Published online Dec 15, 2002. doi: 10.3748/wjg.v8.i6.1088
Revised: August 1, 2002
Accepted: August 9, 2002
Published online: December 15, 2002
AIM: To establish a simple and convenient assay in vitro for the Hepatitis C virus NS3 serine protease based on the recombinant protease and substrate, and to evaluate its feasibility in screening the enzyme inhibitors.
METHODS: Based on the crystallographic structure of hepatitis C virus (HCV) serine protease, a novel single-chain serine protease was designed, in which the central sequence of cofactor NS4A was linked to the N-terminus of NS3 serine protease domain via a flexible linker GSGS. The fusion gene was obtained by two-step PCR that was carried out with three primers and then cloned into the prokaryotic expression vector pQE30, and the recombinant clone was verified by DNA sequencing. The single-chain recombinant protease was expressed when the E.coli was induced with IPTG and the expression conditions were optimized to produce large amount of soluble protease. The recombinant substrate NS5ab that covers the cleavage point NS5A/B was also expressed in E.coli. Both of the protease and substrate were purified by using Ni-NTA agarose metal affinity resin, then they were mixed together in a specific buffer, and the mixture was analyzed by SDS-PAGE. The cleavage system was used to evaluate some compounds for their inhibitory activity on serine protease.
RESULTS: The single-chain recombinant protease was over-expressed as soluble protein when the E.coli was induced at a low dosage of IPTG (0.2 mM) and cultured at a low temperature (15 °C). The protease was purified by using Ni-NTA agarose metal affinity resin (the purity is over 95%). The recombinant substrate NS5ab was expressed in an insoluble form and could refold successfully after purification and dialysis. A simple and convenient assay in vitro was established, in which the purified single-chain serine protease could cleave the recombinant substrate NS5ab into two fragments that were visualized by SDS-PAGE. PMSF had an effect on inhibiting activity of serine protease, while EDTA had not.
CONCLUSION: A simple and convenient assay in vitro for hepatitis C virus NS3 serine protease is based on recombinant substrate NS5ab and single-chain serine protease. This assay can be used in screening of enzyme inhibitors.
-
Citation: Du GX, Hou LH, Guan RB, Tong YG, Wang HT. Establishment of a simple assay
in vitro for hepatitis C virus NS3 serine protease based on recombinant substrate and single-chain protease. World J Gastroenterol 2002; 8(6): 1088-1093 - URL: https://www.wjgnet.com/1007-9327/full/v8/i6/1088.htm
- DOI: https://dx.doi.org/10.3748/wjg.v8.i6.1088
Hepatitis C virus (HCV) is a member of the Flaviviridae and now recognized as the major aetiological agent of post-transfusion non-A non-B hepatitis[1-3]. The viral genome is a 9.6-kb, positive-sense single-stranded RNA molecule that contains a single open reading frame encoding a polyprotein of 3010 to 3030 amino acids[4-6]. The polyprotein undergoes proteolytic processing by both host signal peptidases and viral proteases, giving rise to at least 10 mature proteins, and the cleavage of the 5 nonstructural proteins (NS3, NS4A, NS4B, NS5A and NS5B) were carried out by viral protease NS3/4A[7,8]. Since NS3/4A serine protease is very important for releasing functional proteins from the polyprotein, which are essential for replication and maturation of virus, it is currently being targeted in the development of drugs and diagnostics[9-11].
Previous studies indicate that the NS3/4A serine protease is a heterodimer formed by NS3 protein and its cofactor NS4A[12]. Analysis of the X-ray structures of the truncated NS3 protein complexed with the NS4A-derived peptide revealed that the serine protease domain adopts a chymotrypsin-like fold that means the NS3/4A is a member of chymotrypsin-like serine protease[13,14]. Although the protease domain of NS3 expressed without its cofactor is enzymatic active, its activity is partial and it can only recognize and cleave the NS5A/B site while the other two sites (NS4A/B and NS4B/5A) can not be cleaved[15,16]. That is, the NS4A enhances the cleavage at NS5A/ B site and is absolutely required for cleavage at the NS4B/5A site[17,18]. In order to carry out detailed characterization of this enzyme and develop the antiviral agent, a reproducible and convenient large-scale production of the purified enzyme and its substrate is essential[19]. In this report, we constructed and over-expressed a novel single-chain serine protease in a soluble form in E.coli, and established a simple assay in vitro for HCV serine protease based on the recombinant substrate NS5ab and single-chain protease. In addition, we examined the effect of several well-known protease inhibitors by the established assay system.
Prokaryotic expression vector pQE30, E.coli M15 cells and metal affinity chromatography resins (Ni-NTA agrose) were obtained from Qiagen Inc. (Chatsworth, California). Restriction enzymes (BamHI and HindIII), T4 DNA ligase and Ex.Taq DNA polymerase were purchased from Takara Inc. (Dalian, Liaoning). PCR Pure® PCR purification kit were obtained from Clontech Inc. The prokaryotic expression plasmid pGEX-3X-NS3N that carries the gene of amino terminal 181 amino acids of HCV NS3 protein is a gift from Dr. Chen et al[20].
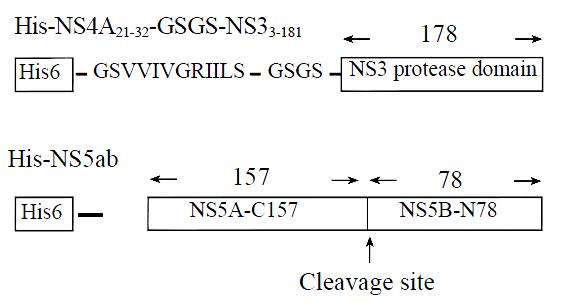
Construction of expression plasmids pQENS3N4A To generate fusion gene of NS4A21-32-GSGS-NS33-181, three primers were designed. Primers P2 (5'-ATTATTTTATCTGGTAGTGGTAGTATCACGGCCTACTCCCAA-3') and P3 (5'-CCCAAGCTTTTAGGACCGCATGGTAGTTTC-3') were used to amplify the NS4A29-32-GSGS-NS33-181 using the plasmid pGEX-3X-NS3N as template. After that the PCR products were used as template to generate NS4A21-32-GSGS-NS33-181 with primers P1 (5'-CGGATCCGGTTCTGTTGTTATTGTTGGTAGAATTATTTTATCTGGT-3') and P3. Underlined sequences represents HindIII and BamHI restriction sites, respectively. The highlighted sequences represents overlapping region of primers P1 and P2. The fusion gene NS4A21-32-GSGS-NS33-181 contained the sequence encoding the NS4A peptide, GSVVIVGRIILS (NS4A a.a 21-32), the linker, glycine-serine-glycine-serine (GSGS)[21], and the NS3 a.a 3-181. The expression vector pQE30 and fusion gene were both digested with BamHI and HindIII, then they were ligated after purification. The resulting plasmid was used to transform competent E.coli M15 and the recombinant clones were selected on LB agar plates with ampicillin (100 mg/mL) and kanamycin (25 mg/mL), and identified by restriction enzyme mapping and sequencing. The resulting recombinant plasmid is termed as pQENS3N4A.
Construction of expression plasmids pQENS5ab To construct the expression plasmid pQENS5ab, a cDNA fragment encoding amino acid residues 2263-2498 in the HCV polyprotein was isolated from the serum of a healthy HCV carrier in China by nested PCR using appropriate oligonulceotides, (1: 5'-AGT(G/A)AT(C/T)CTGGACTCTTTCG-3', 2: 5'-TGCTGGATAGGTTCCTGACGT-3', 3: 5'-TATGGTACCGAGGA(C/T)GAGAGGGAAGTATC-3', 4: 5'-TGCAAGCTTAACTGTGGACGCCT3'), which was inserted a KpnI site at the 5'-end and HindIII site at the 3'-end of the sequence. The PCR product amplified using Ex. Taq (Takara) was cloned into expression plasmid pQE30 that was digested with KpnI and HindIII. The resulting plasmid pQENS5ab encodes the cleavage site of NS5A/B with a N-terminal 20 non-virus encoded amino acids possessing a consecutive stretch of 6 His residues, which allows fusion protein to be purified in a single step by metal chelating affinity chromatography. The cloned DNA fragment was sequenced in order to exclude the introduction of mutations by PCR and also to confirm the in-frameness of the insert.
Expression and purification of recombinant single-chain serine protease in E.coli A single clone from E.coli M15 transformed with the recombinant plasmid pQENS3N4A was used to initiate growth in Luria broth supplemented with 100 μg/mL ampicillin and 25 μg/mL kanamycin. When the absorbance reached a value of 0.8 OD600, IPTG was added to give a final concentration of 0.2 mM and the incubation continued for additional 6 h at 20 °C. Under this condition a high level expression of NS3N4A in a soluble form was observed. The cells were harvested by centrifugation and washed extensively with PBS (20 mM sodium phosphate; pH7.4,140 mM NaCl). The cell pellet was resuspended in lysis buffer (50 mM Tris·HCl; pH 7.4, 10% glycerol, 0.3M NaCl, 2 mM β-mercaptoethnol, 0.5% NP-40) and disrupted by sonication on ice using sonifier (60 s × 10 strokes at 150 W output with 30 s intervals). The homogenate was centrifuged at 15000 × g for 30 min to remove cell debris and was chromatographed on a nickel-agrose column (Qiagen). The column was washed extensively with 10 column volumes of lysis buffer and subsequently washed using 20 column volumes of buffer W (50 mM Tris·HCl; pH7.4, 10% glycerol, 1M NaCl, 20 mM imidazole, 2mM β-mercaptoehanol, 0.5% NP-40) and finally eluted with buffer E (50 mM Tris·HCl; pH 7.4, 10% glycerol, 1 M NaCl, 250 mM imidazole, 2 mM β-mercaptoehanol, 0.5% NP-40). Eluted fractions were subjected to SDS-PAGE and the pooled enzyme was dialyzed against buffer D (50 mM Tris·HCl; pH 7.4, 10% glycerol, 1M NaCl, 2 mM β-mercaptoehanol, 0.5% NP-40) to remove imidazole before stored in aliquots at -20 °C.
Protein concentrations were estimated from UV absorbance at 280 nm and 260 nm and calculated according to the formula: concentration of protein (g/L) = 1.45 × A280 - 0.74 × A260.
Expression, purification and refolding of recombinant substrate NS5ab The transformed cells were grown at 37 °C in Luria broth supplemented with 100 μg/mL ampicillin and 25 μg/mL kanamycin. When cell density reached an OD600 of 0.8-1.0, induction was initiated by the addition of 1 mM IPTG. Cells of 500 mL culture were harvested 5 hours after induction and resuspended in 10 mL lysis buffer (50 mM sodium phosphate; pH7.8, 300 mM NaCl). After disruption by sonication on ice, the inclusion bodies were collected by centrifugation at 12000 × g for 15 min and washed three times in the lysis buffer. The inclusion body was solubilized in 8M urea (50 mM sodium phosphate; pH 8.0, 300 mM NaCl, 8M urea) and was chromatographed on a nickel-agrose column (Qiagen). The purification was performed according to the manual of Qiaexpressionist®. To refold the purified NS5ab protein, the protein was dialyzed against refolding buffer with a stepwise gradient of urea from 6M to 4M, 2M, 1M, 0.5M, 0.25M and finally to TBS (50 mM Tris·HCl; pH 7.4, 140 mM NaCl). The concentration of substrate NS5ab was calculated according to the foregoing formula on the basis of UV absorbance at 280 nm and 260 nm.The purified recombinant substrate NS5ab was stored in aliquots at -20 °C.
In vitro proteolytic assay for HCV NS3 serine protease 20 μg of purified single-chain serine protease was incubated with 20 μg of recombinant substrate NS5ab in 100 μL of 25 mM Tris·HCl (pH7.4), 10% glycerol, 0.5M NaCl, 10mM DTT and 0.5% NP-40 at room temperature. After the reaction was terminated at various time points, there were mixed with same volume of 2 × loading buffer and heated at 90 °C for 10 min, then analyzed by SDS-PAGE and stained with Coomassie brilliant blue R-250 (CBB R-250).
Effects of some known serine protease inhibitors In this assay, the single-chain serine protease was mixed with appropriate amount of PMSF or EDTA at room temperature for 30 min before they were incubated with recombinant substrate NS5ab to pursue the reaction. 45 min later, the reaction was terminated and analyzed with SDS-PAGE and CBB R-250.
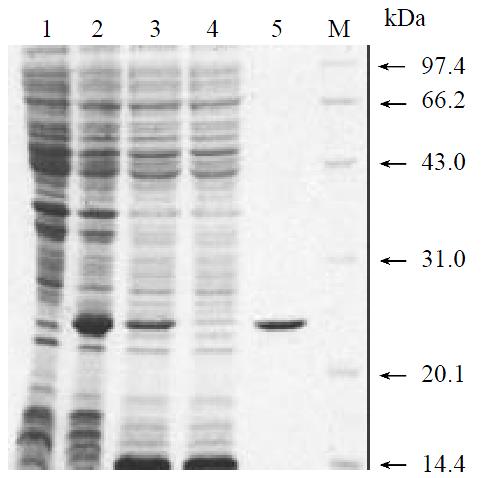
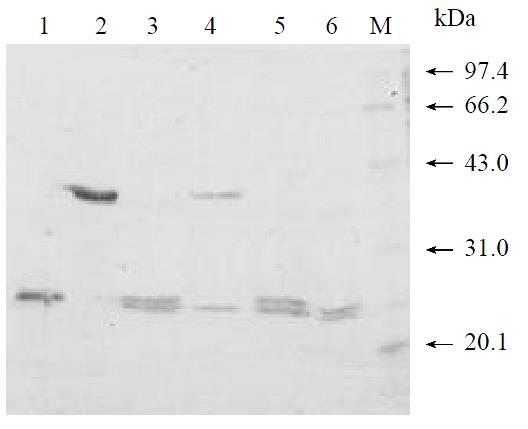
To produce a recombinant HCV NS3N4A protein complex with structural confirmation similar to its biological counterpart, an expression plasmid encoding HCV NS4A central peptide GSGVVIVGRILLS (NS4A a.a. 21-32) covalently jointed to a NS3 N-terminal protease domain via an flexible amino acid linker GSGS was engineered[22]. The cDNA fragment encoding this single-chain protease was inserted into a pQE30 expression vector that provided additional six amino acids and six histidine residues at the amino terminus of the NS4A peptide (Figure 1). Induction of the bacteria harboring this expression plasmid with a low concentration of IPTG (0.2 mM) at low temperature (20 °C) resulted in the production of a 23 kDa recombinant protein (Figure 2, lane 2). Much of the expressed recombinant protein existed in the soluble fraction of the bacterial lysate (Figure 2, lane 3). Further purification of the expressed protein using nickel-affinity chromatography yielded a product with greater than 95% homogeneity as measured by SDS-PAGE and thin layer scanning assay (Figure 2, lane 5). Approximately 7.5 mg of protein was obtained from 500 mL culture following purification.
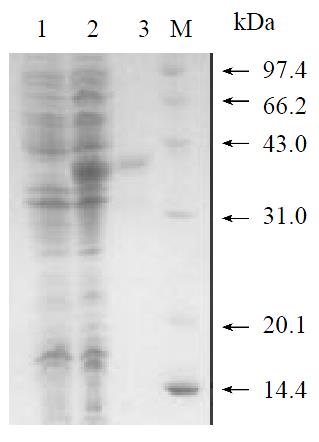
To produce a recombinant substrate of HCV NS3 serine protease, the gene that encoding HCV NS5A C-terminal 157 amino acids residues, cleavage site of NS5A/B, and NS5B N-terminal 78 amino acids residues was isolated by nested RT-PCR from the serum of a healthy HCV carrier in China. The cDNA fragment was inserted into pQE30 expression vector that provided additional 20 amino acids residues (including six histidine residues) at the amino terminal of the viral peptide (Figure 1). Induction of the bacteria harboring expression plasmid pQENS5ab resulted in the production of recombinant protein of 35kDa (Figure 3, lane 2). The majority of the expressed recombinant protein existed in the insoluble fraction of the bacteria lysate. After purification of the recombinant protein NS5ab using nickel-affinity resin, the purified protein was refolded by dialysis method to produce soluble substrate with greater than 90% homogeneity (Figure 3, lane 3). Approximately 7.6 mg of protein per liter of culture was obtained by following purification and refolding.
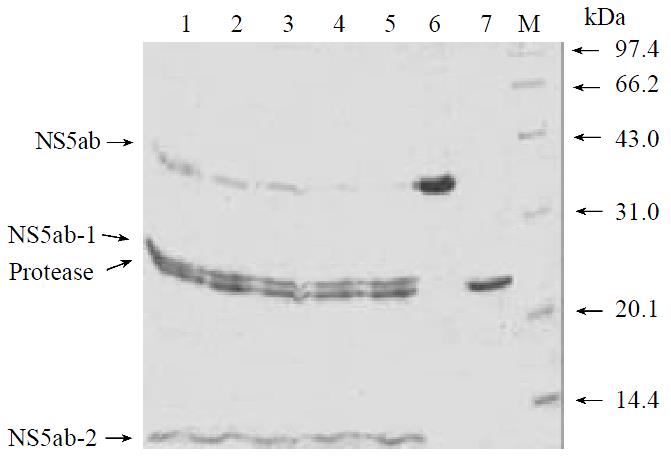
The soluble single-chain serine protease and soluble substrate NS5ab was incubated in a reaction buffer and then investigate whether the enzyme can process the substrate correctly and efficiently[23]. Figure 4 showed that the single-chain serine protease could cleave the substrate NS5ab into two fragments with molecular weight about 24 kDa and 11 kDa respectively. In addition, the amount of products increased and the substrate decreased along with reaction time (Figure 4, lane 1-5). That means the single-chain serine protease has highly active proteolytic activity and the substrate is active, too.
The in vitro trans-cleavage assay system described made it possible to examine the effect of known serine protease inhibitors[24]. Two kinds of known proteinase inhibitors (PMSF and EDTA) were added to the reaction system and the effect visualized by SDS-PAGE and CBB R-250 staining. It was shown that the well-characterized inhibitor PMSF could inhibit proteolytic activity at 5 mM in vitro (Figure 5, lane 4), while the metal chelator EDTA had no observable effect on the protease activity even at a concentration of 5mM (Figure 5, lane 6).
This study aimed at establishing a simple in vitro assay for hepatitis C virus NS3 serine protease, since such an assay is the most important for screening specific antiviral inhibitors from combinatorial chemical library and random biological library[25-27]. To accomplish this object we have to obtain substantial amounts of purified NS3 serine protease with complete functional activity. Previous studies had shown that there are two methods to produce fully active HCV serine protease. The first method is to express full length NS31-631 which was fused to the N-terminal of NS4A1-54 in eukaryotic cells[28], and the second one is to express protease domain in E.coli and mix them with excessive synthetic NS4A core peptide to form heterodimer[29]. Although the two methods work, there still are many shortages. The former can only provide little amount of protein and the enzymatic activity of protein produced by later method is very low[28,29]. According to Yan’s report[30], the reason that poor activity of the heterodimer formed in vitro by synthetic peptide and protease domain lies in that the affinity of synthetic peptide is much lower than that of the full-length NS4A, which makes the complex is unstable. So it is reasonable to express the fusion protein of NS3 protease domain and NS4A in E.coli.
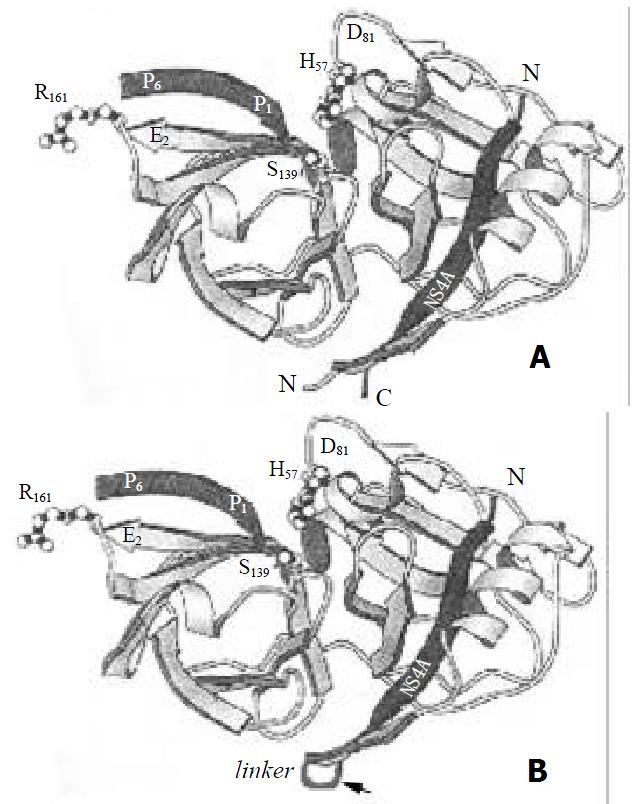
Inoue and his colleague[31] expressed NS3 protease fused at the amino terminal of NS4A in E.coli, which showed ideal stability and activity, but the products existed in inclusion bodies that lead to the poor productivity. In addition, the crystallographic structure of NS3 serine protease domain complexed with synthetic peptide indicated that it might be better for NS4A fused to amino terminal of NS3 protease domain[21,22]. Since the central part of NS4A is embedded in the cleft formed by NS3 protease domain and its C-terminus lies close to the amino terminus of NS3 (Figure 6A), it is reasonable to fuse the NS4A central peptide to amino terminal of NS3 via a flexible linker. This fusion format will change the natural intermolecular interaction to intramolecular interaction (Figure 6B), which will increase the stability of NS4A-NS3N complex. In this study, we constructed a single-chain serine protease that contained central peptide of NS4A fused to amino terminal of NS3 protease domain via GSGS linker, and it could process the recombinant substrate NS5ab efficiently.
The reason that single-chain serine protease can express in a soluble form at high level in E.coli lies in two points: 1) the low concentration of inducer and low temperature culture reduced the synthesis speed of peptide, which enhance the folding of protein[32-34]; 2) the central sequences of NS4A may increase the solubility of the single-chain serine protease. The NS3 domain was in various prokaryotic expression vectors with failures to produce soluble recombinant protein in this study, though various measures had been taken, such as lowering temperature and lowering concentration of IPTG (unpublished materials). In the same condition, the protease domain that fused to the C-terminal of NS4A central segment could express in a soluble form, which means that the central sequences of NS4A may increase the solubility of NS3 protease domain. Previous studies had confirmed that NS3 protease’s N-terminus would be changed from disorded structure to regular structure when it formed a complex with the NS4A peptide, that is why the NS4A central peptide is capable of promoting the folding of protein[35,36]. In addition, the NS4A central peptide is highly hydrophobic, which will reduce the synthesis of peptide in host and be beneficial to protein’s refolding[37].
Based on the recombinant single-chain protease and substrate protein NS5ab, we established a simple and cheap in vitro assay for HCV NS3 protease. In this assay system, the results were analyzed by SDS-PAGE and visualized by Coomassie brilliant blue R-250 staining (CBB R-250), which make it an attractive system because of its convenience, low cost and intuitiveness. To assess the applicability of this assay system in identification of serine protease inhibitors, we examined the effects of two known serine protease inhibitors on the activity of HCV protease. The results confirmed that the PMSF could inhibit the proteolytic activity of HCV serine protease completely at 5 mM, while the metal chelator EDTA had no observable effect on it, which had been observed by other researchers[38,39]. In conclusion, the in vitro assay established here is applicable in evaluating the effect of chemical compounds and biological molecules on the activity of HCV serine protease. Furthermore, this assay can be used in high throughput screening of the potential inhibitors of HCV protease[40,41].
Edited by Zhang JZ
| 1. | 1 Farci P. Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome [Science 1989; 244: 359-362]. J Hepatol. 2002;36:582-585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4996] [Cited by in RCA: 4655] [Article Influence: 129.3] [Reference Citation Analysis (0)] |
| 2. | Alter MJ, Margolis HS, Krawczynski K, Judson FN, Mares A, Alexander WJ, Hu PY, Miller JK, Gerber MA, Sampliner RE. The natural history of community-acquired hepatitis C in the United States. The Sentinel Counties Chronic non-A, non-B Hepatitis Study Team. N Engl J Med. 1992;327:1899-1905. [PubMed] |
| 3. | Kuo G, Choo QL, Alter HJ, Gitnick GL, Redeker AG, Purcell RH, Miyamura T, Dienstag JL, Alter MJ, Stevens CE. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science. 1989;244:362-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2495] [Cited by in RCA: 2343] [Article Influence: 65.1] [Reference Citation Analysis (0)] |
| 4. | Kato N, Hijikata M, Nakagawa M, Ootsuyama Y, Muraiso K, Ohkoshi S, Shimotohno K. Molecular structure of the Japanese hepatitis C viral genome. FEBS Lett. 1991;280:325-328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 31] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 5. | Okamoto H, Okada S, Sugiyama Y, Kurai K, Iizuka H, Machida A, Miyakawa Y, Mayumi M. Nucleotide sequence of the genomic RNA of hepatitis C virus isolated from a human carrier: comparison with reported isolates for conserved and divergent regions. J Gen Virol. 1991;72:2697-2704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 323] [Cited by in RCA: 323] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 6. | Takamizawa A, Mori C, Fuke I, Manabe S, Murakami S, Fujita J, Onishi E, Andoh T, Yoshida I, Okayama H. Structure and organization of the hepatitis C virus genome isolated from human carriers. J Virol. 1991;65:1105-1113. [PubMed] |
| 7. | Grakoui A, Wychowski C, Lin C, Feinstone SM, Rice CM. Expression and identification of hepatitis C virus polyprotein cleavage products. J Virol. 1993;67:1385-1395. [PubMed] |
| 8. | Shimotohno K, Tanji Y, Hirowatari Y, Komoda Y, Kato N, Hijikata M. Processing of the hepatitis C virus precursor protein. J Hepatol. 1995;22:87-92. [PubMed] |
| 9. | Llinàs-Brunet M, Bailey M, Fazal G, Ghiro E, Gorys V, Goulet S, Halmos T, Maurice R, Poirier M, Poupart MA. Highly potent and selective peptide-based inhibitors of the hepatitis C virus serine protease: towards smaller inhibitors. Bioorg Med Chem Lett. 2000;10:2267-2270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 81] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 10. | Dymock BW, Jones PS, Wilson FX. Novel approaches to the treatment of hepatitis C virus infection. Antivir Chem Chemother. 2000;11:79-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 53] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 11. | De Francesco R, Steinkühler C. Structure and function of the hepatitis C virus NS3-NS4A serine proteinase. Curr Top Microbiol Immunol. 2000;242:149-169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 30] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 12. | Bartenschlager R. The NS3/4A proteinase of the hepatitis C virus: unravelling structure and function of an unusual enzyme and a prime target for antiviral therapy. J Viral Hepat. 1999;6:165-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 105] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 13. | Kim JL, Morgenstern KA, Lin C, Fox T, Dwyer MD, Landro JA, Chambers SP, Markland W, Lepre CA, O'Malley ET. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell. 1996;87:343-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 566] [Cited by in RCA: 511] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 14. | Yan Y, Li Y, Munshi S, Sardana V, Cole JL, Sardana M, Steinkuehler C, Tomei L, De Francesco R, Kuo LC. Complex of NS3 protease and NS4A peptide of BK strain hepatitis C virus: a 2.2 A resolution structure in a hexagonal crystal form. Protein Sci. 1998;7:837-847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 190] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 15. | Grakoui A, McCourt DW, Wychowski C, Feinstone SM, Rice CM. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J Virol. 1993;67:2832-2843. [PubMed] |
| 16. | Bartenschlager R, Ahlborn-Laake L, Mous J, Jacobsen H. Kinetic and structural analyses of hepatitis C virus polyprotein processing. J Virol. 1994;68:5045-5055. [PubMed] |
| 17. | Landro JA, Raybuck SA, Luong YP, O'Malley ET, Harbeson SL, Morgenstern KA, Rao G, Livingston DJ. Mechanistic role of an NS4A peptide cofactor with the truncated NS3 protease of hepatitis C virus: elucidation of the NS4A stimulatory effect via kinetic analysis and inhibitor mapping. Biochemistry. 1997;36:9340-9348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 83] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 18. | Barbato G, Cicero DO, Nardi MC, Steinkühler C, Cortese R, De Francesco R, Bazzo R. The solution structure of the N-terminal proteinase domain of the hepatitis C virus (HCV) NS3 protein provides new insights into its activation and catalytic mechanism. J Mol Biol. 1999;289:371-384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 88] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 19. | Steinkühler C, Koch U, Narjes F, Matassa VG. Hepatitis C virus protease inhibitors: current progress and future challenges. Curr Med Chem. 2001;8:919-932. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 51] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 20. | Chen Z, Xu J, Song HB, Zhou YS, Wang HT. Cloning and se-quence analysis of hepatitis C virus NS3 serine protease. Zhonghua Ganzangbing Zazhi. 1998;6:114-115. |
| 21. | Taremi SS, Beyer B, Maher M, Yao N, Prosise W, Weber PC, Malcolm BA. Construction, expression, and characterization of a novel fully activated recombinant single-chain hepatitis C virus protease. Protein Sci. 1998;7:2143-2149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 98] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 22. | Howe AY, Chase R, Taremi SS, Risano C, Beyer B, Malcolm B, Lau JY. A novel recombinant single-chain hepatitis C virus NS3-NS4A protein with improved helicase activity. Protein Sci. 1999;8:1332-1341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 43] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 23. | Koch JO, Bartenschlager R. Determinants of substrate specificity in the NS3 serine proteinase of the hepatitis C virus. Virology. 1997;237:78-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 24. | Mori A, Yamada K, Kimura J, Koide T, Yuasa S, Yamada E, Miyamura T. Enzymatic characterization of purified NS3 serine proteinase of hepatitis C virus expressed in Escherichia coli. FEBS Lett. 1996;378:37-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 25. | Ingallinella P, Fattori D, Altamura S, Steinkühler C, Koch U, Cicero D, Bazzo R, Cortese R, Bianchi E, Pessi A. Prime site binding inhibitors of a serine protease: NS3/4A of hepatitis C virus. Biochemistry. 2002;41:5483-5492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 45] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 26. | Fowler A, Price-Jones M, Hughes K, Anson J, Lingham R, Schulman M. Development of a high throughput scintillation proximity assay for hepatitis C virus NS3 protease that reduces the proportion of competitive inhibitors identified. J Biomol Screen. 2000;5:153-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 9] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 27. | Fukuda K, Vishnuvardhan D, Sekiya S, Hwang J, Kakiuchi N, Taira K, Shimotohno K, Kumar PK, Nishikawa S. Isolation and characterization of RNA aptamers specific for the hepatitis C virus nonstructural protein 3 protease. Eur J Biochem. 2000;267:3685-3694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 77] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 28. | Sali DL, Ingram R, Wendel M, Gupta D, McNemar C, Tsarbopoulos A, Chen JW, Hong Z, Chase R, Risano C. Serine protease of hepatitis C virus expressed in insect cells as the NS3/4A complex. Biochemistry. 1998;37:3392-3401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 55] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 29. | Vishnuvardhan D, Kakiuchi N, Urvil PT, Shimotohno K, Kumar PK, Nishikawa S. Expression of highly active recombinant NS3 protease domain of hepatitis C virus in E. coli. FEBS Lett. 1997;402:209-212. [PubMed] |
| 30. | Yan Y, Li Y, Munshi S, Sardana V, Cole JL, Sardana M, Steinkuehler C, Tomei L, De Francesco R, Kuo LC. Complex of NS3 protease and NS4A peptide of BK strain hepatitis C virus: a 2.2 A resolution structure in a hexagonal crystal form. Protein Sci. 1998;7:837-847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 190] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 31. | Inoue H, Sakashita H, Shimizu Y, Yamaji K, Yokota T, Sudo K, Shigeta S, Shimotohno K. Expression of a hepatitis C virus NS3 protease-NS4A fusion protein in Escherichia coli. Biochem Biophys Res Commun. 1998;245:478-482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 32. | Swartz JR. Advances in Escherichia coli production of therapeutic proteins. Curr Opin Biotechnol. 2001;12:195-201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 292] [Cited by in RCA: 246] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 33. | Weickert MJ, Doherty DH, Best EA, Olins PO. Optimization of heterologous protein production in Escherichia coli. Curr Opin Biotechnol. 1996;7:494-499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 92] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 34. | Hockney RC. Recent developments in heterologous protein production in Escherichia coli. Trends Biotechnol. 1994;12:456-463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 119] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 35. | Kwong AD, Kim JL, Rao G, Lipovsek D, Raybuck SA. Hepatitis C virus NS3/4A protease. Antiviral Res. 1999;41:67-84. [PubMed] |
| 36. | Mossakowska DE. Expression of nuclear hormone receptors in Escherichia coli. Curr Opin Biotechnol. 1998;9:502-505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 37. | Tomei L, Failla C, Vitale RL, Bianchi E, De Francesco R. A central hydrophobic domain of the hepatitis C virus NS4A protein is necessary and sufficient for the activation of the NS3 protease. J Gen Virol. 1996;77:1065-1070. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 80] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 38. | Steinkühler C, Tomei L, De Francesco R. In vitro activity of hepatitis C virus protease NS3 purified from recombinant Baculovirus-infected Sf9 cells. J Biol Chem. 1996;271:6367-6373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 60] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 39. | Shoji I, Suzuki T, Chieda S, Sato M, Harada T, Chiba T, Matsuura Y, Miyamura T. Proteolytic activity of NS3 serine proteinase of hepatitis C virus efficiently expressed in Escherichia coli. Hepatology. 1995;22:1648-1655. [PubMed] |
| 40. | Kakiuchi N, Nishikawa S, Hattori M, Shimotohno K. A high throughput assay of the hepatitis C virus nonstructural protein 3 serine proteinase. J Virol Methods. 1999;80:77-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 20] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 41. | Cerretani M, Di Renzo L, Serafini S, Vitelli A, Gennari N, Bianchi E, Pessi A, Urbani A, Colloca S, De Francesco R. A high-throughput radiometric assay for hepatitis C virus NS3 protease. Anal Biochem. 1999;266:192-197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.4] [Reference Citation Analysis (0)] |