Published online Dec 15, 2002. doi: 10.3748/wjg.v8.i6.1094
Revised: August 3, 2002
Accepted: August 9, 2002
Published online: December 15, 2002
AIM: p73, as a novel member of a family of p53-related transcription factors, shares redundant functions with p53, such as the abilities of inducing apoptosis and suppressing growth. It is well known that p53 can repress HBV expression and transcription efficiently. The aim of this paper is to investigate the transcriptional effect of p73α and p73β on hepatitis B virus (HBV) and to understand the correlation between HBV and p73.
METHODS: To construct an x-gene inactivated HBV plasmid which was cotransfected with p73α or p73β expression vectors into HepG2 cells. After transiently transfection, HBV surface antigen (HBsAg) and HBV e antigen (HBeAg) were detected by ELISA. Viral transcripts synthesized by HBV were evaluated by Northern blotting analysis. The activities of HBV regulatory elements, including enhancer I/X promoter (ENI/Xp) and enhancer II/core promoter (ENII/Cp) were monitored by luciferase assays.
RESULTS: Both p73α and p73β could repress HBsAg and HBeAg expression by downregulating the ENI/Xp and ENII/ Cp activities. But p73β exerted stronger inhibition on the activity of ENI/Xp than p73α, resulting in much lower level of viral transcripts and the antigens expression.
CONCLUSION: p73β as a novel member of p53 family can efficiently inhibit HBV transcription mainly through downregulating the activities of the HBV ENI/Xp regulatory elements.
- Citation: Xu ZH, Zhao MJ, Li TP. p73β inhibits transcriptional activities of enhancer I and X promoter in hepatitis B virus more efficiently than p73α. World J Gastroenterol 2002; 8(6): 1094-1097
- URL: https://www.wjgnet.com/1007-9327/full/v8/i6/1094.htm
- DOI: https://dx.doi.org/10.3748/wjg.v8.i6.1094
p73 gene maps to chromosome 1p36.1, a region which was frequently deleted in several tumors, including neuroblastoma, colorectal cancer and breast cancer[1]. Moreover, it has been found to share significant homology with the tumor suppressor gene p53 within the transactivation domain, DNA binding domain and oligomerization domain. Both p53 and p73 have redundant functions in the regulation of gene expression, because they have amino acid sequence identity reaching to 63% in the DNA binding domain[2]. p73 can activate p53-regulated genes and suppress growth or induce apoptosis, and expression of p73 can be induced by DNA damage as p53 does[3,4]. Although deletion of p73 gene is observed in neuroblastoma and a subtype of T-cell lymphoma, it is rarely mutated in human cancer[5-9], unlike p53 which is mutated in about 50% of human cancers[5,7,10-12]. Other evidence suggests that p73 is important for regulation of normal development[13]. p73 gene is expressed as p73α, a 636 amino acid polypeptide, and p73β, a 499 amino acid polypeptide that is encoded by an alternatively spliced transcript lacking 96 nucleotides corresponding to exon 13[1]. Until now, at least six different p73 proteins (α-ζ) have been found[14].
Hepatitis B virus[15-17] is a causative agent of chronic hepatitis and hepatocellular carcinoma. Upon infection or DNA transfection, four major viral transcripts are detected. The largest 3.5 kb mRNA is composed of precore and pregenomic mRNA, that direct the synthesis of HBV e antigen (HBeAg) and HBV core antigen (HBcAg) respectively. Pregenomic RNA also serves as a template for reverse-transcription to synthesize the viral DNA genome. The largest surface antigen is synthesized from 2.4 kb mRNA, and the middle and major surface antigen are synthesized from 2.1 kb mRNA. The smallest transcript is a 0.7 kb mRNA, which is responsible for HBx protein production[18]. The transcription of these RNAs are governed by the core, S1, S2 and X promoters, respectively. The activities of these promoters are under the control of enhancer I and II[19].
In addition to a function as a tumor suppressor, p53 can defend host cell from the invading virus. p53 actively inhibits viral replication as in the case of SV40 and HBV[20,21]. p53 binds to a sequence adjacent to the replication origin of SV40 and abrogates the helicase activity of T antigen by directly binding to it. It has been reported that p53 can bind specifically to the HBV enhancer I and repress the activity of enhancer I and X promoter, resulting in HBV gene expression downregulated[22]. In addition, p53 can interfere with the life cycle of HBV through down-regulation of the enhancer II and pregenomic/core promoter. Although p53 can not directly bind the enhancer II, it represses the transcriptional activity of HBV through protein-protein interaction[23].
p73, as a novel member of a family of p53-related transcription factors, has attracted more attention during these years. However the relationship between p73 and the hepatitis B virus is not elucidated. Based on the similarity between p53 and p73, we examined whether p73, mainly p73α and p73β, can affect HBV antigens expression and the viral transcription.
p3.8II, kindly provided by Prof. Wang Yuan, is an HBV plasmid which contains terminally redundant HBV genome and can replicate in liver cell. The x-gene inactivated p3.8IIXm was constructed by changing the start condon of the X open reading frame on p3.8II. The pENI/XpLuc and pENII/CpLuc reporter plasmids were constructed by inserting nt1067-1403 and nt1430-1879 which contain enhancer I/X promoter and enhancer II/core promoter respectively in front of the pGL3 basic vector (Promega). The mammalian expression vectors pcDNA3-HA-73α and pcDNA3-HA-73β encode epitope tagged p73 proteins which were kind gifts from Dr. Lu Hua. p53 expression plasmid pRC/CMV hp53 was provided by Dr. Judith Roth. All the constructs were confirmed by restriction enzyme analysis and DNA sequencing.
HepG2 cells were cultured in DMEM supplemented with kanamycin (250 IU/mL), gentamycin (40 IU/mL) and 10% fetal calf serum in 5% CO2 at 37 °C. Transfection was carried out by the calcium phosphate method[24]. Each transfection reaction contained a constant amount of 10 μg DNA per 6 cm dish.
Five days after transfection, cells were washed twice with PBS and were detached from dishes with 10 mM EDTA in PBS. After centrifugation (5000 rpm, 1 min), cells were resuspended in 100 μL 250 mM Tris-HCl (pH 7.5) and lysed by three thawing and freezing cycles. After centrifugation, the supernatant was transferred and stored at 4 °C. The protein concentration was estimated by Bradford method (Biocolor). Culture medium was collected every day after transfection. HBsAg and HBeAg were measured with ELISA kits (Sino-America). All procedures were performed according to the descriptions of the manufacturers.
Total RNA was extracted from transfected cells using TRIzol reagent (Gibco BRL). The RNA samples were treated with RNase-free DNaseI (Pharmacia). For Northern blotting analysis, 20 μg of total cellular RNA per sample was separated on 1% formaldehyde-agarose gel and blotted to a Hybond-N nylon membrane (Amersham) . The membrane was prehybridized in Quick-Hyb buffer (Amersham) for at least 1 h, followed by 2 h hybridization at 65 °C with the probes of α-32p-dCTP labeled 3.2 kb HBV DNA and G3PDH fragments. After hybridization, the membrane was washed in 2% SSC, 0.1% SDS buffer (RT, 20 min), 0.2% SSC, 0.1% SDS (65 °C, 30 min), then exposed to an X-ray film at -70 °C.
Cells were lysed and analyzed with a luciferase assay system (Promega). Corrections of the luciferase activity were made based on the protein concentrations of the lysates. The luciferase activity was measured with a Lumat LB9507 luminometer (Berthold) in 10 μL of the lysate after addition of 100 μL assay reagent.
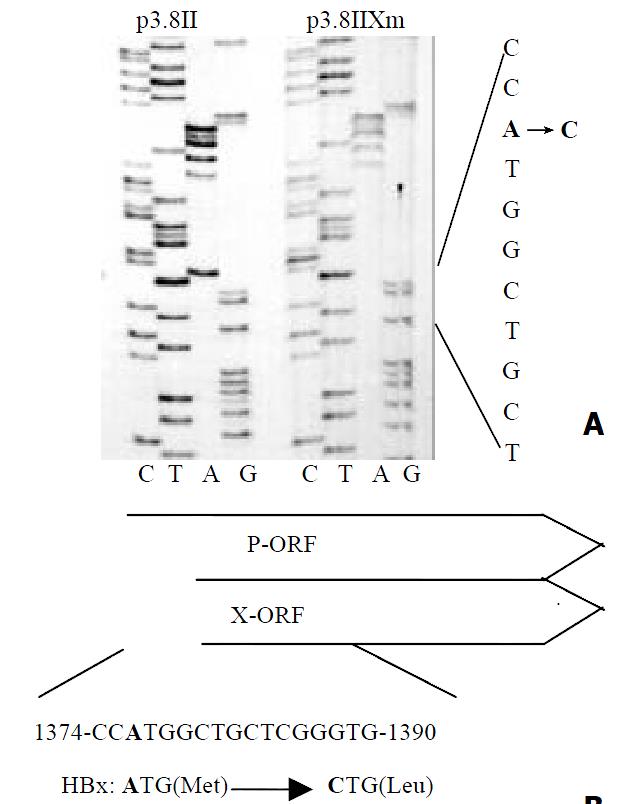
Before investigating whether p73 could affect HBV transcription like p53, we established the X protein-minus HBV mutant p3.8IIXm to avoid the interaction between p73 and the HBV X protein. This HBV mutant had no ATG start codon of the X-ORF (Figure 1). ELISA results revealed that the HBV antigens expression in mutant type of p3.8IIXm was just a half of that in wild type p3.8II.
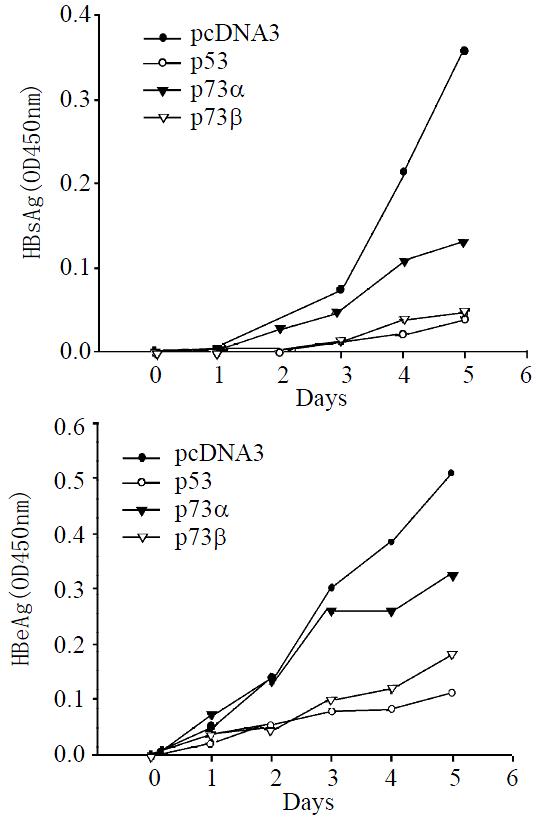
As shown in Figure 2A, P3.8IIXm could secret HBsAg into the medium continuously starting from day 2. Cotransfection of p3.8IIXm with p73β expression plasmid resulted in a reduced level of HBsAg (87% reduction on day 5 post transfection), which was similar to p53 (89% reduction on day 5 post transfection). Cotransfection of p3.8IIXm with p73α expression plasmid exhibited weak repression on HBsAg synthesis (only 63% reduction on day 5 post transfection). The time-dependent alteration in the level of HBeAg (Figure 2B) displayed a similar pattern to that of HBsAg. On day 5 post transfection, the expression of HBeAg was changed at the level of 36% reduction for p73α and 64% reduction for p73β. Also as shown in Figure 2B, p53 could inhibit HBeAg more efficiently (78% reduction on day 5 post transfection) than p73α and p73β. It is concluded that both p73α and p73β can downregulate HBV expression including HBsAg and HBeAg, but p73β can repress the HBV antigens expression more efficiently than p73α.
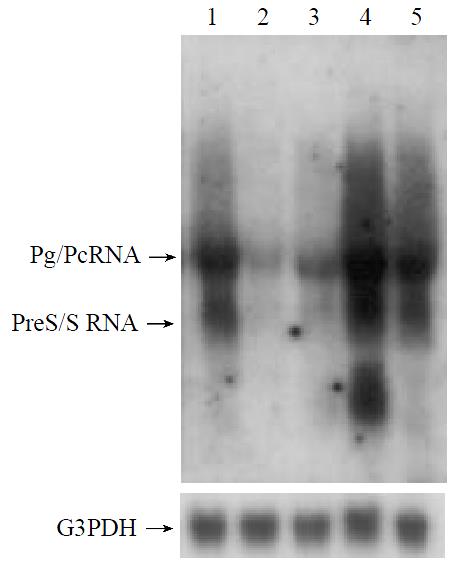
The roles of p73α and p73β in viral transcription were assessed at the HBV RNA levels after cotransfection of p3.8IIXm and p73 expression plasmids. Using Northern blot hybridization with 32P-labeled 3.2 kb HBV fragment as a probe, we detected the pregenomic RNA and precore RNA as well as preS/S mRNA. As Figure 3 shown, cotransfection of p73β resulted in the reduction of the viral transcript level like p53, but p73α seems to exhibit very weak repression on HBV transcription. These results are in accordance with the above ELISA data.
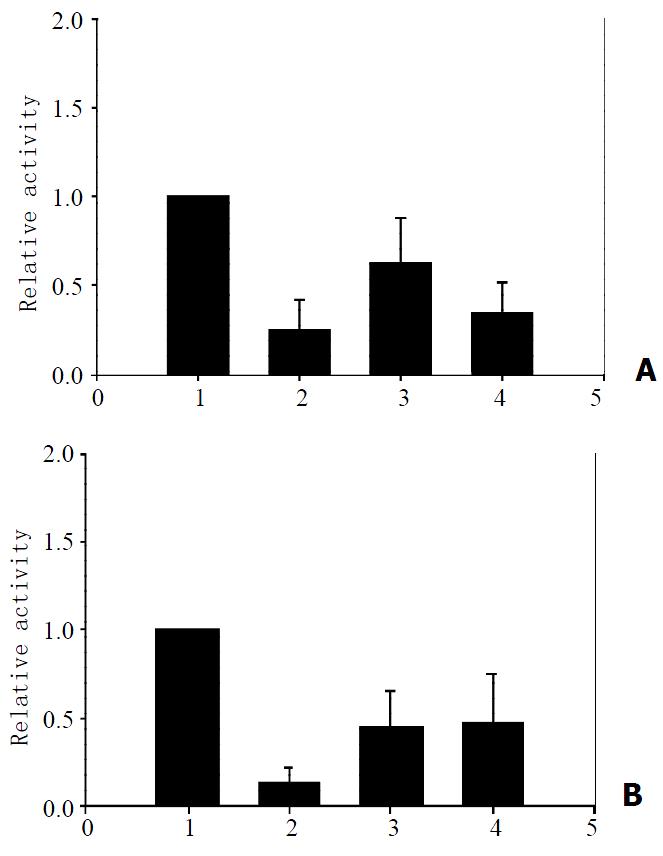
To explain the molecular mechanism of the p73β mediated repression of HBV transcription, we investigated the possibility of p73 regulating the enhancer and promoter activity. pENI/ Xp reporter plasmid containing HBV enhancer I and X promoter was cotransfected into HepG2 cells with p73α and p73β expression plasmids, and the levels of luciferase activity were determined. As shown in Figure 4A, the luciferase activity of the pENI/Xp was decreased with transfection of p73β and p73α, down to 65% and 35% of the control value respectively. Therefore, p73β can repress the activity of enhancer I and X promoter more efficiently than p73α. The activity of HBV enhancer II and core promoter were repressed by p73α and p73β at a similar level, but the effects of inhibition were weaker than that by p53 (Figure 4B). Therefore, it is concluded that p73β represses HBV gene expression mainly through the enhancer I and X promoter.
HBV, an important risk factor of hepatitis and hepatocellular carcinoma, employs a reverse transcription step which is controlled by cellular transcription factors and some cellular signal transduction pathways, including the tumor suppressor gene p53[25,26]. Physiologically activated p53 can repress HBV transcription, and this repression can be abrogated by physiological levels of HBx protein. It is generally believed that p53 can interact with HBx, and the latter destroys p53 function, including the repression of virus transcription. To exclude the interaction between p73 and the X protein, we constructed the X-minus HBV strain p3.8IIXm which could replicate in hepatoma cells without producing X protein.
HBsAg and HBeAg are translated from preS/S mRNA and precore mRNA individually. HBV core promoter is in charge of the transcription of pregenomic RNA as well as precore mRNA. ENI and ENII are the two enhancers in the viral genome. ENI is usually able to upregulate all the HBV promoters, and ENII has a particularly significant stimulatory effect upon the core promoter. In this report, we found that p73β repressed HBsAg and HBeAg expression more efficiently, and it inhibited the viral transcripts level mainly through downregulating the enhancer I and X promoter. In the ENI/Xp construct, there were two sites with homology to the 10 bp half-consence (RRRCWWGYYY) for p53 binding, which had been reported to bind specifically to the HBV enhancer I and repress its activity[22]. Liver-specific enhancer II had also been found to be the target for the p53-mediated inhibition of hepatitis B viral gene expression. p53 could not directly bind to the enhancer II, and it repressed the ENII activity through protein-protein interaction[23]. p73 protein has significant homology with p53, and p73 can interact with the consensus p53-responsive sequences. But that whether the mechanism of repression on enhancer of HBV by p73 and p53 is the same awaits further investigation. In our results, p73α exerted a very weak repression on HBV transcription and antigen expression, while Doitsh et al[27] reported that p73β did not repress but activated HBV transcription, probably due to the high dose of the protein we used in this study.
It was noted that p73 was less active than p53, whereas p73β displayed stronger transcriptional activity than p73α. The difference between p73α and p73β in the potential to regulate transcription may be due to the ability of these proteins to form oligomers. p73β is the truncated form of full-length p73α. While p73β is known to oligomerize with itself efficiently, p73α forms oligomers with a poor efficiency, at least when assayed in the yeast two-hybrid system[2]. Therefore it can be used to explain why p73β exerts more efficient inhibition on HBV transcription than p73α.
Although p73β has been found to interfere with the HBV transcription in this report, whether HBV can inactivate the function of p73 to affect the viral-infected cell fate, and the relationship between the X protein and p73β remain to be investigated.
The authors would like to thank Prof. Wang Yuan for providing p3.8II plasmid, Dr. Lu Hua for pcDNA3-HA-p73α, pcDNA3-HA-p73β, and Dr. Vogelstein for the pRC/CMV-p53.
Edited by Zhang JZ
| 1. | Ikawa S, Nakagawara A, Ikawa Y. p53 family genes: structural comparison, expression and mutation. Cell Death Differ. 1999;6:1154-1161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 112] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 2. | Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A, Minty A, Chalon P, Lelias JM, Dumont X. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809-819. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1224] [Cited by in RCA: 1247] [Article Influence: 44.5] [Reference Citation Analysis (0)] |
| 3. | Steegenga WT, Shvarts A, Riteco N, Bos JL, Jochemsen AG. Distinct regulation of p53 and p73 activity by adenovirus E1A, E1B, and E4orf6 proteins. Mol Cell Biol. 1999;19:3885-3894. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 75] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 4. | Jost CA, Marin MC, Kaelin WG. p73 is a simian [correction of human] p53-related protein that can induce apoptosis. Nature. 1997;389:191-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 731] [Cited by in RCA: 748] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 5. | Ichimiya S, Nimura Y, Kageyama H, Takada N, Sunahara M, Shishikura T, Nakamura Y, Sakiyama S, Seki N, Ohira M. p73 at chromosome 1p36.3 is lost in advanced stage neuroblastoma but its mutation is infrequent. Oncogene. 1999;18:1061-1066. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 91] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 6. | Nimura Y, Mihara M, Ichimiya S, Sakiyama S, Seki N, Ohira M, Nomura N, Fujimori M, Adachi W, Amano J. p73, a gene related to p53, is not mutated in esophageal carcinomas. Int J Cancer. 1998;78:437-440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 7. | Nomoto S, Haruki N, Kondo M, Konishi H, Takahashi T, Takahashi T, Takahashi T. Search for mutations and examination of allelic expression imbalance of the p73 gene at 1p36.33 in human lung cancers. Cancer Res. 1998;58:1380-1383. [PubMed] |
| 8. | Mihara M, Nimura Y, Ichimiya S, Sakiyama S, Kajikawa S, Adachi W, Amano J, Nakagawara A. Absence of mutation of the p73 gene localized at chromosome 1p36.3 in hepatocellular carcinoma. Br J Cancer. 1999;79:164-167. [PubMed] |
| 9. | Alonso ME, Bello MJ, Lomas J, Gonzalez-Gomez P, Arjona D, De Campos JM, Gutierrez M, Isla A, Vaquero J, Rey JA. Absence of mutation of the p73 gene in astrocytic neoplasms. Int J Oncol. 2001;19:609-612. [PubMed] |
| 10. | Yang SM, Zhou H, Chen RC, Wang YF, Chen F, Zhang CG, Zhen Y, Yan JH, Su JH. Sequencing of p53 mutation in established human hepatocellular carcinoma cell line of HHC4 and HHC15 in nude mice. World J Gastroenterol. 1998;4:506-510. [PubMed] |
| 11. | Peng XM, Peng WW, Yao JL. Codon 249 mutations of p53 gene in development of hepatocellular carcinoma. World J Gastroenterol. 1998;4:125-127. [PubMed] |
| 12. | Wang D, SHI JQ. Overexpression and mutations of tumor sup-pressor gene p53 in hepatocellular carcinoma. China Natl J New Gastroenterol. 1996;2:161-164. |
| 13. | Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, Vagner C, Bonnet H, Dikkes P, Sharpe A. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404:99-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 772] [Cited by in RCA: 790] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 14. | Ueda Y, Hijikata M, Takagi S, Chiba T, Shimotohno K. New p73 variants with altered C-terminal structures have varied transcriptional activities. Oncogene. 1999;18:4993-4998. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 112] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 15. | Wands JR, Blum HE. Primary hepatocellular carcinoma. N Engl J Med. 1991;325:729-731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 78] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 16. | Tang ZY. Hepatocellular carcinoma--cause, treatment and metastasis. World J Gastroenterol. 2001;7:445-454. [PubMed] |
| 17. | Feitelson MA, Sun B, Satiroglu Tufan NL, Liu J, Pan J, Lian Z. Genetic mechanisms of hepatocarcinogenesis. Oncogene. 2002;21:2593-2604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 246] [Cited by in RCA: 243] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 18. | Nassal M, Schaller H. Hepatitis B virus replication. Trends Microbiol. 1993;1:221-228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 130] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 19. | Seeger C, Mason WS. Hepatitis B virus biology. Microbiol Mol Biol Rev. 2000;64:51-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1094] [Cited by in RCA: 1115] [Article Influence: 44.6] [Reference Citation Analysis (0)] |
| 20. | Bargonetti J, Friedman PN, Kern SE, Vogelstein B, Prives C. Wild-type but not mutant p53 immunopurified proteins bind to sequences adjacent to the SV40 origin of replication. Cell. 1991;65:1083-1091. [PubMed] |
| 21. | Wang EH, Friedman PN, Prives C. The murine p53 protein blocks replication of SV40 DNA in vitro by inhibiting the initiation functions of SV40 large T antigen. Cell. 1989;57:379-392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 177] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 22. | Ori A, Zauberman A, Doitsh G, Paran N, Oren M, Shaul Y. p53 binds and represses the HBV enhancer: an adjacent enhancer element can reverse the transcription effect of p53. EMBO J. 1998;17:544-553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 81] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 23. | Lee H, Kim HT, Yun Y. Liver-specific enhancer II is the target for the p53-mediated inhibition of hepatitis B viral gene expression. J Biol Chem. 1998;273:19786-19791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 24. | Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning, a Labo-ratory Manual, 2nd edn. New York: Cold Spring Harbor Laboratory Press. 1989;788-791. |
| 25. | Arbuthnot P, Kew M. Hepatitis B virus and hepatocellular carcinoma. Int J Exp Pathol. 2001;82:77-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 203] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 26. | Bréchot C, Gozuacik D, Murakami Y, Paterlini-Bréchot P. Molecular bases for the development of hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC). Semin Cancer Biol. 2000;10:211-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 222] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 27. | Doitsh G, Shaul Y. HBV transcription repression in response to genotoxic stress is p53-dependent and abrogated by pX. Oncogene. 1999;18:7506-7513. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 1.0] [Reference Citation Analysis (0)] |