Published online Jan 15, 2003. doi: 10.3748/wjg.v9.i1.30
Revised: April 11, 2002
Accepted: April 20, 2002
Published online: January 15, 2003
AIM: To clone genes that may predispose us to human gastric cancer and to analyze it’s expression in gastric tissues.
METHODS: Specimens of paired tumor, paratumor and normal gastric mucosa tissues collected from fifteen patients who suffered from stomach antrum adenocarcinoma were used for analysis. Seven out of the fifteen cases were first studied by fluorescent differential display reverse transcription polymerase chain reaction (DDTR-PCR) analysis. The differentially expressed bands of interest were cloned, analyzed by Northern blot, sequencing and RT-PCR. Through BLAST, the sequencing results were compared with GenBank database for homology analysis. In situ hybridization with DIG-labeled cRNA probes was used to analyze the expression of interesting cDNA bands in paraffin embedded paired normal gastric mucosa and cancer tissues isolated from 30 gastric adenocarcinoma patients.
RESULTS: DDRT-PCR showed that one of the interesting cDNA bands, which was named W2, expressed much higher in all seven tested tumor and paratumor samples than in their normal counterparts, it was sub-cloned into a pGEM-T Easy vector. Two subclones were subsequently obtained. One of the subclone, GCRG224, was studied further. The sequencing result showed that GCRG224 consisted of 1159 base pairs and had one open reading frame (ORF). It located at human chromosome 11q14. No homologue was found in GenBank database with GCRG224-ORF. This nucleotide sequence data were submitted to GenBank with accession No. AF438406. RT-PCR showed that GCRG224 expressed higher in 11/15 gastric cancer tissues than in non-tumor tissues. However, the result of Northern blot analysis showed a higher GCRG224 expression in the non-tumor tissue than in the tumor one. Human multiple tissue Northern blot analysis revealed that GCRG224 also expressed in human normal colon tissue, and peripheral blood leukocyte. In situ hybridization analysis showed that only 5/30 adenocarcinoma, 3/18 dysplasia and 6/18 intestinal metaplasia showed higher GCRG224 expression level than the normal gastric glands. However, GCRG224 was over-expressed predominantly in 26/30 cases of normal mucosal epithelium.
CONCLUSION: A novel gene named GCRG224 was identified from human gastric mucosal tissue. It overexpressed in almost all gastric mucosal epithelium but only a small portion of cancer and precancerous leisions. The role of GCRG224 expression in gastric epithelium needs further study.
- Citation: Wang GS, Wang MW, Wu BY, You WD, Yang XY. A novel gene, GCRG224, is differentially expressed in human gastric mucosa. World J Gastroenterol 2003; 9(1): 30-34
- URL: https://www.wjgnet.com/1007-9327/full/v9/i1/30.htm
- DOI: https://dx.doi.org/10.3748/wjg.v9.i1.30
Gastric cancer is one of the most commonly diagnosed malignancies and remains an important cause of mortality world wide[1]. Considerable evidence supports the pivotal role of genetic factors in the pathogenesis of gastric cancer[2-7]. However, the mechanism of the process of multistage carcinogenesis is still unknown for gastric cancer. Abundant clinical and histopathogical data suggest that most, if not all, intestinal-type gastric cancer arise from precancerous leisions, which indicated intestinal-type gastric cancer to be an excellent model for studying the genetic alterations involved in the development of human neoplasms. It would be desirable, therefore, to screen directly from human intestinal-type gastric cancer and its precursor leisions the differentially expressed genes that are closely related to human gastric cancer.
In this study, differential display reverse transcription polymerase chain reaction (DDRT-PCR) analysis was used to identify and characterize differentially expressed genes in human gastric cancer tissues in comparison with their surrounding paratumor and nontumor counterparts. One novel cDNA with a complete cds was identified. We designated this gene as gastric cancer related gene 224 (GCRG224). Expression of GCRG224 in gastric tissues was analyzed using RT-PCR and In situ hybridization (ISH).
Fresh primary intestinal-type gastric adenocarcinoma, paratumor tissue which is 1.0 cm away from the tumor mass and their surrounding noncancerous stomach mucosal tissues obtained from 15 patients (11 male, 4 female, with average age 54 ± 15 years) undergoing surgery were used for reverse transcription polymerase chain reaction (RT-PCR) analysis. 7 cases (4 male, 3 female, with average age 51 ± 18 years) out of the 15 patients were used for differential display analyses. Paraffin embedded gastric adenocarcinoma and their corresponding normal gastric mucosal tissues obtained from 30 advanced gastric adenocarcinoma patients (21 male, 9 female, with average age 59 ± 10 years) were used for ISH analysis. All tissues were histologically confirmed by pathologists.
Total RNA was extracted from tissues using TRIzol reagent (Life Technologies, Inc., Rockville, Maryland). The fluorescent DDRT-PCR procedure was performed essentially as described previously[8]. The primers used in the assay included 4 T12MN primers and 20 arbitrary oligonucleotide primers (ARP-1 to ARP-20) (Genomyx Corporation, Foster City, CA).
The cDNA fragment of interest were subcloned into pGEM-T Easy vector (Promega Corporation, Madison, WI) and confirmed by EcoR I (Life Technologies, Bethesda, MD) digestion according to the manufacturer’s instruction. Sequence analysis was performed with CEQTM 2000 DNA sequencer (Beckman Coulter, Fullerton, CA). The sequenced cDNA were analyzed via the BLAST program for homology matches in the GenBank database[9].
Samples containing 20 μg of total nontumor and tumor RNAs were fractionated on an 1.2% agarose gel containing 5.5% formaldehyde and 1 × MOPS and transferred to a nylon membrane. Fix the RNA to the membrane by baking at 80 °C for 2 hours. Human multiple tissue Northern (MTN) blot II membrane (Clontech Laboratories, Inc., Palo Alto, CA) was used for analysis. Anti-sense cRNA probe labeled with digoxigenin was generated from a digested cDNA insert using Dig Northern Starter Kit (Roche Diagnostic Corporation, Indianapolis, IN) by means of in vitro transcription. The membrane was prehybridized and then hybridized according to the manufacturer’s protocol. The results were detected using chemiluminescent detection.
First-strand cDNAs were synthesized by SuperscriptII Rnase H- reverse transcriptase (Life Technologies, Bethesda, MD) using oligo d(T) primers according to the protocal. PCR primers were designed using Primer Premier software (Premier biosoft international, Palo Alto, CA). The PCR primers used were: forward primer: 5’AAGGGTCACCTCTGTTCAAAGTG3’, reverse primer: 5’GCAGGGTTTATGGGCTCAATAG3’, Product length: 929 bp. G3PDH was used as internal control. Reactions were carried out in 10 μl solutions containing 100 ng cDNA, 0.2 μM of each primer, 50 μM of each dNTP, and 0.5 U of AmpliTaq DNA polymerase (Life Technologies, Bethesda, MD). The amplification conditions were (25 cycles): 1 min at 94 °C, 40 sec at 56 °C, 1 min at 72 °C.
Specimens were fixed in 10% neutral buffered formalin and embedded in paraffin wax. A series of 5 μm sections were cut for analysis. The antisense cRNA probe was prepared according to procedure described in Northern blot analysis section. ISH was performed as described previously[10,11]. Negative control (no probe applied) was performed. Slides were examined using an Olympus microscope. Images were captured using a cooled CCD camera (JVC, Japan). Cytoplasmic staining was noted.
Any appreciable blue staining was considered positive, and graded as ± if very light blue were barely detectable, 1+ if light blue staining were seen diffusely throughout the cytoplasm, 2+ if easily seen fine staining were present throughout the cytoplasm, and 3+ when dark blue were observed. Generally, more than 100 cells (nontumor or tumor) were quantified in each measurement and at least one measurement was taken per slide. H&E slides were then reviewed to determine diagnosis and to map the location of the various histological patterns and correlate with the staining patterns observed in the ISH preparations.
The study protocol was approved by the Institutional Review Board of the hospital under the guidelines of the 1975 Declaration of Helsinki. Written informed consents were obtained from patients.
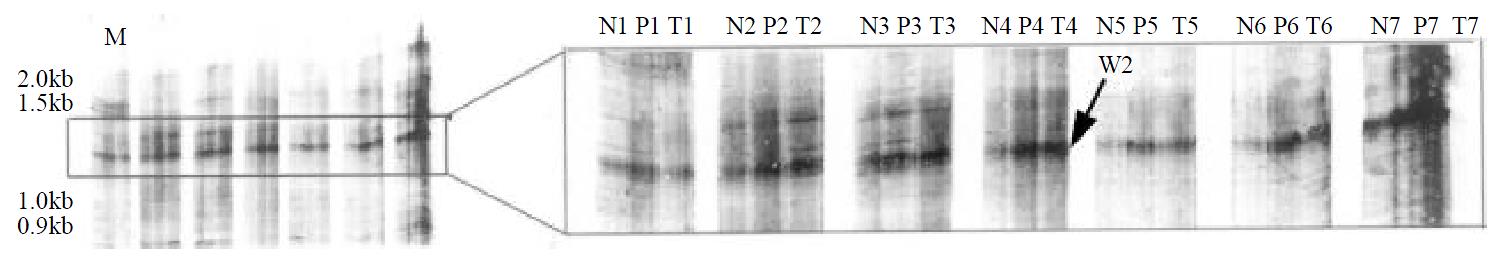
In order to run the assay efficiently, among 80 combinations of primer pairs (4 T12MN primers vs 20 ARP primers), 4 pairs able to amplify more bands in one set of paired tissues were chosen and used to amplify cDNA from 7 sets of paired tissues. One primer pair (T12GG vs ARP-3: 5’GACCATTGCA3’) was found to amplify genes expressed differentially in 7 of 7 sets of the paired tissues. As shown in Figure 1, in comparison with results in the normal tissues, a cDNA fragment (arrowed) was found to be more abundant in the tumor and paratumor samples in all tested patients. The results of mRNA differential display were reproducible. This differentially expressed cDNA band was named as W2.
Two subclones, GCRG213 and GCRG224, were idnetified from the subcloning procedure. GCRG213 consisted of 1194 base pairs, while GCRG224 consisted of 1159 base pairs and had one open reading frame (ORF). The GCRG224-ORF consists of 35 amino acids with an estimated molecular weight of 3.8 kDa. BLAST analysis revealed that GCRG224 nucleotide sequence had 98% homologue with Homo sapiens clone RP11-718B12, but the deduced amino acid sequence of GCRG224-ORF had no homology with any known peptide in the GenBank database. GCRG224 nucleotide sequence data was submitted to GenBank with accession No.AF438406. Further bioinformatic study in the GenBank database showed that GCRG224 is located at human chromosome 11q14.
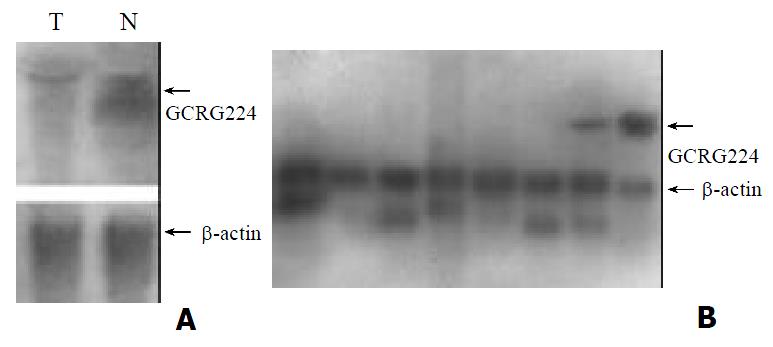
To confirm the expression pattern of GCRG224 in human gastric cancer, we performed RT-PCR analyses in 15 cases of paired gastric adenocarcinoma, paratumor and non-tumor tissues. GCRG224 expressed higher in 11/15 gastric cancer tissues than non-tumor tissues. However, the result of Northern blot analysis was contrary to that of DDRT-PCR, the non-tumor tissue showed higher GCRG224 expression level than the tumor did (Figure 2A). MTN results showed that GCRG224 also expressed in human normal colon tissues and peripheral blood leukocyte (Figure 2B).
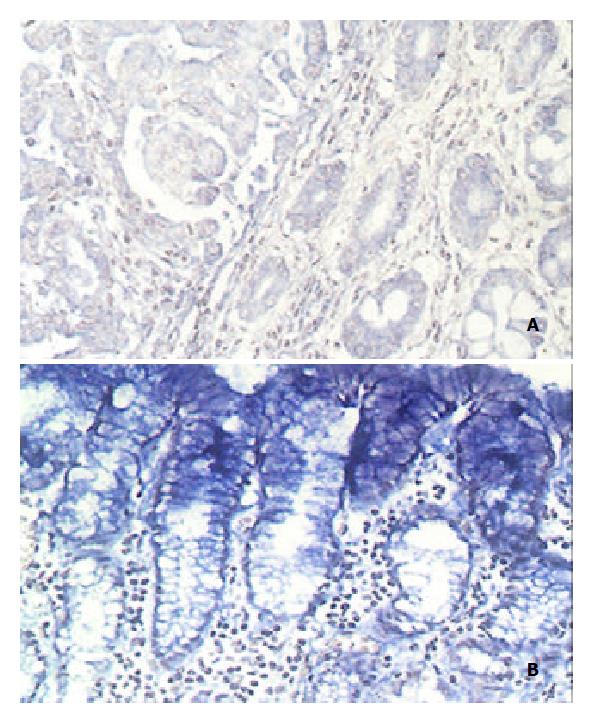
GCRG224 expression was analyzed at mRNA level. The hybridization signal that appears as blue is restricted to the cytoplasmic portion. All normal gastric glands showed ± - 1+ staining. Only 5 out of 30 cases of adenocarcinoma had 2+ staining while the rest had ± staining. Of the 18 cases of intestinal metaplasia (IM) and dysplasia found in the paratumor site, 6/18 IM and 3/18 dysplasia had 2+ staining, respectively. Interestingly, in 26/30 cases the gastric mucosal epithelial cells were stained 2+ -3+ (Table 1, Figure 3).
| gastric mucosal epithelium | Gastric glands | IM | DYS | adenocarcinoma | |
| ± | 0 | 19 | 8 | 13 | 25 |
| 1+ | 4 | 11 | 4 | 2 | 0 |
| 2+ | 15 | 0 | 6 | 3 | 4 |
| 3+ | 11 | 0 | 0 | 0 | 1 |
| Total | 30 | 30 | 18 | 18 | 30 |
Advances in molecular biology have revealed a consistent set of genetic alterations that may correspond to multi-step stomach cancer development. Aberrant expression and amplification of oncogenes such as c-met, c-myc, K-ras, c-erbB-2[12-16], etc., inactivation of tumor suppressor genes such as p53, p16, Rb, DCC, APC, PTEN[17-24], etc, DNA ploidy and microsatellite instability[25-28], abnormal transcript of genes related to metastasis like nm23, CD44, E-cadherin[29,30], etc., are reported common events in the steps of carcinogenesis. Newly found cancer related genes such as COX-2, survivin, metallothionein II and RUNX3, etc. were also expressed abnormally in gastric cancer[31-38]. Recognition of genetic factors had also improved the the treatment of cancer[39-41]. However, very little is currently known about genes that may predispose us to gastric cancer. The primary aim of this study was to identify genes that were closely related to human gastric cancer by means of DDRT-PCR.
DDRT-PCR has been widely applied to identify cancer-related genes[42-47], some of these genes look like to be of clinical value. It might reflect the true patterns of genetic changes in human gastric cancer if we identify gastric cancer-related genes directly from human gastric tissues.
One of the defects when using tissues for DDRT-PCR analysis is that some cDNA fragments amplified in the tumor tissues may actually originate from normal cells present in the tumor tissues, which may affect the results of the analyses. In order to decrease the chance of error in this study, we used 7 sets of tumor, paratumor and non-tumor tissues for the assay, displayed the results simultaneously and chose interesting bands showing distinctive patterns in all sets of specimens. One cDNA fragment which is up-regulated in gastric cancer tissues was identified. However, two subclones were identified from the subsequent subcloning procedure. Sequencing results revealed that these two subclones contained nucleotides of nearly the same size but of totally different sequences. This should be the result of PCR amplification in which arbitrary primers were used.
Because the deduced amino acid sequence of GCRG224-ORF had no homology with any known peptide in the GenBank database, further study was focused on this gene. The result of RT-PCR analysis was consistent with that of DDRT-PCR, but Northern blot analysis revealed an opposite result. Furthermore, ISH analysis also failed to see a general higher expression of GCRG224 in tumor cells. On the other hand, ISH showed an extensively strong expression of GCRG224 in gastric mucosal epithelial cells in almost all tested cases. This indicated GCRG224 might come mainly from normal gastric epithelium other than tumor cells. Although the tissues applied for DDRT-PCR, Northern blot and RT-PCR in this study were examined by pathologists before their usage, they were actually a mixture of different types of cells, the exact percentage of tumor and non-tumor cells in each tissue could not be identified, this resulted in the contradiction we faced. It would be better, therefore, to separate different types of cells using techniques such as microdissection in tissues like gastric mucosa prior to the analyses[48-51].
Correlations between prognoses and the expression of genes such as p53, ras, myc, nm23, etc. were studied extensively in gastric cancer[15-17,52-57]. However, the results turned out to be controversial. It is meaningful to find a biomarker that might predict the diagnosis or prognosis of gastric cancer. The consistent over-expression of GCRG224 in all tested tumor tissues in the DDRT-PCR and RT-PCR analyses prompted us to study further the expression pattern of GCRG224 in gastric mucosal tissues using ISH. However, GCRG224 did not show the expression pattern in gastric cancer and its precancerous leisions as RT-PCR revealed. Only around 20 percent tumor and precancerous leisions showed GCRG224 over-expression comparing with their normal gastric glands. Thus, GCRG224 may not serve as a potential marker for the diagnosis of gastric cancer.
A significant finding in this study is the extensive staining of GCRG224 in gastric epithelium. Up till now, the function of gastric mucosal cells is believed to be a barrier of the stomach as well as secretion of alkali and other ions such as Na+, K+ and Cl-. It will be of great interest to study the role of GCRG224 expression in gastric mucosal epithelium.
Edited by Zhao M
| 1. | Stadtländer CT, Waterbor JW. Molecular epidemiology, pathogenesis and prevention of gastric cancer. Carcinogenesis. 1999;20:2195-2208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 133] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 2. | Becker KF, Keller G, Hoefler H. The use of molecular biology in diagnosis and prognosis of gastric cancer. Surg Oncol. 2000;9:5-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 67] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 3. | Boussioutas A, Taupin D. Towards a molecular approach to gastric cancer management. Intern Med J. 2001;31:296-303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 4. | Palli D. Epidemiology of gastric cancer: an evaluation of available evidence. J Gastroenterol. 2000;35 Suppl 12:84-89. [PubMed] |
| 5. | Yasui W, Oue N, Kuniyasu H, Ito R, Tahara E, Yokozaki H. Molecular diagnosis of gastric cancer: present and future. Gastric Cancer. 2001;4:113-121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 73] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 6. | Maltoni M, Volpi A, Nanni O, Bajorko P, Belletti E, Vecci AM, Liverani M, Danesi S, Calistri D, Ricotti L. Gastric cancer: epidemiologic and biological aspects. Forum (Genova). 1998;8:199-207. [PubMed] |
| 7. | Meltzer SJ. Tumor genomics vs. tumor genetics: a paradigm shift. Gastroenterology. 2001;121:726-729. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 1] [Article Influence: 0.0] [Reference Citation Analysis (0)] |
| 8. | Wang GS, Wang MW, You WD, Wang HF, Feng MF. [Fluorescent mRNA differential display technique]. Zhongguo Yingyong Shenglixue Zazhi. 2000;16:373-376. [PubMed] |
| 9. | Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389-3402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53569] [Cited by in RCA: 52305] [Article Influence: 1868.0] [Reference Citation Analysis (0)] |
| 10. | Komminoth P, Merk FB, Leav I, Wolfe HJ, Roth J. Comparison of 35S- and digoxigenin-labeled RNA and oligonucleotide probes for in situ hybridization. Expression of mRNA of the seminal vesicle secretion protein II and androgen receptor genes in the rat prostate. Histochemistry. 1992;98:217-228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 84] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 11. | Komminoth P. Digoxigenin as an alternative probe labeling for in situ hybridization. Diagn Mol Pathol. 1992;1:142-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 71] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 12. | Kubicka S, Claas C, Staab S, Kühnel F, Zender L, Trautwein C, Wagner S, Rudolph KL, Manns M. p53 mutation pattern and expression of c-erbB2 and c-met in gastric cancer: relation to histological subtypes, Helicobacter pylori infection, and prognosis. Dig Dis Sci. 2002;47:114-121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 43] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 13. | Hiyama T, Haruma K, Kitadai Y, Masuda H, Miyamoto M, Tanaka S, Yoshihara M, Shimamoto F, Chayama K. K-ras mutation in helicobacter pylori-associated chronic gastritis in patients with and without gastric cancer. Int J Cancer. 2002;97:562-566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 57] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 14. | Yokozaki H, Yasui W, Tahara E. Genetic and epigenetic changes in stomach cancer. Int Rev Cytol. 2001;204:49-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 72] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 15. | Nakajima M, Sawada H, Yamada Y, Watanabe A, Tatsumi M, Yamashita J, Matsuda M, Sakaguchi T, Hirao T, Nakano H. The prognostic significance of amplification and overexpression of c-met and c-erb B-2 in human gastric carcinomas. Cancer. 1999;85:1894-1902. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 16. | Gürel S, Dolar E, Yerci O, Samli B, Oztürk H, Nak SG, Gülten M, Memik F. The relationship between c-erbB-2 oncogene expression and clinicopathological factors in gastric cancer. J Int Med Res. 1999;27:74-78. [PubMed] |
| 17. | Gürel S, Dolar E, Yerci O, Samli B, Oztürk H, Nak SG, Gülten M, Memik F. Expression of p53 protein and prognosis in gastric carcinoma. J Int Med Res. 1999;27:85-89. [PubMed] |
| 18. | Sato K, Tamura G, Tsuchiya T, Endoh Y, Usuba O, Kimura W, Motoyama T. Frequent loss of expression without sequence mutations of the DCC gene in primary gastric cancer. Br J Cancer. 2001;85:199-203. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 44] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 19. | He XS, Su Q, Chen ZC, He XT, Long ZF, Ling H, Zhang LR. Expression, deletion [was deleton] and mutation of p16 gene in human gastric cancer. World J Gastroenterol. 2001;7:515-521. [PubMed] |
| 20. | Liu DH, Zhang XY, Fan DM, Huang YX, Zhang JS, Huang WQ, Zhang YQ, Huang QS, Ma WY, Chai YB. Expression of vascular endothelial growth factor and its role in oncogenesis of human gastric carcinoma. World J Gastroenterol. 2001;7:500-505. [PubMed] |
| 21. | Liu XP, Tsushimi K, Tsushimi M, Kawauchi S, Oga A, Furuya T, Sasaki K. Expression of p21(WAF1/CIP1) and p53 proteins in gastric carcinoma: its relationships with cell proliferation activity and prognosis. Cancer Lett. 2001;170:183-189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 22. | Fei G, Ebert MP, Mawrin C, Leodolter A, Schmidt N, Dietzmann K, Malfertheiner P. Reduced PTEN expression in gastric cancer and in the gastric mucosa of gastric cancer relatives. Eur J Gastroenterol Hepatol. 2002;14:297-303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 23. | Kang YH, Lee HS, Kim WH. Promoter methylation and silencing of PTEN in gastric carcinoma. Lab Invest. 2002;82:285-291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 182] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 24. | Ebert MP, Fei G, Kahmann S, Müller O, Yu J, Sung JJ, Malfertheiner P. Increased beta-catenin mRNA levels and mutational alterations of the APC and beta-catenin gene are present in intestinal-type gastric cancer. Carcinogenesis. 2002;23:87-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 116] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 25. | Laghi L, Ranzani GN, Bianchi P, Mori A, Heinimann K, Orbetegli O, Spaudo MR, Luinetti O, Francisconi S, Roncalli M. Frameshift mutations of human gastrin receptor gene (hGARE) in gastrointestinal cancers with microsatellite instability. Lab Invest. 2002;82:265-271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 26. | Sakata K, Tamura G, Endoh Y, Ohmura K, Ogata S, Motoyama T. Hypermethylation of the hMLH1 gene promoter in solitary and multiple gastric cancers with microsatellite instability. Br J Cancer. 2002;86:564-567. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 35] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 27. | Baba H, Korenaga D, Kakeji Y, Haraguchi M, Okamura T, Maehara Y. DNA ploidy and its clinical implications in gastric cancer. Surgery. 2002;131:S63-S70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 28. | Russo A, Bazan V, Migliavacca M, Tubiolo C, Macaluso M, Zanna I, Corsale S, Latteri F, Valerio MR, Pantuso G. DNA aneuploidy and high proliferative activity but not K-ras-2 mutations as independent predictors of clinical outcome in operable gastric carcinoma: results of a 5-year Gruppo Oncologico dell'Italia Meridonale (GDIM) prospective study. Cancer. 2001;92:294-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 29. | Nesi G, Palli D, Pernice LM, Saieva C, Paglierani M, Kroning KC, Catarzi S, Rubio CA, Amorosi A. Expression of nm23 gene in gastric cancer is associated with a poor 5-year survival. Anticancer Res. 2001;21:3643-3649. [PubMed] |
| 30. | Cai J, Ikeguchi M, Tsujitani S, Maeta M, Liu J, Kaibara N. Significant correlation between micrometastasis in the lymph nodes and reduced expression of E-cadherin in early gastric cancer. Gastric Cancer. 2001;4:66-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 57] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 31. | van Rees BP, Saukkonen K, Ristimäki A, Polkowski W, Tytgat GN, Drillenburg P, Offerhaus GJ. Cyclooxygenase-2 expression during carcinogenesis in the human stomach. J Pathol. 2002;196:171-179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 117] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 32. | Kikuchi T, Itoh F, Toyota M, Suzuki H, Yamamoto H, Fujita M, Hosokawa M, Imai K. Aberrant methylation and histone deacetylation of cyclooxygenase 2 in gastric cancer. Int J Cancer. 2002;97:272-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 57] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 33. | Krieg A, Mahotka C, Krieg T, Grabsch H, Müller W, Takeno S, Suschek CV, Heydthausen M, Gabbert HE, Gerharz CD. Expression of different survivin variants in gastric carcinomas: first clues to a role of survivin-2B in tumour progression. Br J Cancer. 2002;86:737-743. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 111] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 34. | Ebert MP, Günther T, Hoffmann J, Yu J, Miehlke S, Schulz HU, Roessner A, Korc M, Malfertheiner P. Expression of metallothionein II in intestinal metaplasia, dysplasia, and gastric cancer. Cancer Res. 2000;60:1995-2001. [PubMed] |
| 35. | Bai YQ, Yamamoto H, Akiyama Y, Tanaka H, Takizawa T, Koike M, Kenji Yagi O, Saitoh K, Takeshita K, Iwai T. Ectopic expression of homeodomain protein CDX2 in intestinal metaplasia and carcinomas of the stomach. Cancer Lett. 2002;176:47-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 161] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 36. | Li QL, Ito K, Sakakura C, Fukamachi H, Inoue Ki, Chi XZ, Lee KY, Nomura S, Lee CW, Han SB. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002;109:113-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 789] [Cited by in RCA: 835] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 37. | Saitoh T, Mine T, Katoh M. Up-regulation of WNT8B mRNA in human gastric cancer. Int J Oncol. 2002;20:343-348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 38. | Li Zy, Wang Yj, Song Jp, Kataoka H, Yoshii S, Gao Cm, Wang Yp, Zhou Jn, Ota S, Tanaka M. Genomic structure of the human beta-PIX gene and its alteration in gastric cancer. Cancer Lett. 2002;177:203-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 39. | Shinohara H, Morita S, Kawai M, Miyamoto A, Sonoda T, Pastan I, Tanigawa N. Expression of HER2 in human gastric cancer cells directly correlates with antitumor activity of a recombinant disulfide-stabilized anti-HER2 immunotoxin. J Surg Res. 2002;102:169-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 40. | Takehana T, Kunitomo K, Kono K, Kitahara F, Iizuka H, Matsumoto Y, Fujino MA, Ooi A. Status of c-erbB-2 in gastric adenocarcinoma: a comparative study of immunohistochemistry, fluorescence in situ hybridization and enzyme-linked immuno-sorbent assay. Int J Cancer. 2002;98:833-837. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 154] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 41. | Kodera Y, Nakanishi H, Ito S, Yamamura Y, Kanemitsu Y, Shimizu Y, Hirai T, Yasui K, Kato T, Tatematsu M. Quantitative detection of disseminated free cancer cells in peritoneal washes with real-time reverse transcriptase-polymerase chain reaction: a sensitive predictor of outcome for patients with gastric carcinoma. Ann Surg. 2002;235:499-506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 103] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 42. | You H, Xiao B, Cui DX, Shi YQ, Fan DM. Two novel gastric cancer-associated genes identified by differential display. World J Gastroenterol. 1998;4:334-336. [PubMed] |
| 43. | Tórtola S, Marcuello E, Risques RA, González S, Aiza G, Capellà G, Peinado MA. Overall deregulation in gene expression as a novel indicator of tumor aggressiveness in colorectal cancer. Oncogene. 1999;18:4383-4387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 44. | Yoshikawa Y, Mukai H, Hino F, Asada K, Kato I. Isolation of two novel genes, down-regulated in gastric cancer. Jpn J Cancer Res. 2000;91:459-463. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 93] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 45. | Wang X, Lan M, Shi YQ, Lu J, Zhong YX, Wu HP, Zai HH, Ding J, Wu KC, Pan BR. Differential display of vincristine-resistance-related genes in gastric cancer SGC7901 cell. World J Gastroenterol. 2002;8:54-59. [PubMed] |
| 46. | Shiozaki K, Nakamori S, Tsujie M, Okami J, Yamamoto H, Nagano H, Dono K, Umeshita K, Sakon M, Furukawa H. Human stomach-specific gene, CA11, is down-regulated in gastric cancer. Int J Oncol. 2001;19:701-707. [PubMed] |
| 47. | Wang G, Wang M, You W, Li H. [Cloning and primary expression analyses of down-regulated cDNA fragment in human gastric cancer]. Zhonghua Yixue Yichuanxue Zazhi. 2001;18:43-47. [PubMed] |
| 48. | Fend F, Kremer M, Quintanilla-Martinez L. Laser capture microdissection: methodical aspects and applications with emphasis on immuno-laser capture microdissection. Pathobiology. 2000;68:209-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 32] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 49. | Maitra A, Wistuba II, Gazdar AF. Microdissection and the study of cancer pathways. Curr Mol Med. 2001;1:153-162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 50. | Rekhter MD, Chen J. Molecular analysis of complex tissues is facilitated by laser capture microdissection: critical role of upstream tissue processing. Cell Biochem Biophys. 2001;35:103-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 31] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 51. | Cör A, Vogt N, Malfoy B. Microdissection techniques for cancer analysis. Folia Biol (Praha). 2002;48:3-8. [PubMed] |
| 52. | Noda H, Maehara Y, Irie K, Kakeji Y, Yonemura T, Sugimachi K. Increased proliferative activity caused by loss of p21(WAF1/CIP1) expression and its clinical significance in patients with early-stage gastric carcinoma. Cancer. 2002;94:2107-2112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 16] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 53. | Saadat I, Saadat M. Glutathione S-transferase M1 and T1 null genotypes and the risk of gastric and colorectal cancers. Cancer Lett. 2001;169:21-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 88] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 54. | Kuniyasu H, Oue N, Wakikawa A, Shigeishi H, Matsutani N, Kuraoka K, Ito R, Yokozaki H, Yasui W. Expression of receptors for advanced glycation end-products (RAGE) is closely associated with the invasive and metastatic activity of gastric cancer. J Pathol. 2002;196:163-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 250] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 55. | Mori M, Mimori K, Yoshikawa Y, Shibuta K, Utsunomiya T, Sadanaga N, Tanaka F, Matsuyama A, Inoue H, Sugimachi K. Analysis of the gene-expression profile regarding the progression of human gastric carcinoma. Surgery. 2002;131:S39-S47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 60] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 56. | Endo K, Maejara U, Baba H, Tokunaga E, Koga T, Ikeda Y, Toh Y, Kohnoe S, Okamura T, Nakajima M. Heparanase gene expression and metastatic potential in human gastric cancer. Anticancer Res. 2001;21:3365-3369. [PubMed] |
| 57. | Wang CS, Lin KH, Hsu YC. Alterations of thyroid hormone receptor alpha gene: frequency and association with Nm23 protein expression and metastasis in gastric cancer. Cancer Lett. 2002;175:121-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 39] [Article Influence: 1.7] [Reference Citation Analysis (0)] |