修回日期: 2009-06-16
接受日期: 2009-06-23
在线出版日期: 2009-07-18
目的: 分析微小RNA(microRNA, miRNA)在HBV感染到肝硬化再到肝癌进程中表达谱的变化.
方法: 利用miRNA芯片技术检测人正常肝脏、乙型肝炎肝硬化、HBV相关性肝癌组织中miRNA表达谱的差异. 实时定量PCR检测上述3种肝脏组织中芯片结果差异性表达的miRNA, 验证芯片结果的可信性.
结果: 与正常肝脏组织相比, 乙型肝炎肝硬化和HBV相关性肝癌组织中表达上调超过2倍的microRNA有6个, 分别为: hsa-miR-602、hsa-miR-129-5p、has-miR-210、hsa-miR-671-5p、hsa-miR-30b*及hsa-miR-572; 表达下调超过2倍的miRNA有8个: hsa-miR-143、hsa-miR-199a-5p、has-miR-195、hsa-miR-27a、hsa-miR-99a、hsa-miR-519e、has-miR-130a及hsa-miR-597. 这些正常肝脏组织相比有差异表达的miRNA, 在乙型肝炎肝硬化和HBV相关性肝癌组织中表达量无明显差别.
结论: 从HBV感染到肝硬化再到肝癌进程中伴有microRNA表达谱的变化, 且变化主要发生在进程早期.
引文著录: 马兆龙, 杨炼, 陈立波, 黄金明, 王冬冬, 王国斌. miRNA在HBV从感染经由肝硬化到肝癌进程中表达谱的变化. 世界华人消化杂志 2009; 17(20): 2112-2116
Revised: June 16, 2009
Accepted: June 23, 2009
Published online: July 18, 2009
AIM: To investigate the differential expression profile of microRNAs in the different stages of hepatitis B virus (HBV) infection-related hepatocarcinogenesis.
METHODS: MicroRNA microarray was used to detect the differential expression profile of microRNAs in the liver tissue taken from healthy controls as well as patients with HBV-induced cirrhosis or HBV-related hepatocellular carcinoma. Real-time quantitative PCR was performed to verify the differential expression of candidate microRNAs obtained from microarray experiment.
RESULTS: Compared with normal liver tissue, six microRNAs (hsa-miR-602, hsa-miR-129-5p, has-miR-210, hsa-miR-671-5p, hsa-miR-30b*, and hsa-miR-572) were upregulated more than two-fold, and eight microRNAs (hsa-miR-143, hsa-miR-199a-5p, has-miR-195, hsa-miR-27a, hsa-miR-99a, hsa-miR-519e, has-miR -130a and hsa-miR-597) were downregulated in the liver tissue taken from patients with HBV-induced cirrhosis or HBV-related hepatocellular carcinoma. Though these identified microRNAs showed significant differential expression between normal liver tissue and HBV-related hepatocellular carcinoma tissue and cirrhotic tissue, they exhibited no significant differential expression between HBV-related hepatocellular carcinoma tissue and cirrhotic tissue.
CONCLUSION: HBV-related hepatocarcinogenesis is associated with the change in the expression profile of microRNAs.
- Citation: Ma ZL, Yang L, Chen LB, Huang JM, Wang DD, Wang GB. Differential expression profile of microRNAs in the different stages of hepatitis B virus infection-related hepatocarcinogenesis. Shijie Huaren Xiaohua Zazhi 2009; 17(20): 2112-2116
- URL: https://www.wjgnet.com/1009-3079/full/v17/i20/2112.htm
- DOI: https://dx.doi.org/10.11569/wcjd.v17.i20.2112
微小RNA(microRNA, miRNA)是一类约22 nt大小的非编码单链RNA分子, 他们通过与靶基因mRNA的结合调控其他基因mRNA和蛋白的表达, 参与发育、炎症、肿瘤等多种生理、病理过程[1-6]. 在肝细胞癌(hepatocellular carcinoma, HCC)中, 多个miRNA的异常表达已被证明参与了肝癌的恶性生物学行为[7-11]. 由于中国绝大多数肝癌患者与HBV感染密切相关, 但HBV感染→肝硬化→肝癌进程中, 有关miRNA表达变化的研究较少. 我们通过应用miRNA芯片技术检测不同肝脏组织标本中miRNA表达谱的差异, 筛选出在乙型肝炎肝硬化和HBV相关性肝癌中表达明显上调或下调的miRNA, 为进一步阐明miRNA表达变化在HBV感染→肝硬化→肝癌进程中起到的作用提供依据.
收集2007-09/2008-08华中科技大学同济医学院附属协和医院行肝癌切除和门诊肝组织活检病例, 选取其中无其他基础肝病, 30-40岁男性患者的正常肝脏组织、乙型肝炎肝硬化组织和HBV相关性肝癌组织, 所有病例术前均未接受放化疗, 均经病理学确诊. 组织标本离体后立即放入液氮冷藏, 后转入-80℃保存备用.
1.2.1 总RNA的提取及质量检测: 按照TRIzol®试剂(Invitrogen life technologies)说明书的步骤, 逐步提取出组织标本中的总RNA. 应用紫外分光光度计测定总RNA的吸光度A260值和A280值, 用A260值计算其浓度, 并计算A260/A280值检测其纯度; 甲醛变性琼脂糖凝胶电泳法进一步检测总RNA质量.
1.2.2 miRNA芯片检测不同肝脏组织miRNA的表达: 为减少性别、年龄和基础疾病的影响, 在所有肝脏组织标本中, 选择年龄为30-40岁, 排除其他基础疾病的男性患者的正常肝脏、乙型肝炎肝硬化和HBV相关性肝癌组织标本各1例, 进行miRNA芯片检测. 该实验采用丹麦Exiqon公司生产的LNATM miRNA芯片(11.0版), 此芯片使用的是基于锁定核苷酸TM(lock nucleic acids, LNA)专利技术的捕获探针, 该探针可以高灵敏、高特异性与互补的靶RNA结合, 从而准确检测样品中miRNA的表达水平. 每张miRNA芯片中包含有至少1807个特异性探针、435个Exiqon特有的miRPlusTM探针(提供miRBase数据库外的新miRNA信息)、对照探针以及无探针的空白对照, 可以检测Sanger miRBase 11.0数据库中人、小鼠、大鼠全部miRNA(其中人约847条, 小鼠约609条, 大鼠约351条). 为保证结果的可靠性, 上述每个探针在芯片内重复4次, 即每张芯片对同一样本重复检测4次. 分别取5 µg上述3个不同类型肝脏组织样本的总RNA, 采用miRCURYTM Array Labelling kit(Exiqon)标记Hy3TM荧光, 采用RNeasy Mini Kit(Qiagen)浓缩标记样品, 然后采用miRCURYTM Array microarray kit(Exiqon)和Hybridization Chamber Ⅱ(Ambion)进行miRNA芯片杂交, 具体步骤按各试剂说明书进行; 杂交后芯片用Genepix 4000B(Molecular Devices)635 nm波长进行图像扫描, 所得数据用Genepix Pro 6.0(Molecular Devices)分析, 通过原始值减去背景值来做修正, 并用中值做标准化, 分别计算出3种样本中miRNA的标准值及两两之间标准值的比值.
1.2.3 实时定量PCR检测目的miRNA表达量: 取2 µg上述3种不同类型肝脏组织样本的总RNA作为初始模板, 总反应体系为20 µL, 应用Revert AidTM H Minus First Strand cDNA Synthesis Kit(Fermentas)在Gene Amp PCR System 9700(Applied Biosystems)上进行逆转录合成cDNA, 反应条件为: 16℃, 30 min; 42℃, 42 min; 85℃, 5 min. 然后取1 µL上述cDNA为模板, 以U6作为内参照, 每个检测样本做3个复孔, 在Rotor-Gene 3000 Real-time PCR仪(Corbett Research)上进行实时定量PCR, 反应条件为: 95℃预热5 min; 95℃, 10 s; 58℃, 15 s; 72℃, 20 s, 35个循环; +0.5℃/cycle, 5 s, 54个循环. 实验中所用引物均由Invitrogen公司合成. 为减小因RNA浓度定量误差和RNA逆转录效率误差等的影响, 所得数据用2-ΔΔCt公式校正, 用校正后的值判定各样本中目的miRNA的表达量的差异.
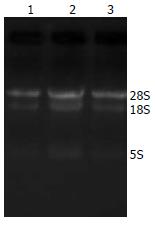
紫外分光光度计检测结果表明, 3种不同类型肝脏组织标本总RNA的A260/A280值分别为1.88、1.85和1.83; 凝胶电泳结果提示各样本的18S和28S条带均较清晰, 且密度之比约为1:2(图1). 上述实验结果表明来自3种不同类型肝脏组织标本的总RNA均有较好的质量, 完全可以满足后续的miRNA芯片检测和实时定量PCR的实验要求.
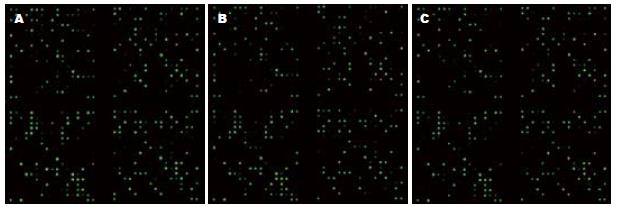
杂交后的miRNA芯片经Genepix 4000B扫描, 所得结果见图2; 再经Genepix Pro 6.0分析及标准化处理后, 芯片结果显示与正常肝脏组织相比, 乙型肝炎肝硬化和HBV相关性肝癌组织中表达上调超过2倍的miRNA有6个, 分别为hsa-miR-602、hsa-miR-129-5p、has-miR-210、hsa-miR-671-5p、hsa-miR-30b*和hsa-miR-572; 表达下调超过2倍的miRNA有8个, 分别为hsa-miR-143、hsa-miR-199a-5p、has-miR-195、hsa-miR-27a、hsa-miR-99a、hsa-miR-519e、has-miR-130a和hsa-miR-597, 具体结果见表1.
| miRNA分子 | 芯片结果 | 实时定量PCR结果 | |||||
| B/A | C/A | C/B | B/A | C/A | C/B | ||
| 上调的miRNA | hsa-miR-602 | 3.234 | 4.134 | 1.278 | 2.703 | 3.486 | 1.290 |
| hsa-miR-129-5p | 2.246 | 6.322 | 2.815 | 2.852 | 4.347 | 1.524 | |
| has-miR-210 | 5.159 | 4.274 | 0.828 | 3.531 | 2.894 | 0.820 | |
| hsa-miR-671-5p | 4.315 | 4.249 | 0.985 | 2.953 | 3.624 | 1.227 | |
| hsa-miR-30b* | 3.571 | 4.005 | 1.122 | 3.841 | 4.896 | 1.275 | |
| hsa-miR-572 | 3.588 | 3.092 | 0.862 | 4.319 | 4.291 | 0.994 | |
| 下调的miRNA | hsa-miR-143 | -3.661 | -5.764 | -1.575 | -2.762 | -3.521 | -1.275 |
| hsa-miR-199a-5p | -3.288 | -5.139 | -1.563 | -2.482 | -2.730 | -1.100 | |
| has-miR-195 | -3.066 | -3.251 | -1.060 | -2.501 | -2.943 | -1.177 | |
| hsa-miR-27a | -2.193 | -3.147 | -1.435 | -3.492 | -3.246 | 1.076 | |
| hsa-miR-99a | -2.137 | -2.746 | -1.285 | -3.581 | -3.484 | 1.028 | |
| hsa-miR-519e | -2.653 | -2.674 | -1.008 | -2.898 | -3.572 | -1.233 | |
| has-miR-130a | -2.016 | -2.740 | -1.225 | -3.805 | -3.872 | -1.018 | |
| hsa-miR-597 | -2.071 | -2.357 | -1.138 | -3.736 | -4.058 | -1.086 | |
以用于上述miRNA芯片检测的3种不同类型肝脏组织标本总RNA为模板, 实时定量PCR检测在芯片结果中差异性表达的miRNA, 结果见表1, 提示实时定量PCR结果与miRNA芯片结果趋势一致, 表明miRNA芯片的结果具有可信性.
miRNA作为一类成熟状态下只有22 nt左右的非编码单链小RNA, 广泛存在于从线虫、植物、动物到人类的多种生物中, 由于其在个体生长发育、细胞增殖凋亡、炎症和肿瘤等多种生理病理过程中都起着十分重要的作用, 因而受到广泛关注. 从Du et al[22]在线虫中发现第1个miRNA(lin-4)开始, 截止2009-03, 已经在103个物种中发现miRNA共约9539个, 其中人类约706个(数据来源于Sanger miRBase 13.0, http://miRNA.sanger.ac.uk/sequences/), 据估计这些miRNA具有调控人类至少30%基因的潜能[12].
大量研究发现, 在多种人类肿瘤都伴有miRNA的表达异常, 如弥漫性大B细胞淋巴瘤中miR-155表达上调[13]、胰腺癌中miR-221表达上调[14]、肺癌中的let-7表达下调[15-16]、结肠直肠癌中的miR-143和miR-145表达下调[17]、慢性淋巴细胞白血病中的miR-15a[18]和miR-16-1表达下调[19]等, 提示miRNA在肿瘤的发生发展中可能发挥类似癌基因或抑癌基因的作用. 在正常情况下, 成熟的miRNA通过调控靶基因mRNA的翻译或其稳定性来参与正常细胞稳态的维持, 故而miRNA的异常表达可能导致其相应靶基因转录后水平的表达异常. 目前对miRNA在肿瘤发生发展中的作用机制还不是太明确, 认为其主要作用机制为: miRNA通过RNA诱导的沉默复合物(RNA-induced silencing complex, RISC)与靶基因mRNA的3'端UTR区互补结合[20-21], 当两者能完全互补配对结合时, 导致靶基因mRNA的降解, 这种现象在植物中多见; 在动物中则多数是通过两者的不完全互补结合, 抑制靶基因mRNA的翻译, 进而影响靶基因蛋白的表达[22].
在排除年龄、性别和基础肝脏疾病的影响下, 我们通过miRNA芯片和实时定量PCR检测发现: 与正常人类肝脏组织相比, 有些miRNA在乙型肝炎肝硬化和HBV相关性肝癌组织中表达明显上调, 如miR-602、miR-129-5p、miR-210、miR-671-5p、miR-30b*和miR-572; 有些则表达明显下调, 如miR-143、miR-199a-5p、miR-195、miR-27a、miR-99a、miR-519e、miR-130a和miR-597, 其中miR-210[23-24]、miR-143[17,25]、miR-195[26]、miR-27a[27]、miR-99a[28]、miR-130a[29]等已被文献报道过在其他肿瘤组织中也有类似结果, 提示这些在不同类型肝脏组织中差异性表达的miRNA可能在HBV感染→肝硬化→肝癌的发生发展过程中起到重要的类似癌基因或抑癌基因的作用. 但上述与正常肝脏组织相比表达上调或下调的miRNA, 在乙型肝炎肝硬化组织和HBV相关性肝癌组织中的表达却无明显差异, 表明miRNA的表达的变化主要发生在HBV感染→肝硬化→肝癌进程中的早期.
通过上述结果我们可以推测出miRNA表达的改变可能为HBV感染→肝硬化→肝癌进程的始动因素, 相关miRNA表达的改变可能促进了该进程的发生发展.
微小RNA(micro RNA, miRNA)是一类约22 nt大小的非编码单链RNA分子, 他们通过与靶基因mRNA的结合调控其他基因mRNA和蛋白的表达, 参与发育、炎症、肿瘤等多种生理、病理过程. 在肝细胞癌(HCC)中, 多个miRNA的异常表达已被证明参与了肝癌的恶性生物学行为. 由于中国绝大多数肝癌患者与HBV感染密切相关, 但HBV感染→肝硬化→肝癌进程中, 有关miRNA表达变化的研究较少.
管冬元, 副教授, 上海中医药大学基础医学院实验中医学教研室; 范小玲, 主任医师, 北京地坛医院综合科
大量研究发现, 在多种人类肿瘤都伴有miRNA的表达异常, 如弥漫性大B细胞淋巴瘤中miR-155表达上调、胰腺癌中miR-221表达上调、肺癌中的let-7表达下调、结肠直肠癌中的miR-143和miR-145表达下调、慢性淋巴细胞白血病中的miR-15a和miR-16-1表达下调等, 提示miRNA在肿瘤的发生发展中可能发挥类似癌基因或抑癌基因的作用.
miRNA表达的改变可能为HBV感染→肝硬化→肝癌进程的始动因素, 相关miRNA表达的改变可能促进了该进程的发生发展.
本文主要应用microRNA芯片技术检测HBV感染到肝癌组织中microRNA表达的差异, 课题思路清晰, 文字表达清楚, 具有很高的学术价值与创新性.
编辑: 李军亮 电编:何基才
| 1. | Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065-7070. [PubMed] [DOI] |
| 2. | He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2005;102:19075-19080. [PubMed] [DOI] |
| 3. | Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029-6033. [PubMed] [DOI] |
| 4. | Bandrés E, Cubedo E, Agirre X, Malumbres R, Zárate R, Ramirez N, Abajo A, Navarro A, Moreno I, Monzó M. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;5:29. [PubMed] [DOI] |
| 5. | Szafranska AE, Davison TS, John J, Cannon T, Sipos B, Maghnouj A, Labourier E, Hahn SA. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442-4452. [PubMed] [DOI] |
| 6. | Lin SL, Chiang A, Chang D, Ying SY. Loss of mir-146a function in hormone-refractory prostate cancer. RNA. 2008;14:417-424. [PubMed] [DOI] |
| 7. | Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537-2545. [PubMed] [DOI] |
| 8. | Jiang J, Gusev Y, Aderca I, Mettler TA, Nagorney DM, Brackett DJ, Roberts LR, Schmittgen TD. Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res. 2008;14:419-427. [PubMed] [DOI] |
| 9. | Huang YS, Dai Y, Yu XF, Bao SY, Yin YB, Tang M, Hu CX. Microarray analysis of microRNA expression in hepatocellular carcinoma and non-tumorous tissues without viral hepatitis. J Gastroenterol Hepatol. 2008;23:87-94. [PubMed] |
| 10. | Budhu A, Jia HL, Forgues M, Liu CG, Goldstein D, Lam A, Zanetti KA, Ye QH, Qin LX, Croce CM. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897-907. [PubMed] [DOI] |
| 11. | Wang Y, Lee AT, Ma JZ, Wang J, Ren J, Yang Y, Tantoso E, Li KB, Ooi LL, Tan P. Profiling microRNA expression in hepatocellular carcinoma reveals microRNA-224 up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific target. J Biol Chem. 2008;283:13205-13215. [PubMed] [DOI] |
| 12. | Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787-798. [PubMed] [DOI] |
| 13. | Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci U S A. 2005;102:3627-3632. [PubMed] [DOI] |
| 14. | Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, Frankel WL, Morgan DL, Postier RG, Brackett DJ, Schmittgen TD. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120:1046-1054. [PubMed] [DOI] |
| 15. | Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635-647. [PubMed] [DOI] |
| 16. | Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753-3756. [PubMed] [DOI] |
| 17. | Michael MZ, O' Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882-891. [PubMed] |
| 18. | Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci U S A. 2004;101:11755-11760. [PubMed] [DOI] |
| 19. | Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999-3004. [PubMed] [DOI] |
| 20. | Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631-640. [PubMed] [DOI] |
| 21. | Tang F, Hajkova P, O'Carroll D, Lee C, Tarakhovsky A, Lao K, Surani MA. MicroRNAs are tightly associated with RNA-induced gene silencing complexes in vivo. Biochem Biophys Res Commun. 2008;372:24-29. [PubMed] [DOI] |
| 22. | Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645-4652. [PubMed] [DOI] |
| 23. | Camps C, Buffa FM, Colella S, Moore J, Sotiriou C, Sheldon H, Harris AL, Gleadle JM, Ragoussis J. hsa-miR-210 Is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res. 2008;14:1340-1348. [PubMed] [DOI] |
| 24. | Giannakakis A, Sandaltzopoulos R, Greshock J, Liang S, Huang J, Hasegawa K, Li C, O'Brien-Jenkins A, Katsaros D, Weber BL. miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biol Ther. 2008;7:255-264. [PubMed] |
| 25. | Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, Bednarikova M, Nenutil R, Vyzula R. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72:397-402. [PubMed] [DOI] |
| 26. | Flavin RJ, Smyth PC, Laios A, O'Toole SA, Barrett C, Finn SP, Russell S, Ring M, Denning KM, Li J. Potentially important microRNA cluster on chromosome 17p13.1 in primary peritoneal carcinoma. Mod Pathol. 2009;22:197-205. [PubMed] [DOI] |
| 27. | Zhu H, Wu H, Liu X, Evans BR, Medina DJ, Liu CG, Yang JM. Role of MicroRNA miR-27a and miR-451 in the regulation of MDR1/P-glycoprotein expression in human cancer cells. Biochem Pharmacol. 2008;76:582-588. [PubMed] [DOI] |
| 28. | Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI. Mature miR-184 as Potential Oncogenic microRNA of Squamous Cell Carcinoma of Tongue. Clin Cancer Res. 2008;14:2588-2592. [PubMed] [DOI] |
| 29. | Sorrentino A, Liu CG, Addario A, Peschle C, Scambia G, Ferlini C. Role of microRNAs in drug-resistant ovarian cancer cells. Gynecol Oncol. 2008;111:478-486. [PubMed] [DOI] |