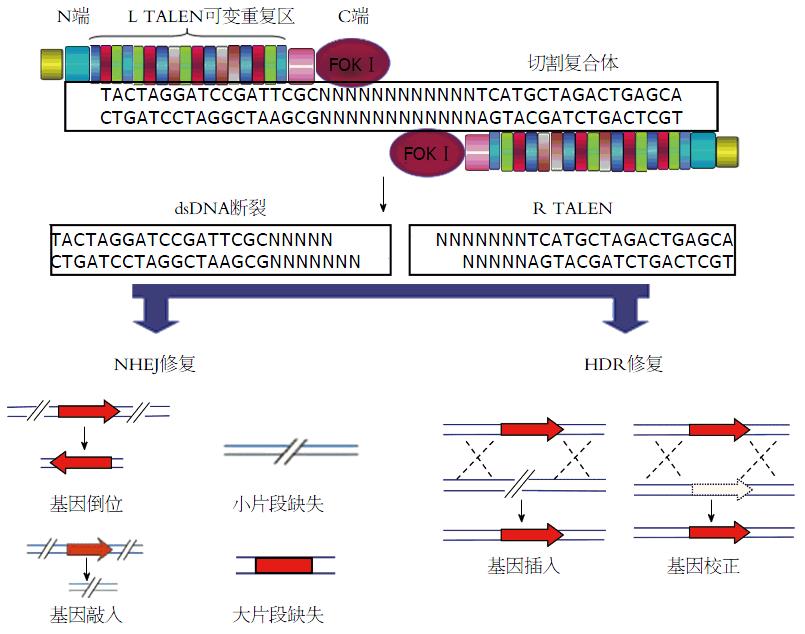
TALENs靶向基因敲除技术是一种崭新的分子生物学工具, 被认为是基因敲除技术发展的里程碑. TALENs的设计和构建是基于植物病原体黄单胞菌分泌的一种转录激活样效应因子(transcription activator-like effector, TALE)可以特异性识别并结合DNA序列的原理[40]. 一个典型TALEN由一个包含核定位信号的N端结构域、一个包含可识别特定DNA序列的典型串联TALE重复序列的中央结构域以及一个具有FokⅠ核酸内切酶功能的C端结构域组成(图3). 一般来说, 人工TALENs元件识别的特异性DNA序列长度则一般为14-20 bp, 用于靶位点的识别、结合并指导DNA切割[41-43].
TALENs技术的原理是通过DNA识别模块将TALENs元件靶向特异性的DNA位点并结合, 然后在FokⅠ核酸酶的作用下完成特定位点的剪切, 并借助于细胞内固有的HDR或NHEJ完成特定序列的插入/倒置、删失及基因融合[44-49](图3). TALE蛋白的核酸结合域的氨基酸序列与其靶位点的核苷酸序列存在恒定的对应关系, 利用类转录激活蛋白的序列模块, 可组装成特异结合任意DNA序列的模块蛋白. TALE蛋白中的DNA结合域与FokⅠ核酸内切酶的切割域融合, 在特异的位点打断目标基因, 进而在该位点进行基因敲入、基因敲出或点突等DNA操作[50-53].
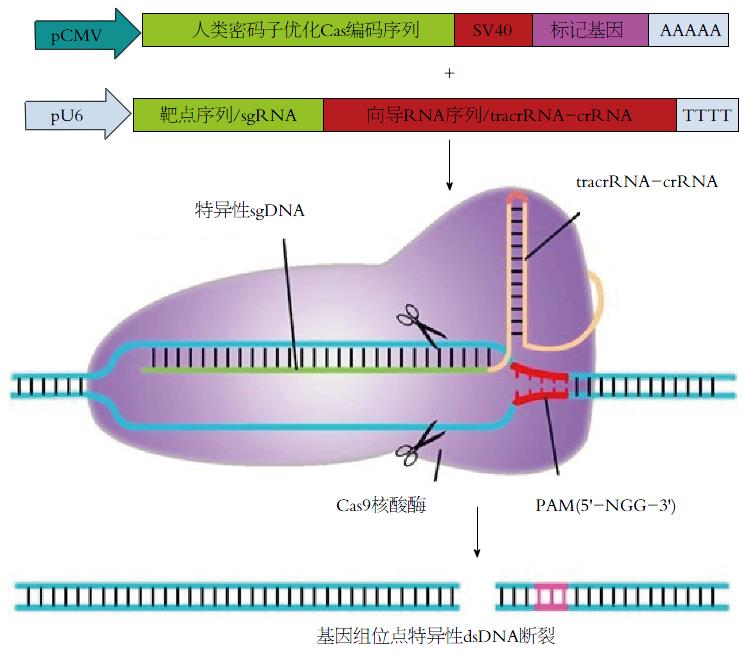
2007年来, 大量的研究报道了一种全新的人工核酸内切酶CRISPR/Cas9系统的出现, 来源于细菌与古细菌的一种获得性免疫系统, 这个系统中, 由RNA起引导作用, 对外来DNA进行靶向切割[54-59]. CRISPR/Cas9核酸酶可以在哺乳动物细胞中进行基因组编辑, 实现对靶基因的敲除、突变、插入以及基因抑制和激活等[60]. 外来DNA会被整合到此类细菌基因组的CRISPR序列中, 并由protospacer序列一一隔开, 在Ⅱ型系统中, CRISPR序列首先被转录成pre-CRISPR序列, 后被加工为短的crRNA, 与CRISPR序列一同被转录的还有反式激活crRNA(trans-activating crRNA, tracrRNA)[61-64]. 在实际工作中, crRNA与靶序列结合, 与tracrRNA形成二聚体后结合CRISPR相关蛋白(CRISPR-associated protein, Cas)9, 并引导Cas9蛋白对DNA进行靶向切割(图4).
从分子生物学技术的角度分析, ZFNs、TALENs及CRISPR/Cas等3种基因组编辑技术都是利用DSBs后, 细胞内发生的NHEJ或HDR修复过程, 将特异性DNA的靶向识别及核酸内切酶细胞内表达以实现基因组DNA序列的改变, 并均可用于与基因组定点修饰相关的基因敲入、基因敲除、DNA删失及基因融合等基因定点修饰的各类操作[65].
作为最早被广泛应用的基因组编辑技术, ZFNs的优点是技术平台较完善, 可利用资源丰富. 但也存在设计繁琐、对目标DNA序列及其上下游序列依赖性高、脱靶率高、细胞毒性大等诸多限制性因素. TALENs技术在TALENs模块进行组装时需要进行大量基因克隆和测序操作, 绝大多数科研实验室都难以独立完成, 对商业化实验室依赖性高, 也限制了其广泛应用. 与ZFNs、TALENs相比, CRISPR/Cas系统组装上更方便, 该系统在改变识别靶位点时, 只需改变一段长为20 bp的可转录为crRNA的引导DNA序列, 其gRNA的设计和合成工作量远远小于TALENs和ZFNs技术的DNA识别模块的构建过程, 且毒性远远低于ZFNs技术, 但也存在着对靶基因区上下游序列依赖性等缺点, 目前只能应用于上游有PAM序列的靶位.
由于新一代基因组编辑技术在肿瘤相关基因的敲除、敲入、缺失及修复等方面的操作中具有的精准的优点, 研究人员将之利用消化系肿瘤的机制阐明和诊疗手段的建立上并取得了一定的进展. 例如, 朱显军等[66]利用TALENs技术构建靶向MYH9基因敲除载体, 转染胃癌MGC803单克隆细胞株, 实现了该细胞株的细胞周期G2/M期阻滞并促进了细胞早期的凋亡. Huang等[67]和Li等[68]利用ZFNs技术靶向敲除了人肝癌细胞株SMMC-7721的在肿瘤侵袭和转移过程中诱导基质金属蛋白酶功能的免疫球蛋白超家族成员HAb18G/CD147基因, 成功的在肿瘤细胞株中降低了黏附、侵袭、迁移和克隆形成能力. Olsen等[69]利用ZFNs技术靶向敲除了人胰腺癌细胞株BxPC-3的β-catenin基因, 发现该基因的敲除并不影响细胞株的正常形态和生长, 进而证明WNT/β-catenin通路在BxPC-3细胞株中并不是不可或缺的. Madsen等[70]利用ZFNs技术靶向敲除了胆囊癌细胞株Capan-1的Core 1酶的分子伴侣COSMC基因, 发现COSMC基因的缺失增加了细胞株对NK介导的抗体依赖的细胞毒性和细胞毒性T淋巴细胞介导的细胞杀伤能力. Xue等[71]运用CRISPR/Cas9基因组编辑技术成功地将抑癌基因p53和pten进行双突变, 构建了肝癌动物模型. Liu等[72]利用CRISPR/Cas9介导的基因敲除技术对p53基因进行突变, 解析了一类抗食管癌新药APR-246的抗食管癌效应和分子机制. Kanda等[73]利用建立了基于CRISPR/Cas9系统的高效EB病毒(epstein-barr virus, EBV)附加体DNA的克隆克隆策略, 成功构建了包含EBV附加体DNA的细菌人工染色体, 并在胃癌细胞株中成功实现了EBV重建.
消化系肿瘤的死亡率居人类恶性肿瘤的第1位, 且总体呈上升趋势. 随着基因组编辑技术研究的不断深入. 基因编辑新技术将成为更好认识和攻克人类消化系肿瘤的一个重要突破口和新视角. 将有望在消化系肿瘤早期诊断、鉴别诊断、临床分期、个性化治疗指导、靶向治疗和预后评估等方面发挥重要的作用. 尽管该技术应用于基因治疗的尝试才刚开始, 但已显示出巨大的应用潜力, 越来越多的研究将通过深入探讨基因组编辑的功能来寻找新的肿瘤治疗方案. 总之, 在组织细胞学、分子生物学等基础学科不断发展的基础上, 随着基因组编辑技术的不断进步, 基因组编辑技术及消化系肿瘤基因治疗的研究将有更广阔的前景.
消化系肿瘤发生涉及到细胞生长因子、信号转导、细胞周期、细胞凋亡、细胞分化、肿瘤血管形成等众多的与肿瘤的发生发展密切相关基因表达的改变或DNA序列的改变. 肿瘤发生的分子生物学机制尚未完全阐明, 基因组编辑新技术将成为更好认识和攻克人类消化系肿瘤的一个重要突破口和新视角.
锌指核酸酶(zinc finger nucleases, ZFNs)技术繁琐的操作, 高成本的劣势限制了该技术的临床应用, 在ZFNs理论基础上发展、类转录激活因子效应物核酸酶(transcription activator-like effector nucleases, TALENs)技术尽管可实现ZFNs大部分功能, 且具有便宜、快速的优势, 但由于TALENs分子比ZFNs大得多, 严重制约了其进入细胞的效率.
2016-05 Nature Biotechnology发表了题为"DNA-guided genome editing using the Natronobacterium gregoryi Argonaute"的论文, 该文作者革新了一种不同于CRISPR的适合在人类细胞中进行基因组编辑的新技术: NgAgo-gDNA; 该技术通过NgAgo核酸酶通过DNA而不是RNA作为介导寻找替换目标极大地克服了CRISPR"脱靶效应".
本文综述了TALENs、ZFNs以及成簇的规律间隔的短回文重复序列及相关基因在内的多种基因组编辑技术的原理及其在消化系肿瘤研究中的应用最新进展.
基因编辑新技术为认识和攻克消化系肿瘤提供了突破口和新视角, 有望在消化系肿瘤早期诊断、鉴别诊断、临床分期、个性化治疗指导、靶向治疗和预后评估等方面发挥重要的作用.
基因组编辑: 指在核酸酶等介导下实现对基因组DNA特定区域"编辑", 以实现对特定靶DNA片段的定点、定向敲除、敲入等分子生物学技术;
同源介导修复(HDR): 是细胞内一种修复DNA双链损伤的机制, 是利用细胞内的同源染色体对应的DNA序列作为修复的模板进行DNA修复的生物学过程.
韩双印, 主任医师, 郑州大学人民医院消化内科; 许洪卫, 主任医师, 大连大学附属新华医院胃肠微创外科中心, 大连大学消化病研究所
基因组编辑技术是位点特异性基因靶向操作的重要工具, 该技术的应用将在消化系肿瘤早期诊断、鉴别诊断、临床分期、个性化治疗指导、靶向治疗和预后评估等方面发挥重要的作用.
手稿来源: 邀请约稿
学科分类: 胃肠病学和肝病学
手稿来源地: 贵州省
同行评议报告分类
A级 (优秀): 0
B级 (非常好): B
C级 (良好): C
D级 (一般): 0
E级 (差): 0
编辑: 于明茜 电编:胡珊
| 1. | Alecu L, Tulin A, Enciu O, Bărbulescu M, Ursuţ B, Obrocea F. Gastrointestinal Stromal Tumors - Diagnosis and Surgical Treatment. Chirurgia (Bucur). 2015;110:525-529. [PubMed] |
| 2. | GBD 2013 Mortality and Causes of Death Collaborators. Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;385:117-171. [PubMed] [DOI] |
| 3. | Li J, Zhang H, Chen Z, Su K. Clinico-pathological characteristics and prognostic factors of gastrointestinal stromal tumors among a Chinese population. Int J Clin Exp Pathol. 2015;8:15969-15976. [PubMed] |
| 4. | Herszényi L, Tulassay Z. Epidemiology of gastrointestinal and liver tumors. Eur Rev Med Pharmacol Sci. 2010;14:249-258. [PubMed] |
| 5. | Zhu RX, Seto WK, Lai CL, Yuen MF. Epidemiology of Hepatocellular Carcinoma in the Asia-Pacific Region. Gut Liver. 2016;10:332-339. [PubMed] [DOI] |
| 6. | Søreide K, Sandvik OM, Søreide JA, Giljaca V, Jureckova A, Bulusu VR. Global epidemiology of gastrointestinal stromal tumours (GIST): A systematic review of population-based cohort studies. Cancer Epidemiol. 2016;40:39-46. [PubMed] [DOI] |
| 7. | Park JY, Jang SH. Epidemiology of Lung Cancer in Korea: Recent Trends. Tuberc Respir Dis (Seoul). 2016;79:58-69. [PubMed] [DOI] |
| 8. | Veisani Y, Delpisheh A. Survival rate of gastric cancer in Iran; a systematic review and meta-analysis. Gastroenterol Hepatol Bed Bench. 2016;9:78-86. [PubMed] |
| 9. | Smith RA, Andrews K, Brooks D, DeSantis CE, Fedewa SA, Lortet-Tieulent J, Manassaram-Baptiste D, Brawley OW, Wender RC. Cancer screening in the United States, 2016: A review of current American Cancer Society guidelines and current issues in cancer screening. CA Cancer J Clin. 2016;66:96-114. [PubMed] [DOI] |
| 10. | Valle I, Tramalloni D, Bragazzi NL. Cancer prevention: state of the art and future prospects. J Prev Med Hyg. 2015;56:E21-E27. [PubMed] |
| 11. | Meerzaman D, Dunn BK, Lee M, Chen Q, Yan C, Ross S. The promise of omics-based approaches to cancer prevention. Semin Oncol. 2016;43:36-48. [PubMed] [DOI] |
| 12. | Farid SG, Morris-Stiff G. "OMICS" technologies and their role in foregut primary malignancies. Curr Probl Surg. 2015;52:409-441. [PubMed] [DOI] |
| 13. | Nakagawa H, Wardell CP, Furuta M, Taniguchi H, Fujimoto A. Cancer whole-genome sequencing: present and future. Oncogene. 2015;34:5943-5950. [PubMed] [DOI] |
| 14. | Zhang X, Li L, Wei D, Yap Y, Chen F. Moving cancer diagnostics from bench to bedside. Trends Biotechnol. 2007;25:166-173. [PubMed] [DOI] |
| 15. | Trujillo E, Davis C, Milner J. Nutrigenomics, proteomics, metabolomics, and the practice of dietetics. J Am Diet Assoc. 2006;106:403-413. [PubMed] [DOI] |
| 16. | Katoh M, Katoh M. Bioinformatics for cancer management in the post-genome era. Technol Cancer Res Treat. 2006;5:169-175. [PubMed] [DOI] |
| 17. | Chakravarti D, Cho JH, Weinberg BH, Wong NM, Wong WW. Synthetic biology approaches in cancer immunotherapy, genetic network engineering, and genome editing. Integr Biol (Camb). 2016;8:504-517. [PubMed] [DOI] |
| 18. | Wang D, Ma N, Hui Y, Gao X. The application of CRISPR/Cas9 genome editing technology in cancer research. Yi Chuan. 2016;38:1-8. [PubMed] [DOI] |
| 19. | Shuvalov O, Petukhov A, Daks A, Fedorova O, Ermakov A, Melino G, Barlev NA. Current genome editing tools in gene therapy: new approaches to treat cancer. Curr Gene Ther. 2015;15:511-529. [PubMed] [DOI] |
| 20. | Avesson L, Barry G. The emerging role of RNA and DNA editing in cancer. Biochim Biophys Acta. 2014;1845:308-316. [PubMed] [DOI] |
| 21. | Kmiec EB. Is the age of genetic surgery finally upon us? Surg Oncol. 2015;24:95-99. [PubMed] [DOI] |
| 22. | Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol. 2013;14:49-55. [PubMed] [DOI] |
| 23. | White MK, Khalili K. CRISPR/Cas9 and cancer targets: future possibilities and present challenges. Oncotarget. 2016;7:12305-12317. [PubMed] [DOI] |
| 24. | Torres-Ruiz R, Rodriguez-Perales S. CRISPR-Cas9: A Revolutionary Tool for Cancer Modelling. Int J Mol Sci. 2015;16:22151-22168. [PubMed] [DOI] |
| 25. | Friedmann T, Roblin R. Gene therapy for human genetic disease? Science. 1972;175:949-955. [PubMed] [DOI] |
| 27. | Maeder ML, Gersbach CA. Genome-editing Technologies for Gene and Cell Therapy. Mol Ther. 2016;24:430-446. [PubMed] [DOI] |
| 28. | Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25-35. [PubMed] [DOI] |
| 29. | Rouet P, Smih F, Jasin M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc Natl Acad Sci USA. 1994;91:6064-6068. [PubMed] [DOI] |
| 30. | Smih F, Rouet P, Romanienko PJ, Jasin M. Double-strand breaks at the target locus stimulate gene targeting in embryonic stem cells. Nucleic Acids Res. 1995;23:5012-5019. [PubMed] [DOI] |
| 31. | Choulika A, Perrin A, Dujon B, Nicolas JF. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:1968-1973. [PubMed] [DOI] |
| 32. | Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat Rev Mol Cell Biol. 2003;4:712-720. [PubMed] [DOI] |
| 33. | Kim S, Lee MJ, Kim H, Kang M, Kim JS. Preassembled zinc-finger arrays for rapid construction of ZFNs. Nat Methods. 2011;8:7. [PubMed] [DOI] |
| 34. | Bhakta MS, Henry IM, Ousterout DG, Das KT, Lockwood SH, Meckler JF, Wallen MC, Zykovich A, Yu Y, Leo H. Highly active zinc-finger nucleases by extended modular assembly. Genome Res. 2013;23:530-538. [PubMed] [DOI] |
| 35. | Gonzalez B, Schwimmer LJ, Fuller RP, Ye Y, Asawapornmongkol L, Barbas CF. Modular system for the construction of zinc-finger libraries and proteins. Nat Protoc. 2010;5:791-810. [PubMed] [DOI] |
| 36. | Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA. Rapid "open-source" engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31:294-301. [PubMed] [DOI] |
| 37. | Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Curtin SJ, Blackburn JS, Thibodeau-Beganny S, Qi Y. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA). Nat Methods. 2011;8:67-69. [PubMed] [DOI] |
| 38. | Gupta A, Christensen RG, Rayla AL, Lakshmanan A, Stormo GD, Wolfe SA. An optimized two-finger archive for ZFN-mediated gene targeting. Nat Methods. 2012;9:588-590. [PubMed] [DOI] |
| 39. | Gersbach CA, Gaj T, Barbas CF. Synthetic zinc finger proteins: the advent of targeted gene regulation and genome modification technologies. Acc Chem Res. 2014;47:2309-2318. [PubMed] [DOI] |
| 40. | Mak AN, Bradley P, Cernadas RA, Bogdanove AJ, Stoddard BL. The crystal structure of TAL effector PthXo1 bound to its DNA target. Science. 2012;335:716-719. [PubMed] [DOI] |
| 41. | Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. [PubMed] [DOI] |
| 42. | Morbitzer R, Römer P, Boch J, Lahaye T. Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors. Proc Natl Acad Sci USA. 2010;107:21617-21622. [PubMed] [DOI] |
| 43. | Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B. Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol. 2011;29:699-700. [PubMed] [DOI] |
| 44. | Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol. 2011;29:149-153. [PubMed] [DOI] |
| 45. | Morbitzer R, Elsaesser J, Hausner J, Lahaye T. Assembly of custom TALE-type DNA binding domains by modular cloning. Nucleic Acids Res. 2011;39:5790-5799. [PubMed] [DOI] |
| 46. | Weber E, Gruetzner R, Werner S, Engler C, Marillonnet S. Assembly of designer TAL effectors by Golden Gate cloning. PLoS One. 2011;6:e19722. [PubMed] [DOI] |
| 47. | Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng G, Zhang F. A transcription activator-like effector toolbox for genome engineering. Nat Protoc. 2012;7:171-192. [PubMed] [DOI] |
| 48. | Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30:460-465. [PubMed] [DOI] |
| 49. | Schmid-Burgk JL, Schmidt T, Kaiser V, Höning K, Hornung V. A ligation-independent cloning technique for high-throughput assembly of transcription activator-like effector genes. Nat Biotechnol. 2013;31:76-81. [PubMed] [DOI] |
| 50. | Mussolino C, Morbitzer R, Lütge F, Dannemann N, Lahaye T, Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283-9293. [PubMed] [DOI] |
| 51. | Holkers M, Maggio I, Henriques SF, Janssen JM, Cathomen T, Gonçalves MA. Adenoviral vector DNA for accurate genome editing with engineered nucleases. Nat Methods. 2014;11:1051-1057. [PubMed] [DOI] |
| 52. | Kleinstiver BP, Wolfs JM, Kolaczyk T, Roberts AK, Hu SX, Edgell DR. Monomeric site-specific nucleases for genome editing. Proc Natl Acad Sci USA. 2012;109:8061-8066. [PubMed] [DOI] |
| 53. | Wolfs JM, DaSilva M, Meister SE, Wang X, Schild-Poulter C, Edgell DR. MegaTevs: single-chain dual nucleases for efficient gene disruption. Nucleic Acids Res. 2014;42:8816-8829. [PubMed] [DOI] |
| 54. | Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709-1712. [PubMed] [DOI] |
| 55. | Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331-338. [PubMed] [DOI] |
| 56. | Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167-170. [PubMed] [DOI] |
| 57. | Fineran PC, Charpentier E. Memory of viral infections by CRISPR-Cas adaptive immune systems: acquisition of new information. Virology. 2012;434:202-209. [PubMed] [DOI] |
| 58. | Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262-1278. [PubMed] [DOI] |
| 59. | Garneau JE, Dupuis MÈ, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67-71. [PubMed] [DOI] |
| 60. | Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602-607. [PubMed] [DOI] |
| 61. | Chylinski K, Makarova KS, Charpentier E, Koonin EV. Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42:6091-6105. [PubMed] [DOI] |
| 62. | Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62-67. [PubMed] [DOI] |
| 63. | Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales AP, Li Z, Peterson RT, Yeh JR. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481-485. [PubMed] [DOI] |
| 64. | Kleinstiver BP, Prew MS, Tsai SQ, Nguyen NT, Topkar VV, Zheng Z, Joung JK. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat Biotechnol. 2015;33:1293-1298. [PubMed] [DOI] |
| 65. | Song M, Kim YH, Kim JS, Kim H. Genome engineering in human cells. Methods Enzymol. 2014;546:93-118. [PubMed] [DOI] |
| 66. | 朱 显军, 邓 海军, 叶 耿泰, 沈 智勇, 李 风萍, 郭 伟洪, 杨 庆斌, 刘 浩, 李 国新. TALEN介导的MYH9基因沉默及对细胞周期与凋亡的影响. 南方医科大学学报. 2016;36:375-380. |
| 67. | Huang Q, Li J, Xing J, Li W, Li H, Ke X, Zhang J, Ren T, Shang Y, Yang H. CD147 promotes reprogramming of glucose metabolism and cell proliferation in HCC cells by inhibiting the p53-dependent signaling pathway. J Hepatol. 2014;61:859-866. [PubMed] [DOI] |
| 68. | Li HW, Yang XM, Tang J, Wang SJ, Chen ZN, Jiang JL. Effects of HAb18G/CD147 knockout on hepatocellular carcinoma cells in vitro using a novel zinc-finger nuclease-targeted gene knockout approach. Cell Biochem Biophys. 2015;71:881-890. [PubMed] [DOI] |
| 69. | Olsen PA, Solberg NT, Lund K, Vehus T, Gelazauskaite M, Wilson SR, Krauss S. Implications of targeted genomic disruption of β-catenin in BxPC-3 pancreatic adenocarcinoma cells. PLoS One. 2014;9:e115496. [PubMed] [DOI] |
| 70. | Madsen CB, Lavrsen K, Steentoft C, Vester-Christensen MB, Clausen H, Wandall HH, Pedersen AE. Glycan elongation beyond the mucin associated Tn antigen protects tumor cells from immune-mediated killing. PLoS One. 2013;8:e72413. [PubMed] [DOI] |
| 71. | Xue Z, Wu M, Wen K, Ren M, Long L, Zhang X, Gao G. CRISPR/Cas9 mediates efficient conditional mutagenesis in Drosophila. Genes Genomes Genetics (Bethesda). 2014;4:2167-2173. [PubMed] [DOI] |