INTRODUCTION
The gut-enriched Krüppel-like factor (GKLF), also named as epithelial zinc finger (EZF) or KLF4, is a recently identified transcription factor with three C2H2 zinc fingers in the carboxyl terminus[1-3]. The expression of GKLF is enriched in the epithelial cells of the gastrointestinal tract and skin[1,2]. Although the physiological role of GKLF is not clear, experimental evidences[1] suggest that GKLF is potentially a negative regulator of cell proliferation and involves in the differentiation process of epithelial cells. In the gene targeting experiment[4], GKLF knockout mice showed the impairment of skin barrier function, indicating that GKLF is both vital to and selective for the barrier acquisition.
GKLF expression is decreased in the intestinal adenomas of mice and colon adenomas of patients[5,6]. Experimental data[7] also suggest that down-regulation of GKLF may play a role in the initiation stage of tumorigenesis of colon. In oral dysplastic epithelium and squamous cell carcinoma, GKLF expression was increased and diffusely distributed[8,9]. In breast cancer, GKLF expression was increased and associated with the stages of tumor progression in the breast[9]. These observations indicated that GKLF expression is regulated during neoplastic progression in a tissue and tumor type-specific fashion.
GKLF expression was enriched in esophageal epithelia[2]. In esophageal cancer cell lines, abundant GKLF expression was also observed[10,11]. The regulating effect of GKLF on the squamous epithelium-specific keratin 4 and Epstein-Barr virus ED-L2 promoters[10] suggests GKLF plays an important role in the differentiation of esophageal squamous epithelium. The human GKLF gene was mapped onto chromosome 9q31[3], which is a hot site of chromosome abnormality in esophageal cancer[12,13]. GKLF gene aberration was observed in colon cancer in which the function of GKLF was demolished[14]. These evidences led us to the research on the expression of GKLF in esophageal cancer, and consequent effect on the cancer cells. To our knowledge, this is the first report on the down-regulation of GKLF in esophageal cancer, and the first evidence of association of GKLF function with cell adhesion ability.
MATERIALS AND METHODS
Tissue samples and cell culture
Fresh surgical specimens of 17 pairs of esophageal squamous cell carcinoma and corresponding normal tissues were immediately frozen and stored in liquid nitrogen after resection. Squamous esophageal cancer cell line EC9706 was kindly provided by Dr. Mingrong Wang in the lab. The cell was grown in M199 (Gibco BRL, Gaithersburg, MD) supplemented with 15% FCS. Primary culture of fibroblast was derived from resected normal esophageal tissue of an esophageal cancer patient, and maintained in McCoy 5A (Gibco BRL, Gaithersburg, MD) supplemented with 10% FBS. Cells were incubated at 37 °C with 5% CO2. For the serum deprivation experiments, cells were seeded into 25 mL flasks with 4 ml full medium and cultured until 50% confluency. The content of FCS in the cell media was reduced to 0.5%. Cells were then incubated continuously at 37 °C, in 5% CO2 for various periods.
RNA preparation and RT-PCR
Total RNA was extracted by TRIzol method (Gibco BRL, Gaithersburg, MD). Up to 5 μg of total RNA was reverse transcripted in 20 μL volume of reaction with both oligo (dT) and random primers, using SuperScript Preamplification System (Gibco BRL, Gaithersburg, MD) according to the instruction.
PCR was carried out in 30 μL total reaction mixture which contains 10 mmol/L Tris-HCL, pH 8.3, 50 mmol/L KCL, 1.75 mmol/L MgCl2, 0.1% Triton X-100, 200 μmol/L each of dGTP, dATP, dTTP, and dCTP, and 200 nmol/L each of the forward and reverse primers; 0.5-2 μL of cDNA template after RNase-free DNase I digestion was added. The primers used in the PCR reaction were synthesized according to the published cDNA sequences encoding human GKLF and GAPDH (GeneBank Accession numbers AF105036 and NM_002046, respectively). For the measurement of GKLF mRNA expression in tissue and cell samples, forward primer 5'-GTCGGACCACCTCGCCTTACACAT-3' and reverse primer 5'-GGTCTTCCCTCCCCCAACTCACG-3' of GKLF were used which amplify the region containing 3' noncoding sequence of GKLF cDNA (+1374 to +1749) and the expected PCR product was 376 bp; For the measurement of GKLF mRNA expression in cells after transfection of GKLF cDNA, forward primer 5'-CCACCGGCCGGCTGCACACGACT-3' and reverse primer 5'-TCATCTGAGCGGGCGAATTTCCATCCACA-3' of GKLF were used which amplify the coding sequence of GKLF cDNA (+864 to +1 298) and the expected PCR product was 435 bp. GAPDH primers (forward primer 5'-GGCAAATTCCATGGCACCGTCAAG-3' and reverse primer 5'-GCAATGCCAGCCCCAGCGTCAAA-3') were added in the same PCR reaction, and the expected PCR product of 746 bp was used as internal control. PCR was set up for 27 cycles: 30 s at 94 °C, 30 s at 56 °C 45 s at 72 °C.
GKLF cDNA cloning and sequencing
The full length GKLF cDNA coding sequence was amplified from reverse transcripted cDNA of EC9706. The forward primer 5'-CTGCTTCGGGCTGCCGAGGACCTTCTGGG-3' and reverse primer 5'-GGCAGTGTGGGTCATATCCACTGTCTGGGA-3' of GKLF were used and the expected PCR product was 1511 bp (-67 to + 1444 nucleotides). The cycling condition was 30 s at 94 °C, 30 s at 63 °C 1 min at 72 °C for 30 cycles. The PCR product was electrophoresed onto 0.9% agarose and 1.5 kb fragment was purified with the purification system (Qiagen, Hilden, Germany). The fragment was then cloned into PCR cloning vector pMD18T (TaKaRa, Dalian, China) and sequenced commercially (TaKaRa, Dalian, China).
Transfection
DNA fragment containing GKLF cDNA (-67 to + 1444 nucleotides) was cut out from pMD18T vector by BamHI and HindIII digestion and subcloned into pCDNA3.1 expression vector (Invitrogen, San Diego, CA) in antisense orientation. EC9706 cells were transfected with antisense GKLF cDNA as well as pCDNA3.1 vector using the Lipofectamine method according to the manufacture's protocol (Gibco BRL, Gaithersburg, MD). Briefly, cells were incubated with 1 mL of the transfection mixture containing 4.0 μg plasmid DNA and 25 μL Lipofectamine reagents in serum-free medium for 6 h, followed by adding 1 mL fresh media supplemented with 30% FCS. The medium was exchanged with fresh complete media after overnight incubation.
RNA content determination
For semi-quantification of GKLF mRNA expression, the PCR products were electrophoresed onto 1.2% agarose gel containing ethidium bromide. The images were analyzed with Multi-Analyst software (Bio-Rad, Hercules, CA) after scanned and the densities of GKLF and GAPDH bands were determined. The ratios of GKLF to GAPDH were compared in each paired sample.
Cell proliferation assay
Cells were inoculated into 96 well plates (2000/well) 36 h after transfection and incubated for further 24 h at 37 °C in CO2 incubator. CellTiter 96 AQuous One Reagent (Promega, Madison, WI) was added into each well and OD490 was measured according to manufacture’s method. The cells were washed three times with PBS and incubated in complete media for another 24 h. The assay was repeated and the ratio of OD490 to that of the first assay was calculated as the relative cell proliferate rate.
Cell adhesion assay
Cells were detached from culture flasks by trypsinisation 48 h after transfection and inoculated into 96 well plates (5000/well). After 2 h incubation, unattached cells were then removed by washing 3 times with PBS. CellTiter 96 AQuous One Reagent was added into each well and OD490 was measured. The percentage adhesion was determined by comparing the OD490 of attached cells to that of corresponding cells without washing.
Statistics
Results are expressed as mean ± S.E.M. The statistical significance was determined with paired or grouped t-test.
RESULTS
GKLF expression in esophageal cancer
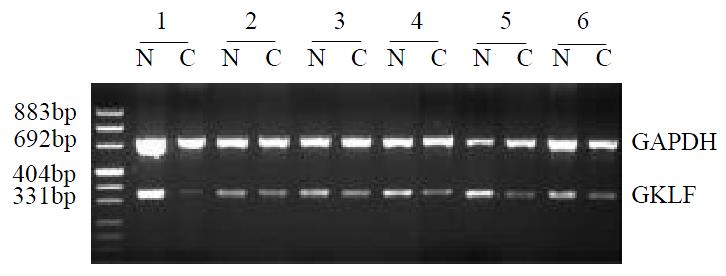
The GKLF mRNA expression was determined semi-quantitively using RT-PCR method. In 17 pairs of tissue specimens, GKLF expression was detected in all samples as an expected PCR product of 376 bp in length, and 14 of them showed decreased expression in cancer sample compared to corresponding normal tissue. Paired t-test has a significant difference (P < 0.05) between cancer sample and normal tissue. Representative cases of GKLF expression detected by RT-PCR are shown in Figure 1.
Figure 1 Semi-quantitative RT-PCR of GKLF in esophageal squamous cancer patients.
RNA from specimens of normal appearing mucosa (N) and cancer (C) were extracted and GKLF as well as GAPDH were amplified. This figure showed the rep-resentative results from several individual patients.
GKLF cDNA coding sequence
GKLF gene resides at 9q31 of human genome[3], which is a frequent site of LOH in esophageal cancer[15,16]. Gene mutation of GKLF coding sequence was also reported in a colon cancer cell line[14]. In this Laboratory, the genomic alterations were observed in squamous esophageal cancer cell line EC9706 which includes 9q region by comparative genomic hybridization (unpublished data). To find out if GKLF cDNA was mutated in esophageal cancer cell line EC9706, the full length cDNA of coding region of GKLF was amplified by RT-PCR from cDNA of EC9706. An 1.5 kb length of PCR product was detected as expected. The single band was cloned into pMD18T vector and sequenced. The sequence conformed to cDNA of GKLF (GeneBank Accession number AF105036) completely without any mutation.
Serum deprivation induction of GKLF
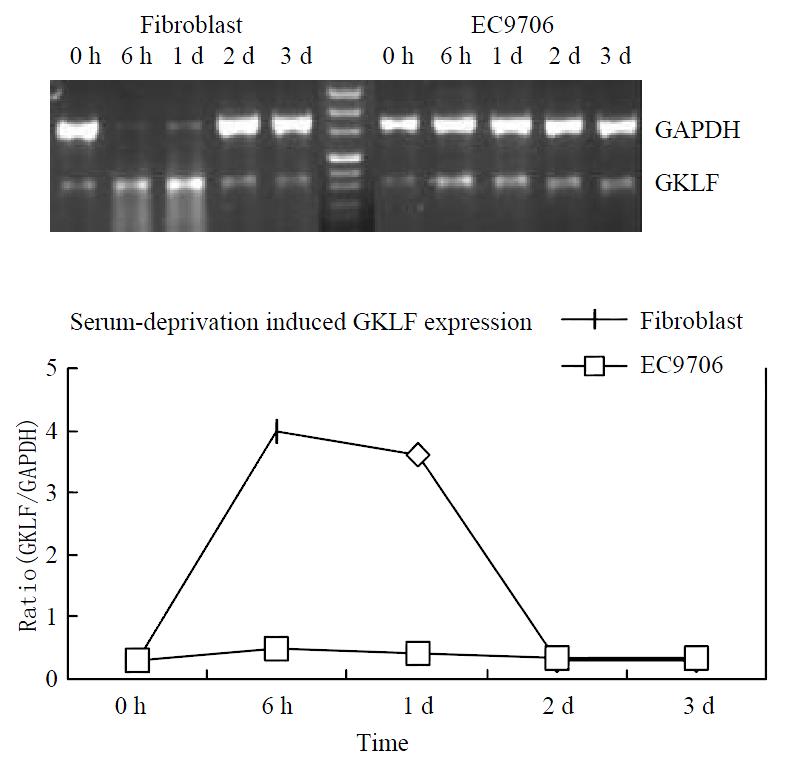
GKLF expression was induced in murine fibroblast by serum deprivation[1]. As GKLF expression was down-regulated in esophageal squamous cell carcinoma, we were interested in observing the inducibility of GKLF expression in esophageal cancer by serum deprivation. The cell line EC9706 was serum deprived and GKLF mRNA expression was detected by RT-PCR at 0 h, 6 h, 1 d, 2 d and 3 d. A primary culture of human fibroblast was also serum deprived as a positive control. Figure 2 showed the inducibility of GKLF by serum deprivation in both EC9706 and fibroblast. Although GKLF was induced in EC9706, the level was greatly decreased compared to that in human fibroblast.
Figure 2 Serum deprivation induced GKLF expression.
Both human primary cultured fibroblast and an esophageal squa-mous cancer cell line EC9706 was underwent serum deprivation. GKLF expression was measured semi-quantitively by RT-PCR. The magnitude of GKLF expression was calcu-lated as ratio of GKLF to GAPDH.
Cell growth and adhesion properties in esophageal cancer cells with low GKLF expression
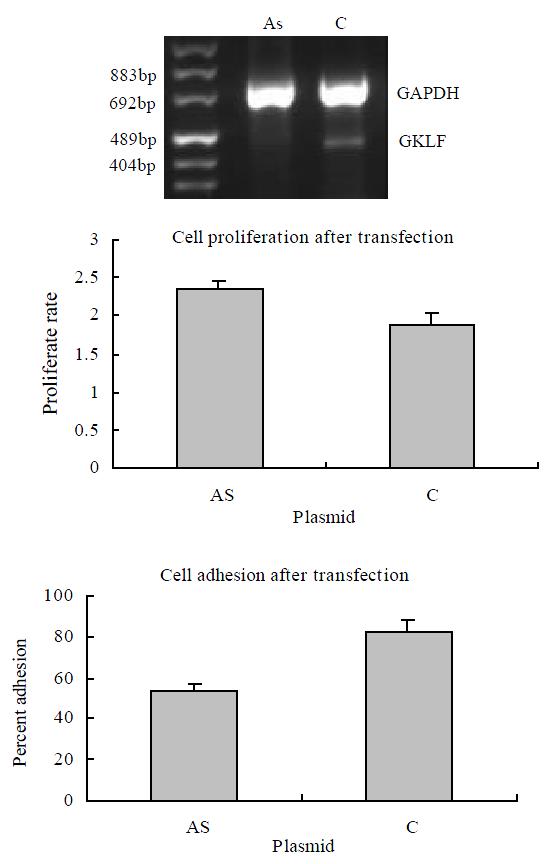
It was reported that forced overexpression of GKLF inhibited DNA synthesis[1] and decreased cell proliferation[17]. We transiently transfected the esophageal cancer cell line EC9706 with antisense GKLF expression vector, which caused low expression of GKLF in the transfected cells (shown in Figure 3). The cell proliferative rate was assayed as described above. Compared to the controlled cells, the low GKLF expression cells grew at a greater rate with a significant difference (P < 0.05). As we noticed that cell adhesion ability changed in individual stable transfectants of EC9706 with GKLF expression vectors (data not shown), cell adhesion assay was also carried out in these transiently transfected cells with low GKLF expression. In these cells, the adhesion ability was decreased significantly compared to the controlled cells (P < 0.05) (Figure 3).
Figure 3 Cell growth and adhesion in GKLF transfected EC9706 cells.
The cells were transiently transfected with antisense GKLF expression plasmid (AS) and pCDNA3.1 as control (C). The GKLF expression levels detected by RT-PCR were shown in the upper panel. The cell growth rates were shown in the middle panel; each data point represents mean ± S.E.M. of 7 repeats; there was a significant difference between AS and C (P < 0.05). The cell adhesions were shown in the lower panel; each data point represents mean ± S.E.M. of 4 repeats; AS had significant difference compared to C (P < 0.05).
DISCUSSION
Alterations of GKLF expression in cancer cells are tissue and tumor type-specific. Our current findings showed that GKLF was down-regulated in esophageal squamous cell carcinoma. It is known that down-regulation of GKLF occurred in intestinal adenoma and colonic adenoma[6] while up-regulation of GKLF occurred in breast carcinomas and oral squamous cell carcinoma[9]. The mechanism of altered GKLF expression and its difference among different tumor type is not clear yet. The results from colon cancer cells[6] showed that no involvement of DNA methylation in the down-regulation of GKLF, while mutated CDX2 expression in some colon cancer cell lines may help to explain the low levels of GKLF expression[18].
Serum deprivation induces growth arrest and apoptosis in normal cells while the inducibility of serum deprivation is usually decreased in cancer cells. Experimental evidence showed that p21[19] and cyclin D1[20] involved in the serum deprivation induces growth arrest and apoptosis. In some kind of cancer cells, serum deprivation did not induce the expression of p21[21]. As a transcription factor, GKLF involves in the p53-p21 regulating pathway[22]. It also regulates the cell cycle factors such as cyclin D1[23]. Decreased inducibility of GKLF by serum deprivation in EC9706 cells may suggest a role of GKLF in decreased response of cancer cells to serum deprivation.
GKLF overexpression inhibits proliferation of NIH3T3 and of HT-29 colon cancer cells[1,7]. Similar to the report[7], we found that antisense expression of GKLF promotes cellular proliferation of EC9706 cells. Combined with the finding that GKLF was down-regulated in esophageal cancer tissues, the result suggested that GKLF might play a role in the initiation and/or progression of esophageal cancer. It was shown that GKLF exerted its effect on cell proliferation at G1 phase of cell cycle[7,24]. That GKLF mediates the p53-p21 regulating pathway[22] could be a mechanism of its cell cycle effect as this pathway is important for the regulation of G1/S transition[25]. Cyclin D1-RB pathway is another possible effecting pathway of GKLF for its cell cycle regulation, since GKLF directly down-regulates cyclin D1 expression[23] while cyclin D1 facilitates progression through the G1 phase which plays a role in the initiation of esophageal cancer[26]. Overexpression of cyclin D1 was observed in the early phase of esophageal tumors in zinc-deficient rat model[27], suggesting an important role of zinc proteins in regulating cyclin D1 expression, and conforming to our proposition that down-regulation of GKLF plays a role in the initiation and/or progression of esophageal cancer.
The basic mechanism of tumour cell metastasis is reduced expression of adhesion molecules which results in an increased migratory ability of the cancer cells[28]. We showed here that alteration of GKLF expression in EC9706 cell decreased its adhesion, implicating that GKLF involves in the metastasis of esophageal cancer. Abnormal expression of such cell adhesion molecules as cadherin, integrin and mucin has definite relationship with cancer metastasis[29-31]. It is known that GKLF could regulate MUC5B gene transcription directly[32]. GKLF also regulates laminin expression[33,34] while laminin is a component of extracellular matrix which plays a key role in cell adhesion and migration. More frequent allelic loss at 9q region where the GKLF gene located was reported in esophageal squamous cell carcinoma patients with metastasis[15], suggesting a clinically significant role of GKLF gene in esophageal cancer.
GKLF belongs to a family of Krüppel-like factors that is the most abundant transcription factors in human cells[35]. It targets to the specific DNA sequence[36] which represents one of the most abundant classes of conserved motifs of intergenic regions over the human genome[37], besides to the common DNA sequences through which this family of factors binds and acts[36,38]. More and more target genes are being discovered[39,40], that will expedite our understandings of GKLF mediated action in both normal and malignant cells.
In summary, the results of this study showed that GKLF is down-regulated in esophageal squamous cell cacinoma, and GKLF deregulation in esophageal cancer cell line EC9706 sho wes a manner of decrease inducibility to serum deprivation. Furthermore, down-regulation of GKLF in EC9706 cells results in the proliferation and decreased adhesion of cancer cells, suggesting that down-regulation of GKLF may contribute to malignant phenotype of esophageal cancer.