Published online Aug 15, 1999. doi: 10.3748/wjg.v5.i4.324
Revised: July 3, 1999
Accepted: July 19, 1999
Published online: August 15, 1999
AIM: To study the significance of p53 gene in hepatocarcino genesis through analyzing codon 249 mutations of p53 gene in non-neoplastic liver tissues.
METHODS: Codon 249 mutation was detected using single-strande d conformational polymorphism analysis and allele-specific PCR in liver tissues from 10 cases of chronic hepatitis, 5 cases of cirrhosis and 20 cases of HCCs.
RESULTS: The detection rate of codon 249 mutation in chronic hepatitis, cirrhosis and pericancerous tissues was 70% (7/10), 100% (5/5) and 70% (14/20), respectively by AS-PCR. These mutations could not be detected b y SSCP analysis. The detection rates were 65% (13/20) and 45% (9/20) in cancerous tissues by AS-PCR and SSCP analysis.
CONCLUSION: Codon 249 mutations of p53 gene were very popular in non-neoplastic liver tissues though the number of those mutant cells was only in subsection. Those mutations in cancerous tissues might take place in the stage before the formation of tumor.
- Citation: Peng XM, Yao CL, Chen XJ, Peng WW, Gao ZL. Codon 249 mutations of p53 gene in non-neoplastic liver tissues. World J Gastroenterol 1999; 5(4): 324-326
- URL: https://www.wjgnet.com/1007-9327/full/v5/i4/324.htm
- DOI: https://dx.doi.org/10.3748/wjg.v5.i4.324
Since AGG to AGT mutations in codon 249 of the p53 gene resulting in an arginine to serine substitution were observed in up to 50%-60% of HCC from Southern Africa and some provinces of China[1,2], the significance of p53 gene and the specific mutations of codon 249 in hepatocarcinogenesis has been widely accepted[3,4]. It is controversial, however, whether the specific mutations of codon 249 are involved in the initiation of HCC. Some authors believed that codon 249 AGT transversions were only involved in the differentiation stage of HCC because they had found that these mutations were only observed in HCC of later stage or bad differentiation[5,6]. Obviously, the crucial point to solve this problem is to clarify whether codon 249 AGT transversions take place in non-neoplastic tissues, such as hepatitis or cirrhotic tissues. Direct DNA sequencing, single-stranded conformational polymorphism (SSCP) and restriction fragment length polymorphism (RFLP) of PCR products were the main methods to detect gene mutations in the past. With lower sensitivity, these methods can not be used t o demonstrate mutations in tissues other than massive, uniform tumor tissues. Recently, AS-PCR, a 100-fold more sensitive assay, has been used in the detection of codon 249 AGT transversions[7]. For these reasons, the occurring time of codon 249 mutation was investigated using AS-PCR to detect codon 249 mutations of p53 gene in non-neoplastic liver tissues from Chinese patients in this study.
Specimens Ten pieces of liver biopsy specimens of chronic hepatitis B, and 5 pieces of autopsy liver specimens were collected from our third affiliated hospital, and 20 pieces of surgically dissected specimens of HCCs were from our cancer hospital. All patients were Chinese living in Southern China. Ca ncerous and pericancerous tissues of surgically dissected specimens of HCCs were separated by pathologists genomic DNA was extracted by digesting with proteinase K and followed by phenol-chloroform extraction. All extract products were stored at -70 °C.
Reagents All primers used in this research were designed according to the sequence of HSp53G from GenBank, and are shown in Table 1.
| Code | Oligonucleotide sequence | Position |
| S1 | 5′-GGCGACAGAGCGAGATTCCA | 13890-13909 |
| A1 | 5′-GATTCTCTTCCTCTGTGCGC | 14534-14515 |
| S2 | 5′-TGGGCGGCATGAACCGGAGT | 14055-14074 |
| S3 | 5′-TGGGCGGCATGAACCGGAGG | 14055-14074 |
| A2 | 5′-GGGTCAGCGGCAAGCAGAGG | 14175-14156 |
PCR Using primers S1 and A2 from intron sequences upper and lower flank of exon 7, 286 bp fragment was amplified. Procedures were decribed in brief as follows: 0.5 μg genomic DNA was added to 30 μL of PCR mixture. The mixture was denatured at 98 °C for 5 min, and then added to 1.5 U Taq polymerase at 80 °C. The amplification was carried out for 30 cycles composed of 40 s at 94 °C, 50 s at 64 °C, 1 min at 72 °C, and another 10 min at 72 °C after the last cycle. Amplified products were visualized by running them on 2% agarose gel and staining with ethidium bromide.
SSCP Twenty μL of PCR products was precipitated at -20 °C for 1 h by adding 2.5 vol of alcohol and 0.1 vol of 4 M sodium acetate. Pellets were re-suspended in 10 μL of formamide dye mixture (95% formamide; 20 mmol/L EDTA; 0.05% bromphenol blue). Samples were heated at 95 °C for 5 min, chilled on ice and immediately loaded (5 μL)on 6% polyacrylamide gel. Gels were run at 40 W for 4 h at room temperature. Silver staining was used to visualize the bands, HepG2215 cell line without codon 249 mutation served as negative control.
DNA sequencing SSCP positive products were purified using low-melting agarose. DNA sequencing was carried out using automatic sequencing system.
AS-PCR Kirby′s- semi-nested PCR protocol was modified by nested PCR for AS-PCR analysis in order to promote the specificity of the amplified products. About 0.25 μg genomic DNA was added to 50 μL of PCR I mixture containing primers S1 and A1. The amplification was carried out for 30 cycles composed of 40 s at 94 °C, 40 s at 55 °C, and 1 min at 72 °C. Five μL PCR I product (1:100) was added to a flesh tube, and 45 μL of PCR II mixture containing primers S2 (allele-specific primer with T replacement in the third base pair of codon 249) or S3 and A2 was added. The amplification was per formed just as above. Amplified products were analyzed by running on 2% agarose gel and visualized by staining with ethidium bromide. 121 bp band was observed.
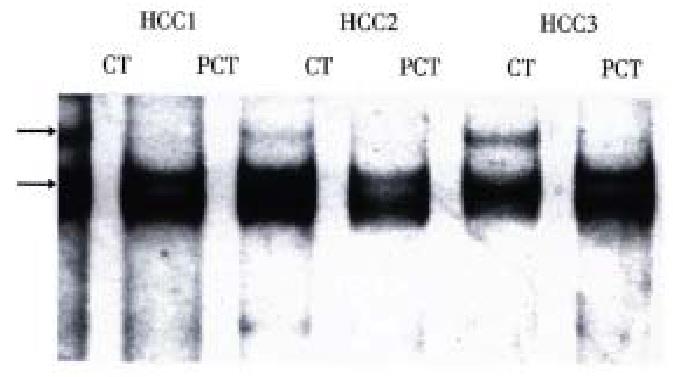
Exon 7 was successfully amplified in all tissue samples with nucleotide number as expected. PCR/SSCP analysis of tissue samples showed that SSCP positive results were only observed in cancerous tissues of some HCCs (Table 2), and that the pattern of band shift was identical (Figure 1). All SSCP positive amplified products were identified to be codon 249 AGT transversions by DNA sequencing.
| Tissue types | Case number | SSCP positive cases | Positive rates (%) |
| Chronic hepatitis | 10 | 0 | 0 |
| Cirrhosis | 5 | 0 | 0 |
| Pericancerous tissues | 20 | 0 | 0 |
| Cancerous tissues | 20 | 9 | 45 |
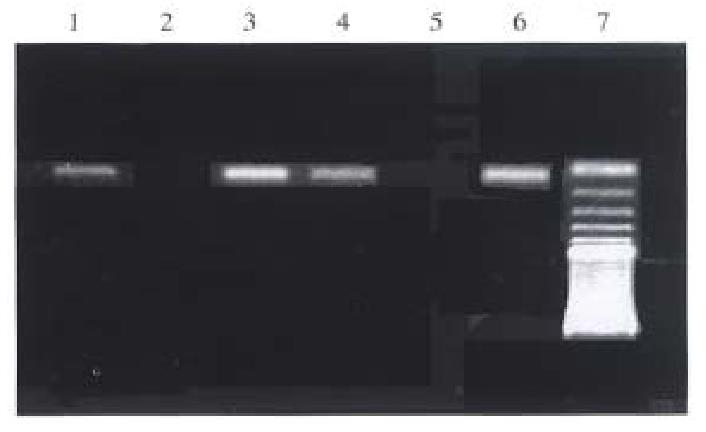
All genomic DNA samples were satisfactorily amplified when universal primers were used in the first step of nested PCR. Ideal bands were obtained in the second amplification when either primer S2 or primer S3 was used. Typical results are shown in Figure 2. The results of codon 249 mutation detection in DNA are summarized in Table 3. The corresponding rate of the detection of codon 249 mutation between pericancerous tissue and cancerous tissue was 95% (19/20). All the 9 cases of HCCs with positive SSCP were positive for AS-PCR both in cancerous tissue and pericancerous tissue.
| Tissues types | Cases | Detective rates of codon 249 mutation (%) |
| Chronic hepatitis | 10 | 70 |
| Cirrhosis | 5 | 100 |
| Pericancerous tissues | 20 | 70 |
| Cancerous tissues | 20 | 65 |
Southern China is one of the endemic areas with a high incidence of HCC in the world. Codon 249 AGT transversion is a major pattern of mutations in HCC from this area, with mutant rate up to 32.9% (23/70). The ratio of codon 249 mutation to all kinds of mutations of p53 gene was 95.7% (22/23)[4]. Therefore, cancerous and non-neoplastic tissue samples from this area is ideal for the study on the effect of codon 249 mutation on hepatocarcinogenesis. Codon 249 mutation may make the mutant-type p53 protein lose its own functions, or even gain some new functions. Loss of p53 gene can increase proliferating cells, diploid cells and G1 to G0 ratios, and even made the mutant cells not show apoptosis after exposure to ionizing radiation or therapeutic drugs[8]. Codon 249 mutation may gain a new ability to promote G0 to G1 and/or M to G1 transition in hepatocytes of transgenic mice[9]. Although the loss of p53 gene or codon 249 mutation can change the cell proliferating mode, it would only p lay a minor role in hepatocarcinogenesis, and the researches of this mutation would be of little importance in prevention and treatment of HCC, if the codon 249 mutation only took place in the late stage of HCC.
Some laborious and time-consuming methods, including cloning analysis of RFLP-PCR products were used in detection of codon 249 mutation in non-neoplastic liv er tissues of aflatoxin-exposed cultured human hepatocytes and extracted genomic DNA from individuals exposed to dietary aflatoxin. The results supported the hypothesis that AFB was a causative mutagen in HCC[10]. A simple method, AS-PCR, was developed, and used in the detection of codon 249 mutation as well. Codon 249 AGT transversions were detected by AS-PCR in 83.3% (5/6) of non- neoplastic liver tissues from high-incidence regions, while none from low-incidence regions of HCC. Codon 249 mutation was detected in 70%-100% of 35 subjects of non-neoplastic tissues living in southern China, an endemic areas of HCC using AS-PCR. In comparison of the results between cancerous tissues and pericancerous tissues, the occurrence of mutant codon 249 in cancerous tissues was found not to be an independent event. Thus, codon 249 mutation might take place before or in the initial stage of HCC development.
The cells with mutant p53 gene extensively existed in non-neoplastic liver tissues though these cells might be in subsection because mutation could not be detected by SSCP analysis. It is still unclear why only a small portion of mutant cells develop into cancer, since about 25% patients with chronic hepatitis or cirrhosis may die from HCC and only 33.7% cases of these HCCs may carry codon 249 mutation.
Dr. Xiao-Mou Peng, male, born on October 28, 1963 in Chaling County, Hunan Province, graduated from Department of Medicine, Hunan University of Medical Sciences, now instructor, engaged in the study on viral hepatitis, having 16 papers published.
Edited by MA Jing-Yun
| 1. | Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49-53. [PubMed] |
| 2. | Hsu IC, Metcalf RA, Sun T, Welsh JA, Wang NJ, Harris CC. Mutational hotspot in the p53 gene in human hepatocellular carcinomas. Nature. 1991;350:427-428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1066] [Cited by in RCA: 969] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 3. | Bressac B, Kew M, Wands J, Ozturk M. Selective G to T mutations of p53 gene in hepatocellular carcinoma from southern Africa. Nature. 1991;350:429-431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 972] [Cited by in RCA: 872] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 4. | Peng XM, Peng WW, Zhou YP. Studies on mutations of p53 gene in HCCs. Zhongshan Yike Daxue Xuebao. 1997;18:245-248. |
| 5. | Slagle BL. p53 mutations and hepatitis B virus: cofactors in hepatocellular carcinoma. Hepatology. 1995;21:597-599. [PubMed] |
| 6. | Murakami Y, Hayashi K, Hirohashi S, Sekiya T. Aberrations of the tumor suppressor p53 and retinoblastoma genes in human hepatocellular carcinomas. Cancer Res. 1991;51:5520-5525. [PubMed] |
| 7. | Kirby GM, Batist G, Fotouhi-Ardakani N, Nakazawa H, Yamasaki H, Kew M, Cameron RG, Alaoui-Jamali MA. Allele-specific PCR analysis of p53 codon 249 AGT transversion in liver tissues from patients with viral hepatitis. Int J Cancer. 1996;68:21-25. [PubMed] |
| 8. | Yin L, Ghebranious N, Chakraborty S, Sheehan CE, Ilic Z, Sell S. Control of mouse hepatocyte proliferation and ploidy by p53 and p53ser246 mutation in vivo. Hepatology. 1998;27:73-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 23] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 9. | Nishida N, Fukuda Y, Ishizaki K, Nakao K. Alteration of cell cycle-related genes in hepatocarcinogenesis. Histol Histopathol. 1997;12:1019-1025. [PubMed] |
| 10. | Aguilar F, Hussain SP, Cerutti P. Aflatoxin B1 induces the transversion of G--> T in codon 249 of the p53 tumor suppressor gene in human hepatocytes. Proc Natl Acad Sci USA. 1993;90:8586-8590. [PubMed] |