Published online Mar 15, 2003. doi: 10.3748/wjg.v9.i3.572
Revised: October 10, 2002
Accepted: October 18, 2002
Published online: March 15, 2003
AIM: To study the preparation and cleavage activity of anti-transforming growth factor (TGF)β1 U1 small nuclear (sn) RNA chimeric hammerhead ribozymes in vitro.
METHODS: TGFβ1 partial gene fragment was cloned into T-vector at the downstream of T7 promoter. 32p-labeled TGFβ1 partial transcripts as target RNA were transcribed in vitro and purified by denaturing polyacrylamide gel electrophoresis (PAGE). Anti-TGFβ1 ribozymes were designed by computer, then synthetic ribozyme fragments were cloned into the U1 ribozyme vector pZeoU1EcoSpe containing U1 snRNA promoter/enhancer and terminator. 32p-labeled U1 snRNA chimeric ribozyme transcripts were gel-purified, incubated with target-RNAs at different conditions and autoradiographed after running denaturing PAGE.
RESULTS: Active U1snRNA chimeric ribozyme (U1Rz803) had the best cleavage activity at 50 °C; at 37 °C, it was active, Km = 34.48 nmol/L, Kcat = 0.14 min-1; while the point mutant ribozyme U1Rz803m had no cleavage activity, so these indicated the design of U1Rz803 was correct.
CONCLUSION: U1Rz803 prepared in this study possessed the perfect specific catalytic cleavage activity. These results indicate U1 snRNA chimeric ribozyme U1Rz803 may suppress the expression of TGFβ1 in vivo, therefore it may provide a new avenue for the treatment of liver fibrosis in the future.
-
Citation: Lin JS, Song YH, Kong XJ, Li B, Liu NZ, Wu XL, Jin YX. Preparation and identification of anti-transforming growth factor β1 U1 small nuclear RNA chimeric ribozyme
in vitro . World J Gastroenterol 2003; 9(3): 572-577 - URL: https://www.wjgnet.com/1007-9327/full/v9/i3/572.htm
- DOI: https://dx.doi.org/10.3748/wjg.v9.i3.572
The incidence of liver cirrhosis is still high all over the world, especially in China[1-6]. Cirrhotic livers are characterized by extensive fibrosis throughout the entire hepatic parenchyma[7-12]. Many factors inducing liver injury and inflammation will lead to chronic liver disease, and hepatic fibrosis[13-22].
TGFβ1 is an important cytokine in the regulation of the production, degradation, accumulation of extracellular matrix proteins, and that it may play a pivotal role in the fibroproliferative changes that follow tissue damage in many vital organs and tissues, including liver, lung, kidney, skin, heart, and arterial walls[7,23-27]. In the past decade dramatic advances have been made in the understanding of cellular and molecular mechanisms underlying liver fibrogenesis, it is thought that TGFβ1 is of crucial importance in rat hepatic fibrosis in vivo[7,28-34]. Inhibition of TGFβ can not only prevent liver fibrosis, but also preserve organ function[30]. So TGFβ1 has been thought to be an ideal target molecule to prevent the progression of liver fibrosis.
Ribozymes are a class of small catalytic RNA molecules that recognize specific substrate RNA molecules by their complementary nucleotide sequence, cleavaging the substrate RNA as an endoribonuclease at enzymatic rates[35-38]. In the last years ribozyme-mediated inhibition of gene expression in intact cells have been tested many times, but some of them were largely unsuccessful[39-42]. Factors that contributed to ribozyme efficacy in transfected cell are expression level, stability against rapid degradation, correct folding for exposure to target, and subcellular localization of ribozyme and target. U1 snRNA is a highly expressed stable small RNA (164 nucleotides) involved in both splicesome and catalytic processing during pre-mRNA splicing. U1small nuclear RNA expression cassette can provide an excellent vehicle for ribozyme delivery and expression in intact cell because of stability, nuclear localization, highly efficient expression[43-45].
Because TGFβ1 plays a crucial role in liver fibrosis, in this study we designed ribozymes directed against TGFβ1 by computer, then cloned them into U1 snRNA chimeric ribozyme vector, it had been proven that it could cleave target RNA efficiently in vitro through the cleavage reaction, so it indicated that it might suppress intracellular TGFβ1 expression, which would provide a new avenue in treatment of liver fibrosis.
HSC-T6 cell line is a kind gift from Dr. Scott L. Friedman (Dept of Medicine and Division of Liver Diseases, Mount Sinai School of Medicine). pZeoU1EcoSpe was provided by Dr. Harry C. Dietz (Department of Pediatrics, Medicine and Molecular Biology & Genetics, Johns Hopkins University School of Medicine). pGEM-T vector kit, transcription kit were purchased from Promega Company. Trizol kit, DMEM were purchased from Gibco BRL Company. The PCR primers and ribozyme fragments were synthesized in the Beckman oligo-1000 DNA synthesizer. Zeocin was purchased from Invitrogen Company. Newborn calf serum was purchased from Hyclone Company. RT-PCR kit, RNase inhibitor, restriction endonucleases, and T4 DNA ligase were purchased from Takara Company. α32P UTP was purchased from Beijing Ya-Hui Company.
Construction of target RNA in vitro Total RNA was extracted using Trizol Kit (GIBCO BRL) from cultured HSC-T6 cell, an immortalized rat hepatic stellate cells line (HSC), exhibited an activated phenotype[46,47]. The upstream primer P1 (5’-GAATTCATTCAGGACTATCACCTACC-3’) in the untranslated region and the downstream primer P2 (5’-AAGCTTTTCTGGTAGAGTTCTACGTG -3’;) in the open reading frame were selected to amplify a 651-base pair fragment corresponding to bases 279 to 930 of the rat TGF beta 1[48]. The extracted RNA was reversely transcribed and polymerase chain reaction (PCR) amplified using a pair of primers in one step reverse transcriptase (RT) PCR kit. The PCR products were analyzed and purified on 1% (w/v) agarose gels. Purified PCR products were ligated into pGEM-T vector. DNA sequencing results showed that the PCR-amplified fragments were cloned into the molecular cloning sites of pGEM-T vector at the downstream of T7 promoter as pTGFβ1. Target RNA was prepared through in vitro transcription of PCR-amplified products of pTGFβ1, which contained T7 promoter at the upstream of upper primer. The sequence of the primers for transcription was GAATTCTAATACGACTCACTATAGGGAGGCGGA-CTACTACGCCAA and TTCTGGTAGAG TTCTACGTG; TAATACGACTCACTATAG GG represents T7 promoter. Then PCR product was analyzed and purified by 1% (w/v) agarose gels electrophoresis as the template for transcription. In vitro transcription was carried out at 37 °C for 90 min in a 40 µL final volume containing 40 mmol/L of Tris·HCl (pH 7.5), 5 mmol/L of DTT, 2 mmol/L of spermidine, 8 mmol/L of MgCl2, 0.25 mmol/L of ATP, GTP,CTP, 0.05 mmol/L of UTP, 20 µCi alpha 32p-UTP, 80 U T7 RNA polymerase and 2 µg purified PCR product. Target RNA was purified by 6% denaturing gel electrophoresis through cutting off the autoradiograph bands and soaking in NES (0.5 mol/L NH4Ac, 0.1 mol/L EDTA, 0.1% SDS pH 5.4) at 42 °C overnight. the products were precipitated by ethanol, washed twice by 75% ethanol, dissolved in DEPC H2O and reserved under -20 °C.
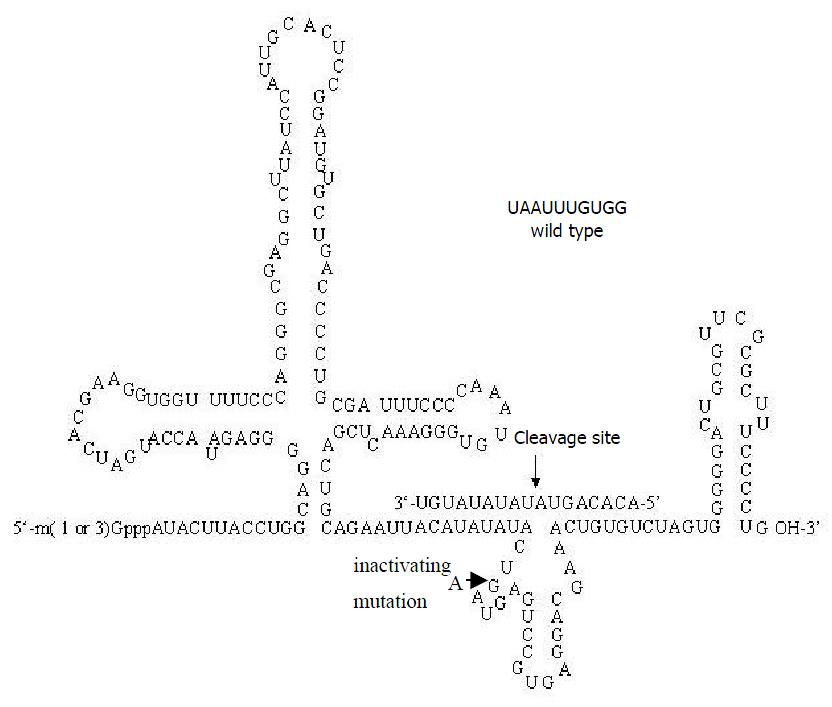
Construction of recombinant plasmid for ribozyme pZeoU1EcoSpe contained the pZeoSV plasmid DNA modified by excising the SV40 promoter, SV40 polyadenylation site, and polylinker at the BamHI sites. In constructing the pZeoU1 EcoSpe, a U1 snRNA expression cassette in pUC13[49,50] was excised with BamHI digestion and ligated into the BamHI sites of the modified pZeoSV. Two rounds of site-directed mutagenesis were then performed to change 4 nt flanking the Sm protein binding site of U1 snRNA, creating unique EcoRI and SpeI restriction sites. The 5’-flanking region of the inserted U1 snRNA expression cassette possessed a promoter/enhancer comparable in strength with the SV40 early promoter[51]. The ribozymes for TGFβ1 were designed according to the computer software pcFOLD combiled by professor Zuker (Canadian Academy of Science). The homologous possibility with the gene of rat was excluded by consulting with RNA sequence of rat cells from NCBI Genbank. The enclosed vector pZeoU1EcoSpe was cut by EcoRI and SpeI restriction en zy mes an d pu rified b y 1% (w/v ) ag arose gels electrophoresis. The synthesized oligonucleotides of ribozyme were mixed with equal molar amounts together, then were cloned into the EcoRI/SpeI sites of pZeoU1EcoSpe to create pU1Rz803. pZeoU1EcoSpe and the reconstruction could be confirmed by DNA sequencing (Figure 1). The oligonucleotides of Rz803 were 5’ AATTACATATATACT(G/A)ATGAGTCCGTGAGGACGAAACTGTGT3’ and 5’ CTAGACACAGTTTCGTCCTCACGGACTCAT(C/T)AGTATATATGT3’; G and C for activated ribozyme, A and T for inactivated ribozyme.
Preparation of ribozymes in vitro The templates used for transcription of U1 snRNA chimeric ribozymes were obtained by PCR amplification of pU1Rz803. The primers used for transcription were as follows: upstream primer: 5’-GAATTCTAATACGACTCACTATAGGGGATACTTACCTGGCAGGGGA-3 ’ ; downstream primer : 5’-CAGGGGAAAGCGCGAACGCA-3’ ; TAATACGACTCACTATAGGG represented T7 promoter. The purification of PCR products was the same as that of the template for target RNA. In vitro transcription and purification of ribozyme were done as described above.
In vitro cleavage reaction of U1Rz803 and U1Rz803m U1Rz803, U1Rz803m and target RNA were quantified by measuring their radioactivate cpm in 1 μL solution. The cleavage reaction was carried out in 5 μL solution containing 50 mmol/L Tris·HCl (pH7.5), 20 mmol/L MgCl2. The molar ratio between ribozyme and target RNA could be estimated according to the cpm value combined with the U number in their RNAs. The initial experiment was: (I) Ribozyme (R): Substrate (S) = 1:1(mol/L) ratio, 37 °C, 120 min; (II) the condition as (I), R:S = 1:5 (mol/L) ratio, 37 °C, 120 min; (III) the condition as (I), U1Rz803 was incubated with target RNA at different temperatures and at different times. 1 μL loading buffer (0.25% Bromophenol Blue, 0.25% Xylene cyanol FF, 20 mmol/L EDTA and saturated Urea) was added to stop the reaction. The result could be analyzed after running a 6% denaturing polyacylamide gel electrophoresis. The cleavage efficiency [CE] was calculated from Bq values of the bands of substrate (S) and products (P) which were cut off from denaturing PAGE. CE = [P / (P + S)]·100%.
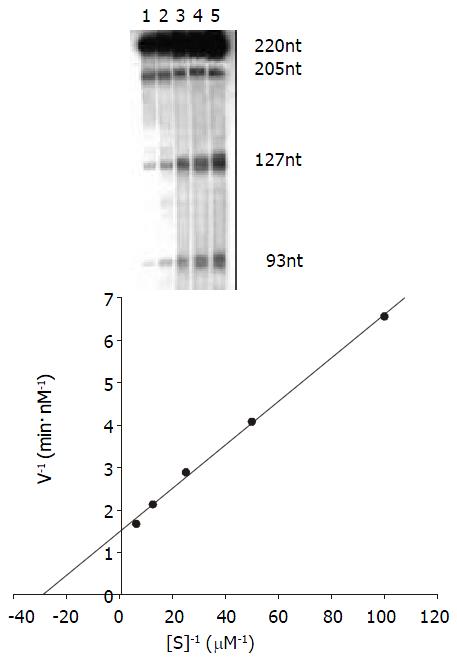
Kinetics studies of the reaction The procedure was described by Uhlenbeck[52]. The Michaelis constant (Km) and Kcat were determined for the ribozyme by performing multiple turnover kinetics experiments. The volume of kinetics reaction is 15 μL. Ribozyme concentration was held constant at 5 nmol/L and substrate concentrations ranged from 10 nmol/l to 160 nmol/ L. The cleavage reaction was done in the same buffer as described above at 37 °C for 20 minutes. The results were analyzed as above. Km and Kcat were calculated by Lineweaver-Burke method (double- riciprocal plot).
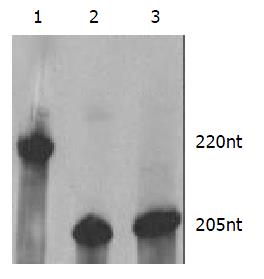
The length of target RNA transcribed from PCR-amplified template should be 220 nt. In this study the ribozymes were embedded into U1 snRNA, but stem-loop structures of U1 snRNA were maintained. Therefore, the transcripts of PCR-amplified template included U1 snRNA and ribozyme, the transcripts of U1snRNA chimeric ribozyme should be 205 nt These results (Figure 2) were inconsistent with our design and proven to be correct.
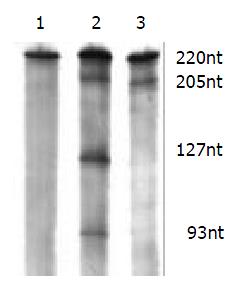
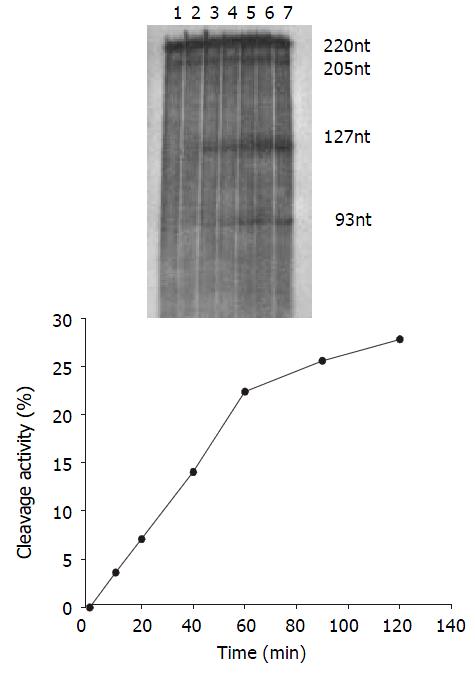
The cleavage result showed that U1Rz803 was capable of cleaving target RNA in vitro. It could cleave target RNA (220 nt) efficiently and exactly to produce two fragment 93 nt/127 nt. while U1Rz803m showed no in vitro cleavage efficacy after 120 min (Figure 3), even at Rz:S = 5:1 (data not shown). At a 1:1 U1Rz803-to-S molar ratio, the cleavage efficiency (CE) was calculated under the condition of 37 °C and 120-minute reaction time, CE = 51.36%. At a 1:5 U1Rz803-to-S molar ratio, CE = 27.81%. This result indicated that the cleavage efficiency increased with increase of ribozyme concentration. The temperature and time would affect the cleavage efficiency.
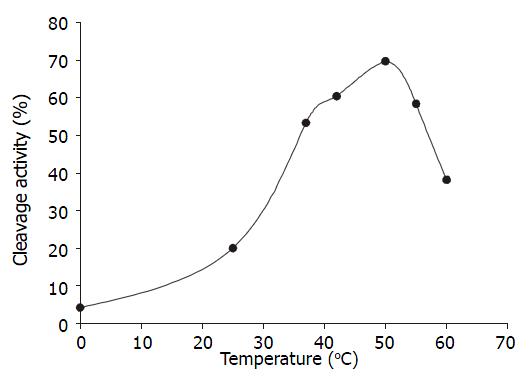
Temperature course When the ratio of U1Rz803 to target RNA was 1:1(molar ratio), the reaction mixture were incubated at different temperature for 90 minutes. The optimal temperature was 50 °C, the cleavage efficacy increased at higher temperatures ranging from 0 °C to 50 °C, but when the temperature was above 50 °C, the cleavage efficacy decreased because the combination of ribozyme and target RNA was weakened (Figure 4).
Time course The cleavage mixture (Rz:substrate = 1:5 mol·L-1) were incubated at 37 °C for different times, it was shown that the reaction product increased with increase in incubation time and it was linear within 60 min, CEmax = 27.81% (Figure 5).
The kinetics of cleavage reaction Under the condition of 37 °C and 20-minute reaction time the cleavage efficiency was calculated at Rz:S = 1:2, 1:4,1:8, 1:16 and 1:32 (mol/L)ratio. Km and Kcat were obtained by the Lineweaver-Burke method (Figure 6) Km = 34.48 nmol/L, Kcat = 0.14 min-1.
Hepatic fibrosis is a common response to chronic liver injury from many causes, including alcohol, persistent viral infection and hereditary metal overload. To date, reversing the causative agent is the only effective therapy to stop or even reverse the liver fibrosis, but the efficacy is limited. Therefore, the development of effective antifibrotic therapies represents a challenge for modern hepatology. With the knowledge on molecular mechanism underlying pathological fibrosis expanding, there are many antifibrotic therapies based on sound biological mechanisms have been carried out. Ueki et al[32] injected a mix of a haemagglutinating virus of Japan (HVJ) liposomes and a plasmid containing the complementary DNA for human hepatocyte growth factor (HGF) into the gluteus muscle of rats treated with dimethylnitrosamine (DMN), a model of persistent liver fibrosis, that could produce the resolution of fibrosis in the cirrhotic liver, but tumorigenicity found in transgenic mice overexpressing HGF[53] and repetitive in vivo transfection are two disadvantages. Qi and Nakamura et al[30,33] prevent liver fibrosis from blockade of TGF beta signal by adenovirus-mediated local expression of a dominant negative type II TGF-beta receptor in the liver of rats treated with DMN, this intervention not only suppressed fibrosis, but also facilitated hepatocyte regeneration, however prolonged period of blocking TGF beta signal could result in unfavorable consequences, such as the inflammation and tissue necrosis. Because TGFβ1 plays a crucial role in liver fibrosis and no report on anti-TGFβ1 ribozyme-mediated cleavage of target RNA for the treatment of liver fibrosis has been published, in this study we designed ribozyme targeting against TGFβ1 and cloned the ribozyme genes into U1 ribozyme vector, prepared U1 snRNA chimeric ribozymes and identified the cleavage activity of ribozymes in vitro.
In the previous study on cleavage activity of U1 snRNA chimeric ribozyme in vitro, ribozymes were prepared through the transcription of synthesized ribozyme genes containing T7/ SP6 promoter, the transcripts only included ribozyme[43]; but ribozyme structure induced by the secondary structure of long flanking sequences would affect ribozyme’s turnover ratio and/ or binding activity as the results of less accurate hybridization and less cleavage. In this study we prepared ribozyme by the PCR-amplified templates. The transcripts included U1 snRNA and ribozyme. Compared with the previous study, it may reflect the cleavage activity of U1Rz803 more accurately in vivo. From our study, we found that the cleavage activity of U1sn RNA chimeric ribozyme was inferior to that of non-modified ribozyme, the result was not shown in this paper. Trimethylguanosine 5’cap, stable stem-loop structures at both end, high GC content of 3’ loop in the structure of U1 snRNA co nfer resistance to exo nu cleases in vivo [54] . Th e hypermethylation of the 5’ cap structure and Sm binding site enable U1 snRNA to accumulate in the nucleoplasm, these make U1 snRNA an effective vector for efficient expression and delivery of ribozyme in the nuclear compartment in vivo. The transcripts of ribozyme labeled by isotope not only provided convenience for us to isolate ribozyme by cutting off autoradiograph bands, but the transcripts were qualified more accurately than non-labeled transcripts of ribozymes. In this study Km and Kcat of U1Rz803 were measured at 37 °C, not at optimal temperature. Because the ribozyme is used in vivo and the temperature in vivo is constant at 37 °C, these results may reflect cleavage activity of U1Rz803 in physiological condition.
Ribozymes have all the properties of antisense RNA with the additional feature of catalytic cleavage. To separate anti-sense from cleavage effect, we created inactive ribozymes by substituting an essential nucleotide of the catalytic core with an inactive one. The cleavage reaction revealed that U1Rz803 possessed the perfect cleavage activity, while U1Rz803m possessed no catalytic activity. It can be used as control to exclude antisense effect of ribozyme in vivo in order that it is proven that the activity of U1Rz803 is due to catalytic cleavage in vivo. The kinetics of U1Rz803 showed that U1Rz803 possessed perfect specific ability of cleaving the TGFβ1 transcripts in vitro. These results made U1Rz803 to be worthy of being studied in intact cell and be developed as a nucleic acid drug in the future. However the in vitro result cannot completely reflect in vivo performance. The secondary and tertiary structure formed by the total TGF beta 1 mRNA transcript in the cell, the subcellular compartment which the ribozyme and target are located in, degradation of ribozyme, the complexes which are formed by ribozyme and ribonucleoprotein within cell and gene delivery system affect expression of ribozyme and cleavage activity of ribozyme. So in vivo effect of the ribozyme should be investigated as soon as possible. Experimental analysis of activity of the anti-TGF beta1 ribozyme in HSC-T6 cell is in progress.
We thanks Mr. F Xu, Mr. G Jiang, Mr.XL Chen, Mr.J Jia, Mr. ZW Wang and Miss W Li for their kind help.
JS Lin and YH Song contributed equally to this study
Edited by Wu XN
| 1. | Zhu YH, Hu DR, Nie QH, Liu GD, Tan ZX. Study on activation and c-fos, c-jun expression of in vitro cultured human hepatic stellate cells. Shijie Huaren Xiaohua Zazhi. 2000;8:299-302. |
| 2. | Du WD, Zhang YE, Zhai WR, Zhou XM. Dynamic changes of type I,III and IV collagen synthesis and distribution of collagen-producing cells in carbon tetrachloride-induced rat liver fibrosis. World J Gastroenterol. 1999;5:397-403. [PubMed] |
| 3. | Huang ZG, Zhai WR, Zhang YE, Zhang XR. Study of heteroserum-induced rat liver fibrosis model and its mechanism. World J Gastroenterol. 1998;4:206-209. [PubMed] |
| 4. | Jia JB, Han DW, Xu RL, Gao F, Zhao LF, Zhao YC, Yan JP, Ma XH. Effect of endotoxin on fibronectin synthesis of rat primary cultured hepatocytes. World J Gastroenterol. 1998;4:329-331. [PubMed] |
| 5. | Cheng ML, Wu YY, Huang KF, Luo TY, Ding YS, Lu YY, Liu RC, Wu J. Clinical study on the treatment of liver fibrosis due to hepatitis B by IFN-alpha(1) and traditional medicine preparation. World J Gastroenterol. 1999;5:267-269. [PubMed] |
| 6. | Nie QH, Cheng YQ, Xie YM, Zhou YX, Cao YZ. Inhibiting effect of antisense oligonucleotides phosphorthioate on gene expression of TIMP-1 in rat liver fibrosis. World J Gastroenterol. 2001;7:363-369. [PubMed] |
| 7. | Friedman SL. Seminars in medicine of the Beth Israel Hospital, Boston. The cellular basis of hepatic fibrosis. Mechanisms and treatment strategies. N Engl J Med. 1993;328:1828-1835. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 793] [Cited by in RCA: 886] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 8. | Wang X, Chen YX, Xu CF, Zhao GN, Huang YX, Wang QL. Relationship between tumor necrosis factor-alphaand liver fibrosis. World J Gastroenterol. 1998;4:18. [PubMed] |
| 9. | Xu LX, Xie XC, Jin R, Ji ZH, Wu ZZ, Wang ZS. Effect of selenium in rat experimental liver fibrosis. Huaren Xiaohua Zazhi. 1998;6:133-135. |
| 10. | Li DG, Lu HM, Chen YW. Studies on anti-liver fibrosis of tetrandrine. Shijie Huaren Xiaohua Zazhi. 1999;7:171-172. |
| 11. | Wu YA, Kong XT. Anti-hepatic fibrosis effect of pentoxifylling. Shijie Huaren Xiaohua Zazhi. 1999;7:265-266. |
| 12. | Liu F, Liu JX, Cao ZC, Li BS, Zhao CY, Kong L, Zhen Z. Relation-ship between TGF-β1, serum indexes of liver fibrosis and hepatic-tissue pathology in patients with chronic liver diseases. Shijie Huaren Xiaohua Zazhi. 1999;7:519-521. |
| 13. | Gu SW, Luo KX, Zhang L, Wu AH, He HT, Weng JY. Relation-ship between ductular proliferation and liver fibrosis of chronic liver disease. Shijie Huaren Xiaohua Zazhi. 1999;7:845-847. |
| 14. | Li BS, Wang J, Zhen YJ, Liu JX, Wei MX, Sun SQ, Wang SQ. Experimental study on serum fibrosis markers and liver tissue pathology and hepatic fibrosis in immuno damaged rats. Shijie Huaren Xiaohua Zazhi. 1999;7:1031-1034. |
| 15. | Wang Y, Gao Y, Huang YQ, Yu JL, Fang SG. Gelatinase a proen-zyme expression in the process of experimental liver fibrosis. Shijie Huaren Xiaohua Zazhi. 2000;8:165-167. |
| 16. | Wang FS, Wu ZZ. Current situation in studies of gene therapy for liver cirrhosis and liver fibrosis. Shijie Huaren Xiaohua Zazhi. 2000;8:371-373. |
| 17. | Wang GQ, Kong XT. Action of cell factor and decorin in tissue fibrosis. Shijie Huaren Xiaohua Zazhi. 2000;8:458-460. |
| 18. | Liu F, Wang XM, Liu JX, Wei MX. Relationship between serum TGF-β1 of chronic hepatitis B and hepatic tissue pathology and hepatic fibrosis quantification. Shijie Huaren Xiaohua Zazhi. 2000;8:528-531. |
| 19. | Li JC, Ding SP, Xu J. Regulating effect of Chinese herbal medicine on the peritoneal lymphatic stomata in enhancing ascites absorption of experimental hepatofibrotic mice. World J Gastroenterol. 2002;8:333-337. [PubMed] |
| 20. | Olaso E, Friedman SL. Molecular regulation of hepatic fibrogenesis. J Hepatol. 1998;29:836-847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 186] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 21. | Pinzani M, Marra F, Carloni V. Signal transduction in hepatic stellate cells. Liver. 1998;18:2-13. [PubMed] |
| 22. | Alcolado R, Arthur MJ, Iredale JP. Pathogenesis of liver fibrosis. Clin Sci (Lond). 1997;92:103-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 130] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 23. | Massagué J. The transforming growth factor-beta family. Annu Rev Cell Biol. 1990;6:597-641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2322] [Cited by in RCA: 2349] [Article Influence: 67.1] [Reference Citation Analysis (0)] |
| 24. | Sporn MB, Roberts AB. Transforming growth factor-beta: recent progress and new challenges. J Cell Biol. 1992;119:1017-1021. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 568] [Cited by in RCA: 583] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 25. | Border WA, Noble NA. Transforming growth factor beta in tissue fibrosis. N Engl J Med. 1994;331:1286-1292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2357] [Cited by in RCA: 2385] [Article Influence: 76.9] [Reference Citation Analysis (0)] |
| 26. | Heldin CH, Miyazono K, ten Dijke P. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465-471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2875] [Cited by in RCA: 2920] [Article Influence: 104.3] [Reference Citation Analysis (0)] |
| 27. | Miyazono K. TGF-beta receptors and signal transduction. Int J Hematol. 1997;65:97-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 49] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 28. | Friedman SL. Molecular regulation of hepatic fibrosis, an integrated cellular response to tissue injury. J Biol Chem. 2000;275:2247-2250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 29. | Bissell DM, Roulot D, George J. Transforming growth factor beta and the liver. Hepatology. 2001;34:859-867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 265] [Cited by in RCA: 284] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 30. | Qi Z, Atsuchi N, Ooshima A, Takeshita A, Ueno H. Blockade of type beta transforming growth factor signaling prevents liver fibrosis and dysfunction in the rat. Proc Natl Acad Sci USA. 1999;96:2345-2349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 234] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 31. | Sanderson N, Factor V, Nagy P, Kopp J, Kondaiah P, Wakefield L, Roberts AB, Sporn MB, Thorgeirsson SS. Hepatic expression of mature transforming growth factor beta 1 in transgenic mice results in multiple tissue lesions. Proc Natl Acad Sci USA. 1995;92:2572-2576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 485] [Cited by in RCA: 485] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 32. | Ueki T, Kaneda Y, Tsutsui H, Nakanishi K, Sawa Y, Morishita R, Matsumoto K, Nakamura T, Takahashi H, Okamoto E. Hepatocyte growth factor gene therapy of liver cirrhosis in rats. Nat Med. 1999;5:226-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 456] [Cited by in RCA: 471] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 33. | Nakamura T, Sakata R, Ueno T, Sata M, Ueno H. Inhibition of transforming growth factor beta prevents progression of liver fibrosis and enhances hepatocyte regeneration in dimethylnitrosamine-treated rats. Hepatology. 2000;32:247-255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 207] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 34. | Ueno H, Sakamoto T, Nakamura T, Qi Z, Astuchi N, Takeshita A, Shimizu K, Ohashi H. A soluble transforming growth factor beta receptor expressed in muscle prevents liver fibrogenesis and dysfunction in rats. Hum Gene Ther. 2000;11:33-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 101] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 35. | Persidis A. Ribozyme therapeutics. Nat Biotechnol. 1997;15:921-922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 36. | Gibson SA, Shillitoe EJ. Ribozymes. Their functions and strategies for their use. Mol Biotechnol. 1997;7:125-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 38. | Muotri AR, da Veiga Pereira L, dos Reis Vasques L, Menck CF. Ribozymes and the anti-gene therapy: how a catalytic RNA can be used to inhibit gene function. Gene. 1999;237:303-310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 39. | von Weizsäcker F, Blum HE, Wands JR. Cleavage of hepatitis B virus RNA by three ribozymes transcribed from a single DNA template. Biochem Biophys Res Commun. 1992;189:743-748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 46] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 40. | Beck J, Nassal M. Efficient hammerhead ribozyme-mediated cleavage of the structured hepatitis B virus encapsidation signal in vitro and in cell extracts, but not in intact cells. Nucleic Acids Res. 1995;23:4954-4962. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 70] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 41. | Xu R, Liu J, Zhou X, Xie Q, Jin Y, Yu H, Liao D. Activity identification of anti-caspase-3 mRNA hammerhead ribozyme in both cell-free condition and BRL-3A cells. Chin Med J (Engl). 2001;114:606-611. [PubMed] |
| 42. | Xu R, Liu J, Xu F, Jiang G, Xie Q, Zhou X, Jin Y, Wang D. Activity identification of chimeric anti-caspase-3 mRNA hammerhead ribozyme in vitro and in vivo. Sci China C Life Sci. 2001;44:618-627. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 43. | Liu R, Li W, Karin NJ, Bergh JJ, Adler-Storthz K, Farach-Carson MC. Ribozyme ablation demonstrates that the cardiac subtype of the voltage-sensitive calcium channel is the molecular transducer of 1, 25-dihydroxyvitamin D(3)-stimulated calcium influx in osteoblastic cells. J Biol Chem. 2000;275:8711-8718. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 44. | Montgomery RA, Dietz HC. Inhibition of fibrillin 1 expression using U1 snRNA as a vehicle for the presentation of antisense targeting sequence. Hum Mol Genet. 1997;6:519-525. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 51] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 45. | Michienzi A, Prislei S, Bozzoni I. U1 small nuclear RNA chimeric ribozymes with substrate specificity for the Rev pre-mRNA of human immunodeficiency virus. Proc Natl Acad Sci USA. 1996;93:7219-7224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 46. | Vogel S, Piantedosi R, Frank J, Lalazar A, Rockey DC, Friedman SL, Blaner WS. An immortalized rat liver stellate cell line (HSC-T6): a new cell model for the study of retinoid metabolism in vitro. J Lipid Res. 2000;41:882-893. [PubMed] |
| 47. | Kim Y, Ratziu V, Choi SG, Lalazar A, Theiss G, Dang Q, Kim SJ, Friedman SL. Transcriptional activation of transforming growth factor beta1 and its receptors by the Kruppel-like factor Zf9/core promoter-binding protein and Sp1. Potential mechanisms for autocrine fibrogenesis in response to injury. J Biol Chem. 1998;273:33750-33758. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 48. | Qian SW, Kondaiah P, Roberts AB, Sporn MB. cDNA cloning by PCR of rat transforming growth factor beta-1. Nucleic Acids Res. 1990;18:3059. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 220] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 49. | Manser T, Gesteland RF. Characterization of small nuclear RNA U1 gene candidates and pseudogenes from the human genome. J Mol Appl Genet. 1981;1:117-125. [PubMed] |
| 50. | Zhuang Y, Weiner AM. A compensatory base change in U1 snRNA suppresses a 5' splice site mutation. Cell. 1986;46:827-835. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 476] [Cited by in RCA: 611] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 51. | Skuzeski JM, Lund E, Murphy JT, Steinberg TH, Burgess RR, Dahlberg JE. Synthesis of human U1 RNA. II. Identification of two regions of the promoter essential for transcription initiation at position +1. J Biol Chem. 1984;259:8345-8352. [PubMed] |
| 52. | Uhlenbeck OC. A small catalytic oligoribonucleotide. Nature. 1987;328:596-600. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 655] [Cited by in RCA: 710] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 53. | Takayama H, LaRochelle WJ, Sharp R, Otsuka T, Kriebel P, Anver M, Aaronson SA, Merlino G. Diverse tumorigenesis associated with aberrant development in mice overexpressing hepatocyte growth factor/scatter factor. Proc Natl Acad Sci USA. 1997;94:701-706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 348] [Cited by in RCA: 334] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 54. | Green MR. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu Rev Cell Biol. 1991;7:559-599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 445] [Cited by in RCA: 561] [Article Influence: 16.5] [Reference Citation Analysis (0)] |