修回日期: 2005-09-20
接受日期: 2005-09-30
在线出版日期: 2005-11-15
目的: 研究小核核糖核蛋白多肽N基因(SNRPN)在肝癌肿瘤HepG2细胞株的表达及基因印迹状态.
方法: 采用RT-PCR方法检测出SNRPN基因在肝癌肿瘤HepG2细胞株中的表达状况, 对HepG2细胞株基因组DNA和cDNA中的SNRPN基因外显子4 nt 1654312位点用RT-PCR为基础的RFLP方法进行基因分型.
结果: HepG2细胞稳定表达SNRPN, SNRPN外显子4 nt 1654312(数字依据NT_026446, SNP rs705)为杂合子(C/T); RT-PCR为基础的RFLP分析表明, SNRPN的双等位基因中只有T等位基因的产生mRNA转录本.
结论: SNRPN基因在HepG2肝癌细胞株中有表达, 其基因印迹状态未丢失.
引文著录: 晏泽辉, 邓国宏, 王宇明. SNRPN基因在HepG2细胞中的印迹状态研究. 世界华人消化杂志 2005; 13(21): 2545-2548
Revised: September 20, 2005
Accepted: September 30, 2005
Published online: November 15, 2005
AIM: To investigate the expression and imprinting style of small nuclear ribonucleoprotein polypeptide N (SNRPN) in hepatic cancer cell line HepG2.
METHODS: Human hepatic cancer cell line HepG2 was cultured in vitro by routine method. The expression of SNRPN gene in the cells was detected by reverse transcription polymerase chain reaction (RT-PCR). The single nucleotide polymorphisms (SNP) of SNRPN at exon 4 nt 1654312 (numbered according to NT_026446, SNP rs705) was genotyped in a genomic DNA sample and a cDNA sample of HepG2 cell line with restriction fragment length polymorphism (RFLP) based on RT-PCR.
RESULTS: SNRPN was stably expressed in HepG2 cells. The heterozygote C/T was found at exon 4 nt 1654312 of SNRPN. In cell lines heterozygous with respect to this SNP, only one of the two alleles (T allele) present in the genomic DNA produced an mRNA transcript.
CONCLUSION: SNRPN mRNA is expressed in HepG2 cells, and there is no loss of imprinting.
- Citation: Yan ZH, Deng GH, Wang YM. Analysis on expression and imprinting style of small nuclear ribonucleoprotein polypeptide N in hepatic cancer cell line HepG2. Shijie Huaren Xiaohua Zazhi 2005; 13(21): 2545-2548
- URL: https://www.wjgnet.com/1009-3079/full/v13/i21/2545.htm
- DOI: https://dx.doi.org/10.11569/wcjd.v13.i21.2545
小核核糖核蛋白多肽N基因(small nuclear ribonucleoprotein polypeptide N, SNRPN)是编码小核核糖核蛋白多肽N的印迹基因[1], 与印迹基因IGF2一样, 它也是仅父源等位基因表达, 其印迹状态的维持与DNA的甲基化[2], 染色质的紧缩[3]以及组蛋白的乙酰化[4,5]密切相关. IGF2迹缺失在肝癌的发生中起到重要作用[6], 与其类似的SNRPN基因在肝癌细胞中的表达及其印迹状态的研究还未见相关报道. 本实验的目的就是利用逆转录PCR(RT-PCR)的方法研究出SNRPN基因在HepG2肝癌细胞株的表达状况, 并以RT-PCR为基础的限制性片段长度多态性分析(restriction fragment length polymorphism, RFLP)的方法检测出SNRPN基因的印迹状态, 探讨印迹基因SNRPN在肝癌细胞株中的印迹状态是否发生改变。
人肝癌来源的传代HepG2细胞株购自中科院上海细胞生物学研究所, 本研究所实验室保存. RPMI 1640培养基干粉、优级胎牛血清(FCS)、胰蛋白酶、HEPES购自Hycolone公司;TriPure、DEPC等购自Roche公司;AMV逆转录酶、RNA酶抑制剂、oligo dT购自Promega公司;DL2000 DNA分子量标准、Taq DNA聚合酶、dNTP、内切酶BstUI、PstI购自Takara公司;氯仿、异丙醇、无水乙醇、Tris碱、EDTA、青霉素、链霉素等其他试剂为国内产品, 购自北京鼎国生物技术公司. PCR引物由北京奥科生物公司合成, SNRPN基因鉴定正向引物S4:5'-CTACTCTTTGAAGCTTCTGCC-3';反向引物AS4:5'-TGAAGATTCGGCCATCTTGC-3'.
1.2.1 HepG2细胞培养: 按常规方法传代培养。HepG2肝癌细胞在含有100 mL/L的胎牛血清、终浓度为1×105 u/L的青霉素和链霉素的RPMI 1640培养基中37℃, 50 mL/L CO2条件下培养数天, 每2 d换液一次, 4 d传代.
1.2.2 核酸的提取: 利用TriPure试剂提取HepG2细胞DNA和总RNA. 用1 mL TriPure试剂收集细胞, 混匀, 室温静置5 min, 以确保核蛋白复合体完全溶解;添加0.2 mL氯仿, 剧烈摇动15 s, 然后再次室温静置15 min;然后4℃, 12000 g, 离心15 min, 溶液分三层. 将无色水样上层转移到一新的离心管中, 加入0.5 mL异丙醇, 混匀, 室温静置15 min, 然后4℃, 12000 g, 离心15 min, 去上清, 添加1 mL 750mL/L酒精洗涤RNA沉淀, 再次4℃, 12000 g, 离心15 min, 去上清, 空气中倒置5-10 min使酒精挥发以除去残留的氯仿, 然后用DEPC水溶解RNA沉淀, 用10 g/L琼脂糖凝胶电泳鉴定RNA提取质量. 另外向中间层和底部红色层中(要将无色水样上层去除干净)添加纯无水乙醇0.3 mL, 混匀, 室温静置5 min, 然后4℃, 2000 g, 离心5 min, 去上清, 添加750mL/L酒精1.5 mL, 室温保留20 min, 偶尔摇动, 再次4℃, 2000 g, 离心5 min, 去上清后空气中自然晾干5-10 min以去除残留的氯仿, TE溶解DNA沉淀. 最后用核酸蛋白定量仪测定HepG2细胞总RNA和基因组DNA含量.
1.2.3 HepG2细胞SNRPN基因分型: 采用PCR-RFLP方法对HepG2细胞SNRPN外显子4 nt 1654312(数字依据NT_026446, SNP rs705)位点进行基因分型. 首先利用S4、AS4做引物, 基因组DNA为模板扩增出包含nt 1654312位点的SNRPN基因片段, 然后利用利用内切酶BstUI(酶切位点:CG↓CG)和PstI(酶切位点:CTGCA↓G)对PCR产物进行双酶切, 最后利用20 g/L琼脂糖凝胶电泳来进行限制性片段长度多态性分析.
1.2.4 RT-PCR及RPLP分析: 参照RT-PCR试剂盒操作程序采用二步法进行逆转录以及PCR扩增。逆转录反应体系和反应条件为:总RNA模板5 μL约2 μg, oligo dT 1 μg, 无RNA酶DEPC处理水3 μL, 混匀后在70℃变性5 min, 冰上冷却5 min后加入5×AMV buffer 5 μL、AMV逆转录酶3 μL、RNA酶抑制剂1.5 μL、dNTP 2 μL, 补加无RNA酶DEPC处理水到总体系25 μL, 然后42℃逆转录60 min, 完成后再70℃加热5 min以灭活逆转录酶. 随后我们以逆转录的cDNA产物为模板按照Sakatani et al[7]所述的方法利用BstUI对SNRPN的nt 1654312位点SNP进行PCR-RFLP分型.
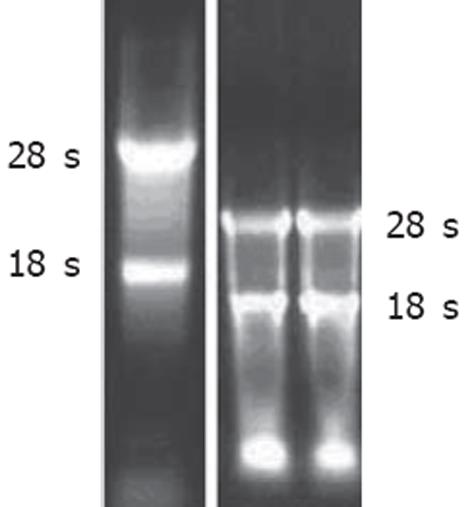
采用核酸蛋白定量仪对所提取的总RNA定量, 产量在50 μg左右(细胞数大约6×106个);采用10 g/L琼脂糖凝胶电泳进行总RNA质量鉴定, 显示所提取的RNA完整性良好(图2).
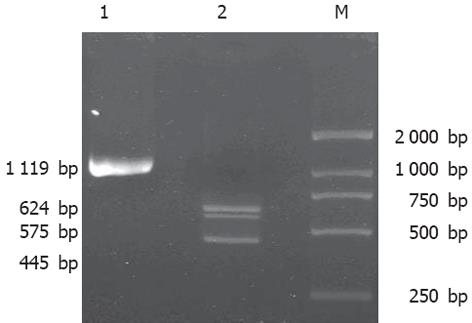
HepG2细胞基因组DNA PCR扩增及BstUI和PstI双酶切后20 g/L琼脂糖电泳分析结果(图3)证明HepG2细胞株SNRPN基因nt1654312位点SNP为C/T杂合子基因型.
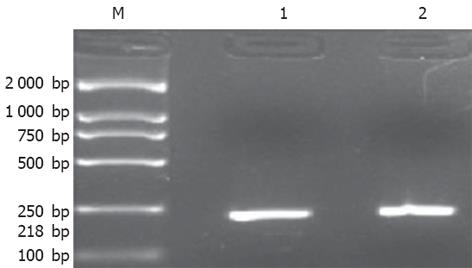
HepG2细胞cDNA扩增及BstUI酶切后30 g/L琼脂糖电泳分析结果(图4)证明HepG2细胞株SNRPN基因印迹状态未丢失, 仅出现一种等位即T等位表达.
基因印迹也称作基因组印迹、配子印迹或亲源印迹, 是近年来发现的一种不遵从孟德尔定律的依靠单亲传递某些遗传学性状的现象, 也就是某些基因呈亲源依赖性的单等位基因表达(父源性或母源性), 其另一等位基因不表达或表达极弱[8-13], 仿佛这些基因的不同亲本来源的一对等位基因上带有某种可供识别的印迹, 故名. 具有这种现象的基因被称为印迹基因. 在人类基因中, 预计印迹基因的总数大约100~200个, 已经明确的印迹基因超过40个. 基因印迹在人类遗传性疾病尤其是肿瘤发生中的作用正引起越来越多的注意[14-18].
SNRPN是编码小核核糖核蛋白多肽N的印迹基因[19-21], 与印迹基因IGF2一样, 它也是仅父源等位基因被表达, 其印迹状态的维持与DNA的甲基化[22-24], 染色质的紧缩[25]以及组蛋白的乙酰化[4,5]密切相关. 鉴于IGF2迹缺失在肝癌的发生中起到重要作用[26-31], 与其类似的SNRPN基因在肝癌细胞中的表达及其印迹状态的研究也是值得我们关注的一个课题. 本实验的目的就是利用RT-PCR的方法研究出SNRPN基因在HepG2肝癌细胞株的表达状况, 并以RFLP分析的方法检测SNRPN的基因印迹状态.
SNRPN是一个印迹基因, 它的两个等位基因根据父系来源的不同有显著的不同表达水平(仅父源等位基因表达). 为了区分不同表达量的等位基因, 我们选择HepG2细胞SNRPN外显子4上nt1654312(数字依据NT_026446来确定)为杂合子C/T的SNP位点作为研究对象, 以研究该C/T SNP(dbSNP rs705)的等位特异性表达情况, 并以cDNA的PCR-RFLP检验双等位的差异性表达。实验发现, C/T杂合子HepG2细胞系中, SNRPN基因依然维持其印迹状态, 逆转录PCR产物酶切实验证实SNRPN等位基因仅仅只有一个等位产生mRNA的转录, 即仅父源的T等位出现转录.
本研究提示SNRPN基因在HepG2肝癌细胞株中稳定表达, 其基因印迹状态未丢失, 更进一步的实验来研究证明印迹基因SNRPN在肝癌组织以及正常人群中的表达和基因印迹丢失情况是必要的, 相信随着对SNRPN研究的深入, 必将为肝癌的发病机制研究提供新的思路.
电编: 张敏 编辑: 菅鑫妍 审读: 张海宁
| 1. | Ozcelik T, Leff S, Robinson W, Donlon T, Lalande M, Sanjines E, Schinzel A, Francke U. Small nuclear ribonucleoprotein polypeptide N (SNRPN), anexpressed gene in the Prader-Willi syndrome critical region. Nat Genet. 1992;2:265-269. [PubMed] [DOI] |
| 2. | Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362-365. [PubMed] [DOI] |
| 3. | Feil R, Khosla S. Genomic imprinting in mammals: an interplay betweenchromatin and DNA methylation? Trends Genet. 1999;15:431-435. [PubMed] [DOI] |
| 4. | Hu JF, Oruganti H, Vu TH, Hoffman AR. The role of histone acetylation in the allelic expression of the imprinted human insulin-like growth factor II gene. Biochem Biophys Res Commun. 1998;251:403-408. [PubMed] [DOI] |
| 5. | Saitoh , S , Wada T. Parent-of-origin specific histone acetylation and reactivation of a key imprinted gene locus in Prader-Willi syndrome. Am J Hum Genet. 2000;66:1958-1962. [PubMed] [DOI] |
| 6. | Ohlsson R, Nystrom A, Pfeifer-Ohlsson S, Tohonen V, Hedborg F, Schofield P, Flam F, Ekstrom TJ. IGF2 is parentally imprinted during human embryogenesis and in the Beckwith-Wiedemann syndrome. Nat Genet. 1993;4:94-97. [PubMed] [DOI] |
| 7. | Sakatani T, Wei M, Katoh M, Okita C, Wada D, Mitsuya K, Meguro M, Ikeguchi M, Ito H, Tycko B. Epigenetic heterogeneity at imprinted loci in normal populations. Biochem Biophys Res Commun. 2001;283:1124-1130. [PubMed] [DOI] |
| 8. | Takahashi Y, Rayman JB, Dynlacht BD. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 2000;14:804-816. [PubMed] |
| 9. | Sun X, Ding H, Hung K, Guo B. A new MALDI-TOF based mini-sequencing assay for genotyping of SNPs. Nucleic Acids Res. 2000;28:68-75. [PubMed] [DOI] |
| 11. | Neumann B, Barlow DP. Multiple roles for DNA methylation in gametic imprinting. Curr Opin Genet Dev. 1996;6:159-163. [PubMed] [DOI] |
| 12. | Wutz A, Smrzka OW, Schweifer N, Schellander K, Wagner EF, Barlow DP. Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature. 1997;389:745-749. [PubMed] [DOI] |
| 13. | Wutz A, Barlow DP. Imprinting of the mouse Igf2r gene depends on an intronic CpG island. Mol Cell Endocrinol. 1998;140:9-14. [PubMed] [DOI] |
| 14. | Jirtle RL. Genomic imprinting and cancer. Exp Cell Res. 1999;248:18-24. [PubMed] [DOI] |
| 15. | Murphy SK, Jirtle RL. Imprinted genes as potential genetic and epigenetic toxicologic targets. Environ Health Perspect. 2000;108:5-11. [PubMed] [DOI] |
| 16. | Jirtle RL, Sander M, Barrett JC. Genomic imprinting and environmental disease susceptibility. Environ Health Perspect. 2000;108:271-278. [PubMed] [DOI] |
| 17. | Murphy SK, Jirtle RL. Imprinting evolution and the price of silence. Bioessays. 2003;25:577-588. [PubMed] [DOI] |
| 18. | Luedi PP, Hartemink AJ, Jirtle RL. Genome-wide prediction of imprinted murine genes. Genome Res. 2005;15:875-884. [PubMed] [DOI] |
| 19. | Tsai TF, Chen KS, Weber JS, Justice MJ, Beaudet AL. Evidence for translational regulation of the imprinted Snurf-Snrpn locus in mice. Hum Mol Genet. 2002;11:1659-1668. [PubMed] [DOI] |
| 20. | Tsai TF, Bressler J, Jiang YH, Beaudet AL. Disruption of the genomic imprint in trans with homologous recombination at Snrpn in ES cells. Genesis. 2003;37:151-161. [PubMed] [DOI] |
| 21. | Geuns E, De Rycke M, Van Steirteghem A, Liebaers I. Methylation imprints of the imprint control region of the SNRPN-gene in human gametes and preimplantation embryos. Hum Mol Genet. 2003;12:2873-2879. [PubMed] [DOI] |
| 22. | Landers M, Bancescu DL, Le Meur E, Rougeulle C, Glatt-Deeley H, Brannan C, Muscatelli F, Lalande M. Regulation of the large (approximately 1000 kb) imprinted murine Ube3a antisense transcript by alternative exons upstream of Snurf/Snrpn. Nucleic Acids Res. 2004;32:3480-3492. [PubMed] [DOI] |
| 23. | Rodriguez-Jato S, Nicholls RD, Driscoll DJ, Yang TP. Characterization of cis- and trans-acting elements in the imprinted human SNURF-SNRPN locus. Nucleic Acids Res. 2005;33:4740-4753. [PubMed] [DOI] |
| 24. | El-Maarri O, Seoud M, Riviere JB, Oldenburg J, Walter J, Rouleau G, Slim R. Patients with familial biparental hydatidiform moles have normal methylation at imprinted genes. Eur J Hum Genet. 2005;13:486-490. [PubMed] [DOI] |
| 25. | Perk J, Makedonski K, Lande L, Cedar H, Razin A, Shemer R. The imprinting mechanism of the Prader-Willi/Angelman regional control center. EMBO J. 2002;21:5807-5814. [PubMed] [DOI] |
| 26. | Song BC, Chung YH, Kim JA, Lee HC, Yoon HK, Sung KB, Yang SH, Yoo K, Lee YS, Suh DJ. Association between insulin-like growth factor-2 and metastases after transcatheter arterial chemoembolization in patients with hepatocellular carcinoma: a prospective study. Cancer. 2001;91:2386-2393. [PubMed] [DOI] |
| 27. | Fan ZR, Yang DH, Cui J, Qin HR, Huang CC. Expression of insulin like growth factor II and its receptor in hepatocellular carcinogenesis. World J Gastroenterol. 2001;7:285-288. [PubMed] [DOI] |
| 28. | Scharf JG, Braulke T. The role of the IGF axis in hepatocarcinogenesis. Horm Metab Res. 2003;35:685-6893. [PubMed] |
| 29. | Li H, Zhang N. Study on the expression and genomic imprinting status of insulin-like growth factor 2 gene in hepatocellular carcinoma. Zhonghua Gan Zang Bing Za Zhi. 2004;12:347-349. [PubMed] |
| 30. | Tsai JF, Jeng JE, Chuang LY, You HL, Wang LY, Hsieh MY, Chen SC, Chuang WL, Lin ZY, Yu ML. Serum insulin-like growth factor-II as a serologic marker of small hepatocellular carcinoma. Scand J Gastroenterol. 2005;40:68-75. [PubMed] [DOI] |
| 31. | Dong ZZ, Yao DF, Yao DB, Wu XH, Wu W, Qiu LW, Jiang DR, Zhu JH, Meng XY. Expression and alteration of insulin-like growth factor II-messenger RNA in hepatoma tissues and peripheral blood of patients with hepatocellular carcinoma. World J Gastroenterol. 2005;11:4655-4660. [PubMed] [DOI] |