Published online Feb 15, 2002. doi: 10.3748/wjg.v8.i1.54
Revised: July 16, 2001
Accepted: August 15, 2001
Published online: February 15, 2002
AIM: To isolate and clone the vincristine-resistine-related genes in gastric cancer SGC7901 cell line and to clarify the multidrug-resistant molecular mechanism of gastric cancer cells.
METHODS: The modified differential-display polymerase chain reaction (DD-PCR) was used to examine the differences in the mRNA composition of Vincristine-resistant gastric cancer SGC 7901 cells (SGC7901/VCR), induced by vincristine sulfate versus SGC7901 cells. The differentially expressed cDNA fragments were confirmed by reverseNorthern analysis, sequencing, BLAST analysis and Northern bolt analysis.
RESULTS: DD-PCR identified that 54 cDNA fragments were preferentially expressed in SGC 7901/VCR cells. When these cDNA fragments were analyzed by reverse Northern blot, 20 were reproducibly expressed at a high level in SGC7901/VCR. Sequencing and BLAST analysis revealed that seven of the genes were known genes: ADP-ribosylation factor 4, Cytochrome oxidase subunit II, Ss-A/Ro ribonucleoprtein autoantigen 60kd subunit, ribosomal protein S13, galaectin-8 gene, oligophrenin 1 mRNA, ribosomal protein L23 mRNA; thirteen of the genes were unknown genes. The length and abundance of the four unknown genes mRNA were further confirmed by Northern blot analysis.
CONCLUSION: The twenty differential known and unknown genes may be related to the vincristine-resistant mechanism in human gastric cancer SGC7901 cell line.
- Citation: Wang X, Lan M, Shi YQ, Lu J, Zhong Y-, Wu HP, Zai HH, Ding J, Wu KC, Pan BR, Jin JP, Fan DM. Differential display of vincristine-resistance-related genes in gastric cancer SGC7901 cell. World J Gastroenterol 2002; 8(1): 54-59
- URL: https://www.wjgnet.com/1007-9327/full/v8/i1/54.htm
- DOI: https://dx.doi.org/10.3748/wjg.v8.i1.54
The primary factor affecting the chemotherapy for gastric cancer is the resistance of cancer cells to anti-tumor drugs. This phenomenon is called multidrug-resistance (MDR). Previous studies have shown that the mechanisms of MDR involved P-glycoprotein (P-gp), multidrug-resistance related protein (MRP 1-5), lung drug-resistance related protein (LRP), and recently discovered breast cancer drug-resistance related protein (BCRP), GSH/GST, PKC, Topo DNA plerosis[1-3], genes related to apoptosis and changes of cellular environment (such as pH, hypoxia and temperature). However, over-expression of the genes encoding these proteins cannot completely account for MDR, nor can the treatment aiming at these mechanisms significantly revert MDR. Thus, it is most likely that MDR involves multi-genes. Little has been known about MDR of gastric carcinoma cells. Preliminary studies have shown specific features in MDR of gastric carcinoma cells[4-19], which cannot be completely explained by any known mechanisms of gastric carcinoma MDR[20-32]. We therefore used mRNA differential display to investigate the differential expression of the genes in MDR gastric cells in order to elucidate the molecular mechanism of gastric cancer MDR and to discover the related genes so as to lay the foundation for the eventual solution of the problem.
Gastric cancer SGC7901 cell (preserved by the Institute), grown in RPMI1640 medium containing 100 g·L-1 fetal bovine serum and cultured in incubator filled with 950 mL·L-1 O2, 50 mL·L-1 CO2 at 37 °C. The procedure of SGC7901/VCR cell induction was as follows[33]: SGC7901 cells were harvested at the mid log-growth phase, and then cultured in the medium containing 0.2 mg·L-1 vincristine; the medium exchanged every two or three days, and then the vincristine concentration was increased by 0.1 mg·L-1 every other week, until it reached 0.8-1.0 mg·L-1. SGC7901/VCR cells with stable phenotype were obtained after three months of continuous culture.
Total RNA of gastric cancer SGC7901 and SGC7901/VCR cells was extracted and underwent RNA formaldehyde denatured agarose gel electrophoresis. The purity and RNA content of the samples were measured by OD260/280. Five μg of total RNA were taken from SGC7901 and SGC7901/VCR espectively, and combined with anchoring primers 5’-AAGCTTTTTTTTTTTA-3’, 5’-AAGCTTTTTTTTTTTG-3’, and 5’-AAGCTTTTTTTTTTTC to undergo RT-PCR. The procedure is as follows: to each 20 μL reaction system, 5 μg total RNA and 1.6 μL anchoring primer were added, and the system was put at 70 °C for 5 min and then into ice bathing. Buffer of 5 × RT 4 μL, 10 mmol·L-1 dNTP (each) 2 μL, RNase inhibitor 20U, M-MuLV retrotranscriptase (MBI) 2 μL (40 U) were added into the reaction system sequentially, and put in H2O to get a total volum of 20 μL. The reaction went at 37 °C for 1h, and for 10min at 70 °C to terminate.
Eight random primers AP1-8 were employed in PCR. Three kinds of cDNA products of retrotranscription were each combined with the eight random primers to form 24 PCR reaction systems.There were 48 PCR reactions for two kinds of cells. In each 20 µL PCR reaction system, there were 2 µL RT product cDNA,1 µL anchoring primer (4 µmol·L-1), 1 µL random primer (4 µL),10 × PCR buffer 2 µL,MgCl2 (25 µmol·L-1) 1.6 µL, dNTP (5 µmol·L-1) 1.6 µL, Taq plus I DNA polymerase (MBI) 0.15 µL (0.4 U), α-32P-dNTP0.5 µL(185 kBq), and ion-free water. The conditions of PCR: 95 °C 3 min; 94 °C 30 s→40 °C 2 min→72 °C 40 s for 40 cycles; and then 70 °C 10 min for termination. PCR product was 95 °C heat denatured for 3 min, and underwent ice-bathing, and 60 g·L-1 urea denatured polyacylamine gel (sequencing gel) electrophoresis. The gel was preserved with membrane after electrophoresis and underwent X-ray film autoradiography at -70 °C in black box for 48 h.
Differential bands were cut from the gel and the DNA was recovered and amplified with secondary PCR.The primers and conditions were identical to that of DD-PCR. The amplified product was separated with agarose gel electrophoresis, purified and retrieved, cloned into PUCm-T vector(Shanghai Shenggong Co.), and underwent restriction enzymatic cleavage identification.
The plasmids were extracted from the bacteria with positive clones of the 46 differential DNA bands; the quantities were determined through 10 g·L-1 agarose gel electrophoresis. One µg plasmid was used to solve in 6(SSC buffer, boiled at 100 °C for 10 min to denature, put into ice-water, and pointed onto two NC membranes, each NC membrane having 46 points, and 1µg plasmid. The location of the points on the two membranes were identical. The NC membranes were dried at room temperature. The samples were alkaline-denatured on the membranes; and the membranes were heated at 80 °C for 2 h to fix the plasmid DNA and preserved at room temperature.
Preparation of radioactive cDNA probe: Total RNA was extracted from SGC7901 and SGC7901/VCR cells by the same method as in DD-PCR. 10 µg total RNA was taken; 2 µL Olig dT18 (0.5 mg·L-1) were added, and put at 70 °C for 5 min, and ice-water bathed for 5 min. Buffer of 5 × RT 10 µL, dNTP(10 mmol·L-1/ each) 2.5 µL, RNase inhibitor 2.5 µL(50 U),α-32P-dATP 5 µL (1850 kBq), α-32P-dCTP 5 µL(1850 kBq), M-MuLV 4 µL(80 U), and water 50 µL were added. The reaction was processed at 37 °C for 1 h,and 70 °C for 5 min to terminate. EDTA of 0.5 mol·L-1(pH 8.0) 2 µL, 100 g·L-1ªª SDS 2 µL and 10 mol·L-1ªª NaoH 1.8 µL were put into the retrotranscription product, and incubated at 68 °C for 30 min. 10 µL 1 mol·L-1 Tris.Cl (pH 7.4) and 3 µL 2 mol·L-1 HCl were added to neutralize at room temperature. Pre-hybridization was made at 42 °C for 6 h, and different cDNA probes were added onto the two membranes to hybridize at 42 °C for 20 h. The membranes were washed, dried and underwent X-ray film autoradiography at -70 °C for 72 h.
The cDNA fragments were sequenced and confirmed through reverse Northern blot to be highly expressed in SGC7901/VCR. BLAST program was used for homologic comparison in genetic library and FASTA was used to analyze the results.
The total RNA of the two kinds of cells was extracted by the method identical to that mentioned above. Sixteen µg total RNA of each sample was denatured; 10 g·L-1 formaldehyde denature agarose gel electrophoresis (16 µg RNA/lane) was performed; the mRNA was transfered onto the NC membranes by capillary blot, and heated at 80 °C for 2 h to fix the mRNA. Plasmids were extracted from positive clone bacteria; restriction endonuclease cleavage and electrophoresis were performed; the target cDNA fragments were recovered and purified; the cDNA probes were labeled by random primer method, and pre-hybridized on the NC membrane for 3 h at 42 °C.The probes were added and hybridized at 42 °C for 20 h. the membranes were washed, dried and undergone for X-ray film autoradiography at -70 °C in black box for 48 h.
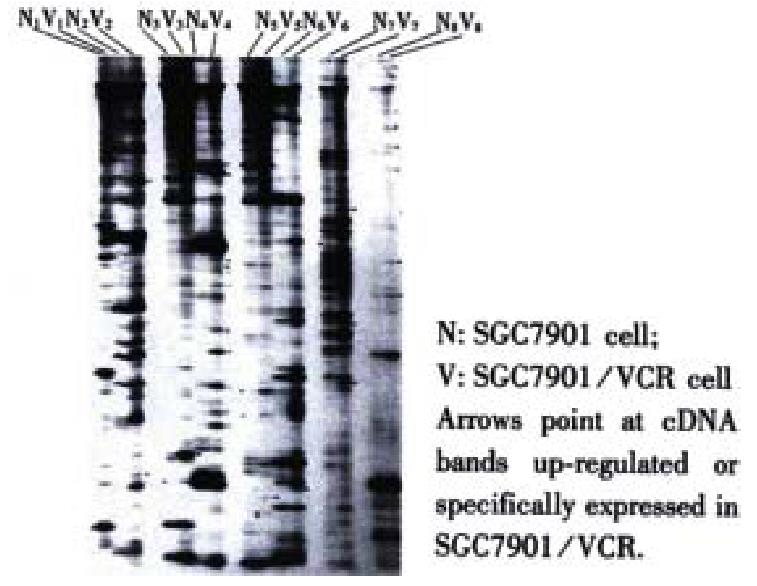
The mRNA of SGC7901 and SGC7901/VCR cells was amplified by RT-PCR, each was divided into 24 groups, and each group was further separated into 150-200 cDNA fragments of diverted length (100-1500 bp) after 60 g·L-1 polyacylamide gel electrophoresis. The comparison showed that the abundance of certain DNA bands in SGC7901/VCR cells was up-regulated, down-regulated, newly generated or lost. A total of 196 differential cDNA fragments were obtained (Figure 1).
Fifty-four of the 196 differential cDNA fragments were specifically or significantly highly expressed in SGG7901/VCR cells. Restriction enzymatic cleavage identification showed that 46 cDNA fragments were successfully cloned(Figure 2).
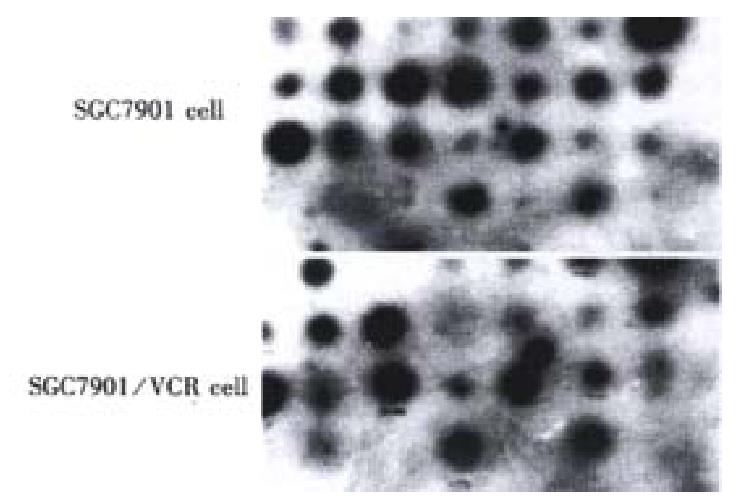
In order to confirm the abundance changes of the differential genes and exclude false positive, reverse Northern blot was performed and the high expression of the 20 cDNA fragments in SGC7901/VCR was further validated (Figure 3).
Sequencing and homologic analysis of the 20 highly expressed genes in SGC7901/VCR cells, confirmed by reverse Northern blot, showed that 7 of the 20 positive clones were highly homologous with known gene sequences (Table 1), and 13 were unknown sequences: GRP-2, GRP-8, GRP-12, GRP-15,GRP-18,GRP-19,GRP-24,GRP-28,GRP-31,GRP-36, GRP-37, GRP-38, GRP-41.
| Serial number | Homologous genes |
| GRP-44(168 bp) | ADP-ribosylation factor4 (ARF4) mRNA (1610 bp) |
| GRP-39(181 bp) | Cytochrome oxidase subunit II mRNA |
| GRP-33(199 bp) | Ss-A/Ro ribonucleoprtein autoantigen 60kd subunit mRNA |
| GRP-42(498 bp) | Ribosomal protein S13 (RPS13) mRNA (530 bp) |
| GRP-6 (390 bp) | Galaectin-8 (LGALS 8) gene, exons4, mRNA |
| GRP-16(197 bp) | Oligophrenin 1 (OPHN1) mRNA (7350 bp) |
| GRP-1 (442 bp) | Ribosomal protein L23 (RPL23) mRNA (490 bp) |
GRP-2
CAGTGACTTTATTTTAATGGGTTT TCAGACATACAGAAAGGGATTCTT TAGATGGGGCTGTGTCACTAGTCA ACCATCTTCACTGTGGAGTCCTAG TCACTATGATTTTGTTTTGTAGAT CATGAGGATTCATTCAAATTGTCT CCTCTTCCACTCCTTCGTAATAGG TTACATGATCTGAAAGTACATCCC TCCTTTAGTACATTGTATTCAAAC AGGTTGCTGCTACTTCTCTACTGT CATTAATCTTTTCATCATCTTCTT ATTCCTCTTAGC——
GRP-8
GTTTGAGAGGATACTCATCTTTTT GGAATCCTGACCTTAGGTTCGGCATGT AGACCAAGTGATGAGAAGTGAATACAT GGAAGAGTTTTTAAGTGTGACTTGAAA AATATGC—--
GRP-12
GGAACCTGGATTCTTTTAATAGTT GTTGAAGCCTCCAGGGGGCCAGGCGGA TCACTTGAGGCCCAGCCTGGCCAACAT AGCGAAACCCTGCCTCTACTAAAACCA CAAAAATCAGCCGGGCATGGTGGCACA CGCTTATAATCCCAGCTACTTGGGACG CTGAGGTGGGATATAGCTTGAACCCGG GAAGGAGACTGCAGTCAGGGAAGCCTA GGGAAGCCTCAGACCAAGGATGATTGA ATAACAAAGAAAAGGTGTAAGTAAAGA TCTCCAACTCTTAGGGAGTACTAATTA GGAAAGTTAAGG GTAGAAAAAGATAA ATTAAGGAAAATAC—--
GRP-15
GAAAGACAGAAAACAAGGGCAAAACAG GAGATGAGGAAATGTTAAAGGATAAAG GAAAGCCAGAGAGTGAGGGAGAGGCAA AAGAAGGAAAGTCAGAGAGGGAGGGAG AGTCAGAGATGGAGGGAGGATCAGAGA GAGAGGGAAAACCAGAGATAGAGGGAA AGCCAGAGAGTGAAGGAGAGCCAGGGA GTGAAACAAGGGCTGCAGGAAAGCGCC CAGCTGAGGATGATGTACCCAGGAAAG C——-
GRP-18
GGAACCTGGATTCTTTTAATAGTTGTT GAAGCCTCCAGGGGGCCAGGCGGATCA CTTGAGGCCCAGCCTGGCCAACATAGC GAAACCCTGCCTCTACTAAAACCACAA AAATCAGCCGGGCATGGTGGCACACGC TTATAATCCCAGCTACTTGGGACGCTG AGGTGGGATATAGCTTGAACCCGGGAA GGAGACTGCAGTCAGGGAAGCCTAGGG AAGCCTCAGACCAAGGATGATTGAATA ACAAAGAAAAGGTGTAAGTAAAGATCT CCAACTCTTAGGGAGTACTAATTAGGA AAGTTAAGGGTAGAAAAAGATAAATTA AGGAAAATACCGTTGAGAAGCTTAAGA CTGGAGATCTGGATCCCTCGAGTCTAG AGTCGACCTGCA-
GRP-19
AAAAGAAGAAAATAAACACCAAAAACA AGGAGAATAAAATGGCAGCAGACTTGA TGCAAAAAATTAAAAAATTAAGAGAAA TTACAGATGCACCTTTTATTGATTGCA AAAAAGCTTTAGAGCAAACAGGTGCTG ATTTAGATAAAGCAGTTGCTTGATTAC AAGAAAACGGAAAAACTAAAGCACTTA AAAAAGCTGATAGAATTGCTGCTGAAG GTTTAGTTTTTGCTACTAAAAATGAAA CACATGGTGTTATAGTAGAATTAAACT CAGAAACAGACTTTGTTG—-
GRP-24
GGAACCTGGATTCTTTTAATAGTTGTTG AAGCCTCCAGGGGGCCAGGCGGATCACT TGAGGCCCAGCCTGGCCAACATAGCGAA ACCCTGCCTCTACTAAAACCACAAAAAT CAGCCGGGCATGGTGGCACACGCTTATA ATCCCAGCTACTTGGGACGCTGAGGTGG GATATAGCTTGAACCCGGGAAGGAGACT GCAGTCAGGGAAGCCTAGGGAAGCCTCA GACCAAGGATGATTGAATAACAAAGAAA AGGTGTAAGTAAAGATCTCCAACTCTTA GGGAGTACTAATTAGGAAAGTTAAGGGT AGAAAAAGATAAATTAAGGAAAATAC—-
GRP-28
CTGTGGCAAATATTGTGTTCCAAATA AAAGACTTGGTTTCCTCAGATCTACG CCATTTTCAATCTTCCCTAACAATAC GTGCATTTTTAACAAAGCTGGTATTT GAATACTTACCTGAGGTAACATGCCT TGGTATAGTTTCTTCTAAAGTTCACT GCAGGCTGGGTGCTGTGGCTCGTGCC TGTAATCCTAGCACTTTGGGAGGCCG AGTCGGGAGAATTGCTTGAGCCCGGG AGTTCAAGACCAGCCTGCGGGACAAA ATGAGACCCCATCCATA—-
GRP-31
CTGTGATTTTTTTTAAGGTCTTAATA TTTGAAGGAAGTCAACAGTCATTTAT TCCGAATTAAACTTGAGGTTAATAAA GTTTCAATTCGTAATTTTTCCAAACC AACCAATGTAAAAACCCAGATTTTCC TGAATTGAGTCATGTAAGGATTTTTG TAAGTG-
GRP-36
ACGGCAACTGATAGCTTTAAGGAGGA AGAGAGAATAGAGAGAGTGTGTGTTG GGGGGTGTGTGTGTGCGTTTAGTCCT ATAAAATGTTTACTATTGATTTTTTC TGTTTACTACCTCTTTCTCAAATAGG TTGGTTGTGAAGATGGATCTGTGAAA CTATTTCAAATTACCCCAGACAAAAT TCAGTTTGAAAGAAATTTTGATCGGC AGAAAAGTAAGCGTCATTTTTCATGGG-
GRP-37
TCATTTTACAAAAGGAACATTAATATT AATATAAGAAAAGAATTTCTTATACGT ACCAATATGGTATCACATTTCAGCTCA ACATCAGACATGCAATAAATGTATACA AGTACTTCAATTTGCATTAGAACATTT TTAAAGAAATACACAATTATTTCTAAA TTATATTTTATATATAACTAAGGCGGT AAAAGCTTAAGACTGGAGATCTGGATC CCTCGAGTCTAGAGTCGACCTGCAGGC ATGCAAGCTTGGCGTAATCATGGTCAT AGCTGTTTCCTGTGTGAAATTGTTATC CGCTCACAATTCCACACAACATACGAG CCGGAAGCATA—-
GRP-38
TAGAACCCTGAGTGGGAACAGAATCTCAA AATAAATAAATGGAAATGAGATCATGGCT GTAAATGGTAATTATATTATGTCAAAAAC ACCTTTAGAGTTACTTTAGAGCTCCCTAA AAATAACAATTAAAAAGTAGCTTGTAGAG GATATTTCCTCTCTTCTCCTTTACATTTA TGTGGTGTTCAAGGTGGACAAAGTACTTT TG—--
GRP-41
TCAGAAATGAGAATGCACTGGAGGCTGGTG ATTACTTCTGGACCCCCTCTTCCCCATCCA TCGTTTCGGCTAAAAGTCATCATAAATTGG GAGTCCTTCCCTTTACTGGTCTAGAAGTTC CCTCAGGAAGCAGCCGTCACTTCTCTCCCT GCTCTTCACTGAGGAGGGAGGGAAGAGGAG CAAGAGAAGACTTTCCGGTTTTCCAAATGG CAGATTTGGTTTCGGGCATGTGAGGAGCCA CATA—-
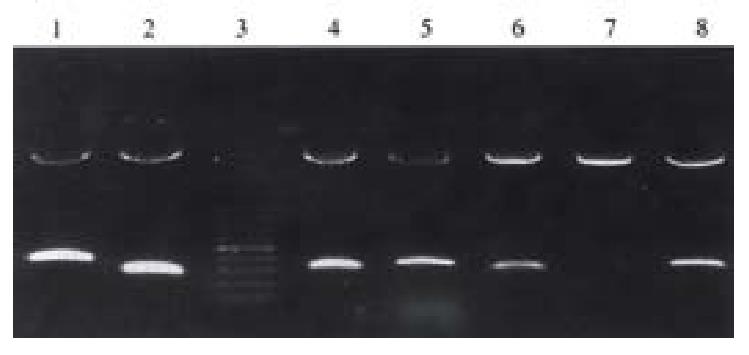
The cDNA fragments of four genes randomly selected from the 13 unknown genes mentioned above were used to produce probes for Northern blot, and the result was in agreement with that of DD-PCR and reverse Northern analysis. All the four genes were specifically (GRP-2) or highly expressed in SGC7901/VCR.
MDR is a major obstacle to tumor chemotherapy. In recent years, progress has been made concerning the mechanism of MDR at the molecular level, which includes the separation and identification of some proteins encoded by MDR-related genes, including P-gp, MRP, LRP and BCRP. All of them are membrane proteins, which engender MDR by reducing the accumulation of chemotherapeutic drugs in the target cells or redistributing the drugs, which consequently reduce the drug concentration in the target organelles. Their expressions differ in different tumors. Certain anti-MDR drugs with some specific MDR mechanisms have been applied clinically. The first generation anti-MDR drugs require that the drug concentration to be high enough to revert MDR, so their side-effects often exceed the tolerance of patients. The second and third generation drugs (PSC833, GF 120918, VX-710, LY335979 etc) cannot revert MDR effectively, either. This phenomenon suggests that tumor cells may tolerate anti-tumor drugs through various mechanisms. The changes at the genetic level are usually more sensitive and prompt than those at the protein level, so screening MDR-related genes at the genetic level has great advantages. One method for screening differential expression genes is mRNA differential display, which contributes substantially in many researches. We employed this technique to screen 20 gene fragments that were highly expressed in gastric cancer MDR cells. This result indicates that MDR involves multi-genes, each playing its own role in the process.Among these genes, seven were separated and identified as known genes.
Human mitochondrial cytochrome C oxidase consists of three subunits. It has essential functions in mitochondrial respiration. Higuchi[39]obtained two highly expressed genes in human head and neck squamous epithelium cancer cells that were cisplatin-resistant, using differential display, and one of the two encodes human mitochondrial cytochrome C oxidase subunit II. It is regarded as a sign of cisplatin-resistance. Denis-Gay et al[40] discovered that the concentration and activity of mitochondrial cytochrome C oxidase did not change in K562 leukemia cells that were adriamycin-resistant, while the concentrations of most other cytochrome oxidases declined significantly. A noteworthy phenomenon recently reported is that methamphetamine or 3,4-methylenedioxymethamphetamine (MDMA) could suppress the activity of mitochondrial cytochrome C oxidase, which implied that they could probably revert MDR[41].
If transferred into tumor 11299 cells, galectin-8 could suppress the formation of the clones by 75%. Galectin-8 is a binding protein of integrin, and has effects against tumor cell adhesion after binding. It can also induce apoptosis, too[43-45].
It is homologous with E. coli ribosomal protein L11. As the drug-resistant mutant strain of E. coli lacks L11, it is postulated that L23 may be related to drug-resistance[46].
Its mRNA (1890 bp) has an open reading frame (ORF), encoding a polypeptide of 417 amino acid residues whose apparent molecular mass is 60 ku. It is the antigen of several autoimmune diseases. In ultraviolet radiation resistant bacteria, an RNA-binding protein was recently discovered to mediate the resistance and to be highly homologous with Ss-A/Ro 60 ku protein. Therefore, Ss-A/Ro ribonucleoprotein autoantigen 60 ku subunit presumably has the similar function in the recovery of cells after UV radiation in higher eukaryotes[47].
Its gene covers 500 kb genomic DNA and is composed of 25 exons. It is highly expressed primarily in fetal brain, encoding a protein whose molecular mass is 91ku. It has a typical Rho GTPase activation protein (rhoGAP) domain, the activation of which induces GAP to inactivate Rho and Ras. Therefore, the inactivation of rhoGAP can activate its target molecules and subsequently influence cell differentiation and migration. Its mutation can result in non-specific X-chromosome linked mental retardation, as its loss of function probably interferes with or affects cell signal transduction pathways, impeding cell migration and axonal growth in the development of cells in the neural system[48,49].
It is a member of the G-protein family, with a molecular mass of about 20 ku. It was first identified to activate cholera toxin ribosyltransferase, and was recently discovered to effect essentially in vesicular transport in cell, and to be an activator of phospholipase D[50]. Human ribosomal protein S13 (RPS13) is a translation initiation factor and is related to cell growth and differentiation[51]. Some of these seven genes are apparently related to MDR, and the relationship of others to MDR remains to be further studied.
Two cDNAs (GRP-24, GRP-41) which are highly expressed in SGC7901/VCR cells, are 100% homologous with human genomic clone fragments. The two genomic clones contain the genes of dynamin, neuroendocrine secretory protein 55 (NESP55), and GANS1. Dynamin is a membrane protein with GTPase activity; its alternative splicing produces six subtypes engaging in endocytosis, exocytosis, pinocytotic vesicle cycle, intracellular and Golgi complex vesicular transport, and cell membrane transport. NESP55 and GANS1 are both G-proteins, involved in cell signal transduction. Further confirmation of homologous genes of the two cDNA fragments is necessary.
We randomly selected 4 cDNA fragments (GRP-2,GRP-19, GRP-28,GRP-31) from the rest 11 unknown genes to do Northern analysis. The result was in agreement with that of reverse Northern analysis; and gave the lengths of the four genes and also confirmed the reliability of reverse Northern blot. The result is conducive to the cloning of the entire length of the genes.
To sum up, we managed to separate and clone 20 significantly highly or specifically expressed genes from drug-resistant gastric cancer SGC7901/VCR cells using DD-PCR. Some known genes are related to drug-resistance and others may enhance drug-resistance. It is necessary to conduct further research on the unknown genes.
Edited by Ma JY
| 1. | Fan K, Fan D, Cheng LF, Li C. Expression of multidrug resistance-related markers in gastric cancer. Anticancer Res. 2000;20:4809-4814. [PubMed] |
| 2. | Jonker JW, Smit JW, Brinkhuis RF, Maliepaard M, Beijnen JH, Schellens JH, Schinkel AH. Role of breast cancer resistance protein in the bioavailability and fetal penetration of topotecan. J Natl Cancer Inst. 2000;92:1651-1656. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 483] [Cited by in RCA: 460] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 3. | Tan B, Piwnica-Worms D, Ratner L. Multidrug resistance transporters and modulation. Curr Opin Oncol. 2000;12:450-458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 288] [Cited by in RCA: 289] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 4. | Liu ZM, Shou NH, Jiang XH. Expression of lung resistance protein in patients with gastric carcinoma and its clinical significance. World J Gastroenterol. 2000;6:433-434. [PubMed] |
| 5. | Zhang LJ, Chen KN, Xu GW, Xing HP, Shi XT. Congenital expression of mdr-1 gene in tissues of carcinoma and its relation with pathomorphology and prognosis. World J Gastroenterol. 1999;5:53-56. [PubMed] |
| 6. | Fan DM, Xiao B, Shi YQ, Ming-Feng , Qiao TD, Chen BJ, Chen Z. A novel cDNA fragment associated with gastric cancer drug resistance was screened out from a library by monoclonal antibody MGr1. World J Gastroenterol. 1998;4:110-111. |
| 7. | Liu Y, Lu MZ, Li QM, Wang YL. Expression of p53 C-myc and P-gp in gastric cancer. Xin Xiaohuabingxue Zazhi. 1997;5:585-586. |
| 8. | You HN, Chen Q, Jiang HP, Li DG, Zhang WZ. The influence of P-glycoprotein expression on the prognosis of gastric carcinoma. Xin Xiaohuabingxue Zazhi. 1997;5:23-24. |
| 9. | Zhou WJ, Pan FQ. Significance of P-gp and P53 in patients with gastric cancer. Huaren Xiaohua Zazhi. 1998;6:318-319. |
| 10. | Xiao B, Shi YQ, Zhao YQ, You H, Liu XL, Fan DM. Expression of Fas gene in gastric cancer cells transducted with Fas gene. Huaren Xiaohua Zazhi. 1998;6:400-403. |
| 11. | Cheng SD, Wu YL, Zhang YP, Qiao MM, Guo QS. Abnormal drug accumulation in multidrug resistant gastric carcinoma cells. Shijie Huaren Xiaohua Zazhi. 2001;9:131-134. |
| 12. | Lage H, Jordan A, Scholz R, Dietel M. Thermosensitivity of multidrug-resistant human gastric and pancreatic carcinoma cells. Int J Hyperthermia. 2000;16:291-303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 36] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 13. | Yin L, Chen K, Li D. [Inherent mdr-1 gene expression in fresh tumor tissue specimens from several high-incidence malignancies]. Zhonghua Zhongliu Zazhi. 1997;19:420-422. [PubMed] |
| 14. | Gürel S, Yerci O, Filiz G, Dolar E, Yilmazlar T, Nak SG, Gülten M, Zorluoğlu A, Memik F. High expression of multidrug resistance-1 (MDR-1) and its relationship with multiple prognostic factors in gastric carcinomas in patients in Turkey. J Int Med Res. 1999;27:79-84. [PubMed] |
| 15. | Anzai H, Kitadai Y, Bucana CD, Sanchez R, Omoto R, Fidler IJ. Expression of metastasis-related genes in surgical specimens of human gastric cancer can predict disease recurrence. Eur J Cancer. 1998;34:558-565. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 23] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 16. | Motoo Y, Su SB, Nakatani MT, Sawabu N. Expression of multidrug resistance gene (mdr-1) mRNA in gastric and colorectal cancers. Anticancer Res. 1998;18:1903-1906. [PubMed] |
| 17. | Yeh KH, Chen CL, Shun CT, Lin JT, Lee WJ, Lee PH, Chen YC, Cheng AL. Relatively low expression of multidrug resistance-1 (MDR-1) and its possible clinical implication in gastric cancers. J Clin Gastroenterol. 1998;26:274-278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 18. | Son YS, Suh JM, Ahn SH, Kim JC, Yi JY, Hur KC, Hong WS, Muller MT, Chung IK. Reduced activity of topoisomerase II in an Adriamycin-resistant human stomach-adenocarcinoma cell line. Cancer Chemother Pharmacol. 1998;41:353-360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 19. | Endo K, Maehara Y, Kusumoto T, Ichiyoshi Y, Kuwano M, Sugimachi K. Expression of multidrug-resistance-associated protein (MRP) and chemosensitivity in human gastric cancer. Int J Cancer. 1996;68:372-377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 20. | Liu XL, Xiao B, Yu ZC, Guo JC, Zhao QC, Xu L, Shi YQ, Fan DM. Down-regulation of Hsp90 could change cell cycle distribution and increase drug sensitivity of tumor cells. World J Gastroenterol. 1999;5:199-208. [PubMed] |
| 21. | Darimont BD. The Hsp90 chaperone complex A potential target for cancer therapy. World J Gastroenterol. 1999;5:195-198. [PubMed] |
| 22. | Xiao B, Shi YQ, Zhao YQ, You H, Wang ZY, Liu XL, Yin F, Qiao TD, Fan DM. Transduction of Fas gene or Bcl-2 antisense RNA sensitizes cultured drug resistant gastric cancer cells to chemotherapeutic drugs. World J Gastroenterol. 1998;4:421-425. [PubMed] |
| 23. | Yin F, Shi YQ, Zhao WP, Xiao B, Miao JY, Fan DM. Suppression of P-gp induced multiple drug resistance in a drug resistant gastric cancer cell line by overexpression of Fas. World J Gastroenterol. 2000;6:664-670. [PubMed] |
| 24. | Shi YQ, Xiao B, Miao JY, Li MF, Qiao TD, Chen BJ, Chen Z, Han JL, Zhou SJ, Fan DM. A novel cDNA fragment associated with gastric cancer drug resistance screened from a library by mAb MGr1. Huaren Xiaohua Zazhi. 1998;6:656-659. |
| 25. | Xiao B, Shi YQ, Zhao YQ, You H, Wang ZY, Liu XL, Yin F, Qiao TD, Fan DM. Transductionof fas gene or bcl-2 antisense RNA sensitizes cultured drug resistant gastric cancer cells to chemotherapeutic drugs. Huaren Xiaohua Zazhi. 1998;6:675-679. |
| 26. | Liu B, Staren E, Iwamura T, Appert H, Howard J. Effects of Taxotere on invasive potential and multidrug resistance phenotype in pancreatic carcinoma cell line SUIT-2. World J Gastroenterol. 2001;7:143-148. [PubMed] |
| 27. | Wang X, Shi Y, Zhao Y. [Differential display of vincristine-resistant proteins in gastric cancer cell line SGC7901]. Zhonghua Zhongliu Zazhi. 2001;23:281-284. [PubMed] |
| 28. | Lin HL, Liu TY, Wu CW, Chi CW. Berberine modulates expression of mdr1 gene product and the responses of digestive track cancer cells to Paclitaxel. Br J Cancer. 1999;81:416-422. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 81] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 29. | Zheng G, Han F, Liu X. [Drug resistance mechanism of doxorubicin-resistant human gastric cancer cells BGC-823/DOX]. Zhonghua Waike Zazhi. 1997;35:325-328. [PubMed] |
| 30. | Varga A, Sokolowska-Kohler W, Presber W, Von Baehr V, Von Baehr R, Lucius R, Volk D, Nacsa J, Hever A. Toxoplasma infection and cell free extract of the parasites are able to reverse multidrug resistance of mouse lymphoma and human gastric cancer cells in vitro. Anticancer Res. 1999;19:1317-1324. [PubMed] |
| 31. | Nakamura T, Oka M, Aizawa K, Soda H, Fukuda M, Terashi K, Ikeda K, Mizuta Y, Noguchi Y, Kimura Y. Direct interaction between a quinoline derivative, MS-209, and multidrug resistance protein (MRP) in human gastric cancer cells. Biochem Biophys Res Commun. 1999;255:618-624. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 56] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 32. | Kellner U, Hutchinson L, Seidel A, Lage H, Danks MK, Dietel M, Kaufmann SH. Decreased drug accumulation in a mitoxantrone-resistant gastric carcinoma cell line in the absence of P-glycoprotein. Int J Cancer. 1997;71:817-824. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 33. | Nitta A, Chung YS, Nakata B, Yashiro M, Onoda N, Maeda K, Sawada T, Sowa M. Establishment of a cisplatin-resistant gastric carcinoma cell line OCUM-2M/DDP. Cancer Chemother Pharmacol. 1997;40:94-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 34. | Wang X, Huang YX, Wen QS, Wang QL. Silver-staing mRNA differ-ential display method and cloning of tumor related genes in HepG2 cell line. Di-si Junyi Daxue Xuebao. 2001;22:843-845. |
| 35. | Wang L, Lu W, Chen YG, Zhou XM, Gu JR. Comparison of gene expression between normal colon mucosa and colon carcinoma by means of messenger RNA differential display. World J Gastroenterol. 1999;5:533-534. [PubMed] |
| 36. | Ji F, Peng QB, Zhan JB, Li YM. Study of differential polymerase chain reaction of C-erbB-2 oncogene amplification in gastric cancer. World J Gastroenterol. 1999;5:152-155. [PubMed] |
| 37. | You H, Xiao B, Cui DX, Shi YQ, Fan DM. Two novel gastric cancer-associated genes identified by differential display. World J Gastroenterol. 1998;4:334-336. [PubMed] |
| 38. | Cui DX, Yan XJ, Su CZ. Differentially expressed genes were isolated in gastric carcinoma by optimised differential display PCR. Shijie Huaren Xiaohua Zazhi. 1999;7:139-144. |
| 39. | Higuchi E. [Up-regulation of human chorionic gonadotropin alpha subunit gene and human mitochondrial cytochrome c oxidase subunit II gene in cis-Diamminedichloroplatinum(II)-resistant human head and neck squamous carcinoma cells]. Hokkaido Igaku Zasshi. 1999;74:231-238. [PubMed] |
| 40. | Denis-Gay M, Petit JM, Mazat JP, Ratinaud MH. Modifications of oxido-reductase activities in adriamycin-resistant leukaemia K562 cells. Biochem Pharmacol. 1998;56:451-457. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 41. | Burrows KB, Gudelsky G, Yamamoto BK. Rapid and transient inhibition of mitochondrial function following methamphetamine or 3,4-methylenedioxymethamphetamine administration. Eur J Pharmacol. 2000;398:11-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 103] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 42. | Gopalkrishnan RV, Roberts T, Tuli S, Kang D, Christiansen KA, Fisher PB. Molecular characterization of prostate carcinoma tumor antigen-1, PCTA-1, a human galectin-8 related gene. Oncogene. 2000;19:4405-4416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 45] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 43. | Hadari YR, Arbel-Goren R, Levy Y, Amsterdam A, Alon R, Zakut R, Zick Y. Galectin-8 binding to integrins inhibits cell adhesion and induces apoptosis. J Cell Sci. 2000;113:2385-2397. [PubMed] |
| 44. | Bassen R, Brichory F, Caulet-Maugendre S, Bidon N, Delaval P, Desrues B, Dazord L. Expression of Po66-CBP, a type-8 galectin, in different healthy, tumoral and peritumoral tissues. Anticancer Res. 1999;19:5429-5433. [PubMed] |
| 45. | Camby I, Belot N, Rorive S, Lefranc F, Maurage CA, Lahm H, Kaltner H, Hadari Y, Ruchoux MM, Brotchi J. Galectins are differentially expressed in supratentorial pilocytic astrocytomas, astrocytomas, anaplastic astrocytomas and glioblastomas, and significantly modulate tumor astrocyte migration. Brain Pathol. 2001;11:12-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 127] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 46. | McElwain KB, Boynton JE, Gillham NW. A nuclear mutation conferring thiostrepton resistance in Chlamydomonas reinhardtii affects a chloroplast ribosomal protein related to Escherichia coli ribosomal protein L11. Mol Gen Genet. 1993;241:564-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 47. | Chen X, Quinn AM, Wolin SL. Ro ribonucleoproteins contribute to the resistance of Deinococcus radiodurans to ultraviolet irradiation. Genes Dev. 2000;14:777-782. [PubMed] |
| 48. | Billuart P, Bienvenu T, Ronce N, des Portes V, Vinet MC, Zemni R, Roest Crollius H, Carrié A, Fauchereau F, Cherry M. Oligophrenin-1 encodes a rhoGAP protein involved in X-linked mental retardation. Nature. 1998;392:923-926. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 313] [Cited by in RCA: 310] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 49. | Ljubimova JY, Khazenzon NM, Chen Z, Neyman YI, Turner L, Riedinger MS, Black KL. Gene expression abnormalities in human glial tumors identified by gene array. Int J Oncol. 2001;18:287-295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 18] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 50. | Lebeda RA, Haun RS. Cloning and characterization of the human ADP-ribosylation factor 4 gene. Gene. 1999;237:209-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 51. | Ito T, Kim GT, Shinozaki K. Disruption of an Arabidopsis cytoplasmic ribosomal protein S13-homologous gene by transposon-mediated mutagenesis causes aberrant growth and development. Plant J. 2000;22:257-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 115] [Article Influence: 4.6] [Reference Citation Analysis (0)] |