Published online Oct 15, 2003. doi: 10.3748/wjg.v9.i10.2344
Revised: May 12, 2003
Accepted: May 19, 2003
Published online: October 15, 2003
AIM: To develop a simple and rapid detection of HBV gene variants and prediction of lamivudine-resistance in patients.
METHODS: Initially, plasmids harboring the wild-type or mutant HBV DNA fragments were used in a model system. The technique was then applied to clinical samples for an analysis of YMDD mutations. The sera were extracted from chronic hepatitis patients who had received lamivudine treatment for more than one year. P region gene of HBV was amplified by polymerase chain reaction. The excess primers and dNTPs in PCR products were removed by cleaning-up reagents. Template-directed dye-terminator incorporation reaction was performed and R110 or TAMRA labeled acyclo-terminator was added on the 3’ end of TDI-primer specifically. Fluorescence polarization value was measured with Victor 2 multilabel counter and the genotypes of HBV were analyzed.
RESULTS: The YMDD genotypes in recombined positive plasmid and 56 serum samples of HBV infected patients were analyzed by using our TDI-FP method and the specificity and sensitivity were confirmed by DNA sequencing. Five of 56 serum samples showed YVDD phenotype (9%), including 1 YMDD and YVDD mixed infection. Four of 56 showed YIDD phenotype (7.1%).
CONCLUSION: This is a simple, rapid, low cost and high throughput assay to detect HBV polymerase gene variants and suitable for large-scale screening and prediction of the lamivudine-resistance in clinical samples.
- Citation: Bai YJ, Zhao JR, Lv GT, Zhang WH, Wang Y, Yan XJ. Rapid and high throughput detection of HBV YMDD mutants with fluorescence polarization. World J Gastroenterol 2003; 9(10): 2344-2347
- URL: https://www.wjgnet.com/1007-9327/full/v9/i10/2344.htm
- DOI: https://dx.doi.org/10.3748/wjg.v9.i10.2344
Hepatitis virus B (HBV) is the causative agent of acute and chronic hepatitis. Approximately 400 million people worldwide are chronically infected with HBV. There are 170 million people infected in China[1,2]. HBV infection can lead to cirrhosis and primary hepatocellular carcinoma[3-5]. To date, interferon alfa and lamivudine are the only two agents approved for chronic hepatitis[6-10]. Because interferon alfa shows significant dose-dependent side effects, lamivudine has emerged as the first therapeutic agent[11-13]. Chinese patients are immunotolerant to interferon alfa because of acquisition of the disease during early childhood. The efficacy of interferon alfa in Chinese is lower than in white patients[14]. However, the efficacy of lamivudine is equal in Chinese and white patients.
Lamivudine is an antiviral nucleoside analogue. It can inhibit HBV reverse transcriptase activity and also act as a viral DNA chain terminator[15]. It is an inactive form before activated in hepatocytes by addition of phosphate group[2]. Lamivudine has two pathways of viral suppression. First, the active triphosphate metabolite is incorporated into newly synthesized HBV DNA to stop the chain extension. Second, it inhibits HBV reverse transcriptase. Its clinical use has resulted in HBeAg seroconversion and undetectable HBV DNA[16]. Unlike some nucleoside analogues, lamivudine is well tolerated and has an excellent safety profile. It has little or no effect on bone marrow, hepatocytes, kidney, or muscle tissues. However, long-term lamivudine treatment has led to emergence of HBV variants resistant to lamivudine therapy in some patients. The major sites of mutation are situated in the highly conserved motif, tyrosine (Y), methionine (M), aspartate (D), aspartate (D) (YMDD), of the catalytic (C) domain of the reverse transcriptase. Mutations consist of an amino acid substitution from M to either valine (A739G, Met552→Val552) or isoleucine (G741T, Met552→Ile552). Those mutations at the YMDD motif could render HBV resistant to lamivudine[17].
Fluorescence polarization assay (FPA) is a well established method used to analyze associations and dissociations between molecules in solution. This technique relates the change in the molecular size of a fluorophore to a change in the fluorescence polarization value (mp unit). The value changes indicate molecular associations or dissociations, so that FPA have been used to examine the interactions between molecules, such as protein-protein interactions, DNA-protein interaction and DNA-DNA hybridization.
Based on the FP detection method and the combined template-directed dye-terminator incorporation technique, a novel SNP (single nucleotide polymorphism) detection system (TDI-FP) has been developed[18]. It relies on the ability of AcycloPolTM, a novel mutant thermostable polymerase from the Archeon family, to extend accurately an annealed probe by a single dye-terminator that is complementary to the opposite strand. It performs an assay to determine the base at a SNP site with many advantages over the other methods, such as homogeneity, low cost, high accuracy, high throughput and ready adaptation to automation. This method has been used to genotyping and SNP detections but has not, to our knowledge, been used to microbial mutant analysis yet.
Routine detection of HBV YMDD mutation after lamivudine treatment will be required to increase the efficacy. In this study, we undertook such a study using TDI-FP to detect the YMDD variants using mutagenesis plasmids and serum samples of 56 patients. This new method is a rapid, accurate and high throughout method to analyses the YMDD mutations. It will be a very useful tool for the clinical diagnosis and monitor of the lamivudine resistance.
Fluorescence polarization values were determined with the VICTOR2 multilabel counter (Perkin-Elmer, USA). The excitation and emission wavelengths were 544 nm and 595 nm for R110 and 485 nm and 535 nm for TAMRA, respectively. The samples were measured in 384 well PCR plates (MSP-3862 MJ Research, USA).
Taq DNA polymerase, DNA extractor kit were obtained from Hua-mei Bio-tech Co., LTD. AcycloPrimeTM-FP detection kit (including AcycloPolTM, AcycloTeminatorsTM labeled with R110 and TAMRA, shrimp alkaline phosphatase, exonuclease I) was product of Perkin-Elmer Co., Ltd.
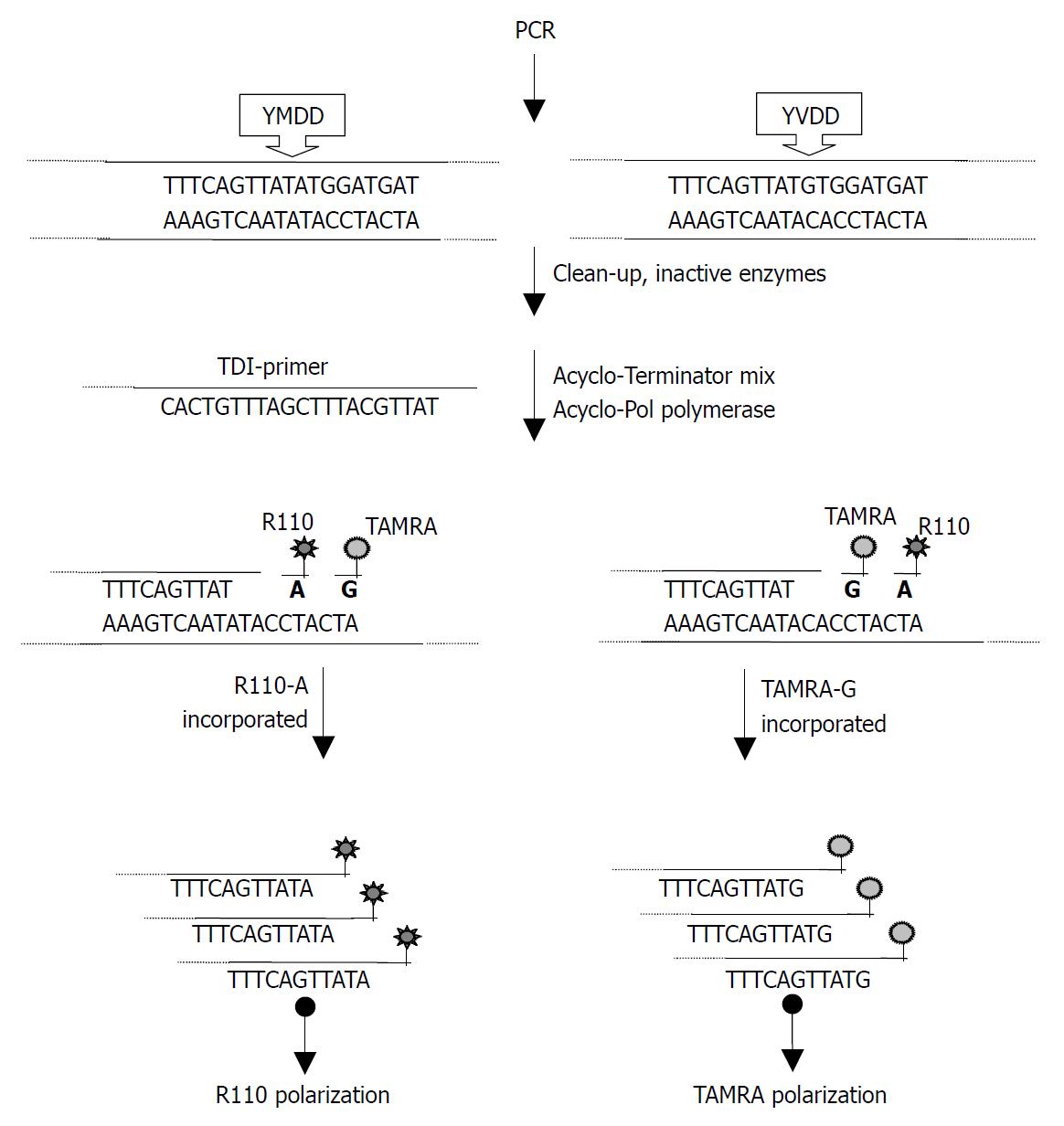
A set of PCR primers was designed to amplify the P region of HBV gene including the YMDD motif encoding sequences. 2 TDI-primers were also designed to encompass 17-21 bp 5’ (A739Gtdi, sense) or 3’ (G741T, antisense) to the nucleotide adjacent to the mutation sites using DNAStar software. 3 sets of primers for the site-directed mutagenesis were also designed using DNAStar software. All the primers, whose sequences are shown in Table 1, were synthesized using 391 DNA synthesizer (Perkin-Elmer, USA) by Bao Tai Ke Biotech Co.
| Primer | Sequence |
| PCRfor | 5’-GCACTTGTATTCCCATCCCATCAT-3’ |
| PCRrev | 5’-GTATACCCAAAGACAAAAGAA-3’ |
| 739tdi | 5’-CACTGTTTAGCTTTCAGTTAT-3’ |
| 741tdi | 5’-CCCAATACCACATCATC-3’ |
| A719Gfor | 5’-CTTTCAGTTATGTGGATGATGTGGTA3’ |
| A719Grev | 5’-CCAGACAGTGGGGGAAAGC3’ |
| G741Tfor | 5’-CTTTCAGTTATATTGATGATGTGGTA3’ |
| G741Trev | 5’-CCAGACAGTGGGGGAAAGC3’ |
| diMutfor | 5’-CTTTCAGTTATGTTGATGATGTGGTA3’ |
| diMutrev | 5’-CCAGACAGTGGGGGAAAGC3’ |
The site-directed HBV YMDD mutants were made using Takara MutanBEST Kit by TaKaRa Biotechnology Co., Ltd. The A719G mutation was created using primers A739Gfor and A739Grev. The G741T mutation was created using primers G741Tfor and G741Trev. The A739G/G741T double mutants were made using primers diMutfor and diMutrev with the plasmid pMD/HBV as a template respectively. (All the sequences of primers are shown in Table 1). The mutagenesis was detected according to the manufacture’s manual and verified by DNA sequencing.
Venous blood (1 mL) was obtained from 56 patients with HBV chronic hepatitis B in Tangdu Hospital and Second Hospital of Jiaotong University, Xi’an.
PCR amplification was performed in a total volume of 50 μL, containing 1.5 U Taq polymerase, PCR buffer, 0.1 mmol/L dNTPs, 1 μL of DNA sample (10 ng plasmid DNA or 100 ng genome DNA), 100 nmol/L PCRfor and PCRrev primer each. The amplification procedure consisted of an initial denaturation and enzyme activation step at 94 °C for 5 min, followed by 40 cycles containing denaturation at 94 °C for 30 s, annealing at 56 °C for 30 s and extension at 72 °C for 1 min. After a final extension step at 72 °C for 10 minutes, the reactions were cooled to 4 °C until further use. The PCR reaction was carried out in a Touchgene gradient termal cycler (TECHNE Co. USA). In order to evaluate the amplification, each 5 μL of PCR products was separated on a 20 g/L agarose gel in 0.5 × TBE buffer. The products were visualized and photographed under ultraviolet (UV) light.
Following PCR amplification, 2 μL clean-up reagent (including shrimp alkaline phosphatase and exonuclease I and reaction buffer) was added into 5 μL of PCR product and incubated at 37 °C for 60 min to remove unincorporated dNTPs and free primers. The enzymes were then heat-inactivated at 80 °C for 30 min.
TDI-FP was performed according to the protocol supplied by the manufacturer (Perkin-Elmer Life Sciences, Inc, USA). The reaction system consisted of 13 μL of AcycloPrime-FP mixture (containing 0.05 μL of AcycloPolTM polymerase, 2 μL of 10 × reaction buffer, 1 μL of Acyclo-Terminator Mix, 0.5 μL mutant detection primer and 9.45 μL of water) and 7 μL of amplified and processed target DNA. After an initial denaturation at 95 °C for 2 min, 25 cycles at 95 °C for 15 sec and at 55 °C for 30 s were performed on a black 384-well PCR plate (MSP-3862, MJ Research). The fluorescence polarization values were measured on the 384-well plate using Victor2 multilabel counter directly.
The PCR products were analyzed by agarose gel electrophoresis (Figure 1). A single band corresponding to a molecular size of 200 bp was detected, which was in concordance with expectation.
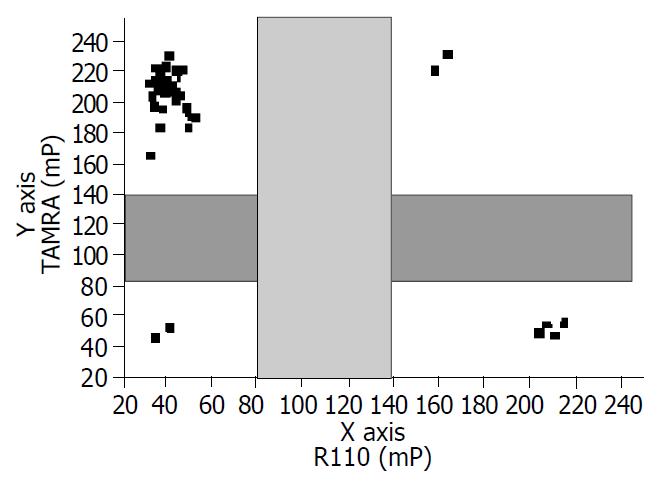
Following PCR amplification, excess primers and dNTPs were removed. In the TDI extension reaction, 729tdi primer and dye-terminators mix (R110-acyGTP/TAMRA-acyATP) were used to detect the A739G mutation (Figure 2). 741tdi primer and R110-acyCTP/TAMRA-acyATP) were used to detect the G741T mutation. The dye-terminator was incorporated and the TDI-primer was extended by one base complementary to the specific mutation site. The fluorescence intensity was measured using Victor 2 multilabel counter and the FP value was calculated by the instrument software automatically. The graph was created from an Excel workbook and the cluster of AcycloPrime-FP data was obtained by plotting the TAMRA polarization vs the R110 polarization. The FP reading of the samples was clustered into four distinct groups (Figure 3). As expected, for HBV YVDD samples, the values for R110-acyGTP were high and the values for TAMRA-acyATP were low, reflecting incorporation of the R110-acyGTP but not TAMRA-acyATP. The genotypes were represented by the cluster in the lower left. Conversely, the YMDD genotypes appeared in the upper right. YMDD and YVDD double mutations or mixed infection samples appeared in the upper right. Negative controls were in the lower left cluster (Figure 3). For the G741T detection, the YIDD samples appeared in the upper left, YMDD samples in the lower right and the negative control in the lower left.
To evaluate the utility and accuracy of the TDI-FP in clinical samples, 56 serum samples from patients with chronic hepatitis B were analyzed. Among the sera analyzed, the wild-type YMDD variant was detected in 46 (82%), the YVDD variant in 5 (8.9%) and the YIDD in 4 (7.1%). In one patient (1.8%), both YMDD and YVDD variants were detected, which might reflect a mixed infection.
As reported before, the major drawback to the therapy of lamivudine is the emergence of drug-resistance due to the HBV DNA mutations. The patients who have lamivudine resistant HBV variants infection are more likely to develop more severe liver damage than those who do not have drug-resistant mutants. The mutation at YMDD (YMDD→YVDD or YMDD→YIDD) is considered to be the major lamivudine-resistance related variants. DNA sequencing is considered as the ideal method for characterizing DNA mutants, but it can not be afforded by most laboratories, especially clinical laboratories due to the cost and equipment requirement. Type-specific PCR, PCR-RLFP and PCR-SSCP have been used to detect HBV YMDD mutants. However, all the methods have drawbacks, e.g. lack of accuracy, difficulty in determining the mutation sites exactly or detecting mixed infection[19].
We reported here, for the first time, the successful application of the TDI-FP to detect the HBV YMDD mutation in hepatitis B patients. As a homogenous assay, the FP value reflected the total sum of free and incorporated dye-terminators. Two major factors affecting the result were faced: (1) The concentration of the first PCR product in TDI extension was too high or too low, which would create misincorporation or lower incorporation of dye-terminators, (2) the excess primers in the first PCR product might lead to misincorporation, while excess dNTPs would interfere the dye-terminator incorporation competitively. We increased the number of thermal cycles to 40-45 to ensure that all PCR components were exhausted and adjusted to the concentration of dNTPs and primer in PCR reaction, which were lower than standard PCR protocol. We also prolonged the digestion time from 30 min to 60 min to completely remove the dNTPs and primers. At the same time, to get best TDI-FP performance, forward and reverse TDI primers were used and compared in our study (data not shown). The predicted reaction conditions (annealing temperature and MgCl2) were tested on several nonessential DNA samples to optimize the system.
DNA diagnostic tests will no doubt be applied more and more in clinical rather than in research laboratories. The main advantages of our technique over the other methods for scoring DNA variation, are high accuracy, high throughput and easy to adaptation automatic. It will improve the analysis and prediction of drug-resistance in clinic.
Edited by Zhang JZ and Wang XL
| 1. | Yan JC, Ma JY, Pan BR, Ma LS. The study of chronic hepatitis B in China. Shijie Huaren Xiaohua Zazhi. 2001;9:611-616. |
| 2. | Su Q, Fu Y, Liu YF, Zhang W, Liu J, Wang CM. Laminin induces the expression of cytokeratin 19 in hepatocellular carcinoma cells growing in culture. World J Gastroenterol. 2003;9:921-929. [PubMed] |
| 3. | Cao XY, Liu J, Lian ZR, Clayton M, Hu JL, Zhu MH, Fan DM, Feitelson M. Differentially expressed genes in hepatocellular carcinoma induced by woodchuck hepatitis B virus in mice. World J Gastroenterol. 2001;7:575-578. [PubMed] |
| 4. | Huang YX, Wu GH. The relationship between hepatitis B and hepatocellular carcinoma. Shijie Huaren Xiaohua Zazhi. 2001;9:682-685. |
| 5. | Wang FS, Xing LH, Liu MX, Zhu CL, Liu HG, Wang HF, Lei ZY. Dysfunction of peripheral blood dendritic cells from patients with chronic hepatitis B virus infection. World J Gastroenterol. 2001;7:537-541. [PubMed] |
| 6. | Han HL, Lang ZW. Changes in serum and histology of patients with chronic hepatitis B after interferon alpha-2b treatment. World J Gastroenterol. 2003;9:117-121. [PubMed] |
| 7. | Lai YC, Hu RT, Yang SS, Wu CH. Coinfection of TT virus and response to interferon therapy in patients with chronic hepatitis B or C. World J Gastroenterol. 2002;8:567-570. [PubMed] |
| 8. | Zhuang L, You J, Tang BZ, Ding SY, Yan KH, Peng D, Zhang YM, Zhang L. Preliminary results of Thymosin-a1 versus interferon-alpha-treatment in patients with HBeAg negative and serum HBV DNA positive chronic hepatitis B. World J Gastroenterol. 2001;7:407-410. [PubMed] |
| 9. | You J, Zhuang L, Tang BZ, Yang WB, Ding SY, Li W, Wu RX, Zhang HL, Zhang YM, Yan SM. A randomized controlled clinical trial on the treatment of Thymosin a1 versus interferon-alpha in patients with hepatitis B. World J Gastroenterol. 2001;7:411-414. [PubMed] |
| 10. | Cheng ML, Wu YY, Huang KF, Luo TY, Ding YS, Lu YY, Liu RC, Wu J. Clinical study on the treatment of liver fibrosis due to hepatitis B by IFN-alpha(1) and traditional medicine preparation. World J Gastroenterol. 1999;5:267-269. [PubMed] |
| 11. | Zoulim F. A preliminary benefit-risk assessment of lamivudine for the treatment of chronic hepatitis B virus infection. Drug Saf. 2002;25:497-510. [PubMed] |
| 12. | Yang SS, Hsu CT, Hu JT, Lai YC, Wu CH. Lamivudine does not increase the efficacy of interferon in the treatment of mutant type chronic viral hepatitis B. World J Gastroenterol. 2002;8:868-871. [PubMed] |
| 13. | Han T, Qian SC, Chen Y, Xiao SX, Wang FM, Han ZC, Lu HM. Treatment of 198 patients with chronic hepatitis B with lamivudine and Yi –Gan-Jian. Shijie Huaren Xiaohua Zazhi. 2002;10:1236-1237. |
| 14. | Lai CL, Dienstag J, Schiff E, Leung NW, Atkins M, Hunt C, Brown N, Woessner M, Boehme R, Condreay L. Prevalence and clinical correlates of YMDD variants during lamivudine therapy for patients with chronic hepatitis B. Clin Infect Dis. 2003;36:687-696. [PubMed] |
| 15. | Cao H, Tao P. [Anti-hepatitis B virus effects of lamivudine and other five drugs in vitro]. Zhonghua Yixue Zazhi. 2001;81:1004-1007. [PubMed] |
| 16. | Dienstag JL, Schiff ER, Wright TL, Perrillo RP, Hann HW, Goodman Z, Crowther L, Condreay LD, Woessner M, Rubin M. Lamivudine as initial treatment for chronic hepatitis B in the United States. N Engl J Med. 1999;341:1256-1263. [PubMed] |
| 17. | Li G, Shu X, Ma HH, Chen W, Chen WS, Chen Q Jiang YS, Yao JL. Detection of HBV, HCV and HBV YMDD mutants by DNA microarray. Shijie Huaren Xiaohua Zazhi. 2003;11:178-181. |
| 18. | Hsu TM, Chen X, Duan S, Miller RD, Kwok PY. Universal SNP genotyping assay with fluorescence polarization detection. Biotechniques. 2001;31:560, 562, 564-568, passim. [PubMed] |
| 19. | Jardi R, Buti M, Rodriguez-Frias F, Cotrina M, Costa X, Pascual C, Esteban R, Guardia J. Rapid detection of lamivudine-resistant hepatitis B virus polymerase gene variants. J Virol Methods. 1999;83:181-187. [PubMed] |