Published online Aug 15, 1998. doi: 10.3748/wjg.v4.i4.320
Revised: January 2, 1998
Accepted: February 14, 1998
Published online: August 15, 1998
AIM: To study the role of HBV especially HBx Open Reading Frame (ORF) in the development of hepatocellular carcinoma (HCC).
METHODS: HBV 3.2 kb fragment was retrieved by digesting recombinant plasmid pBR322-2HBV with EcoR I, and HBx 0.59 kb fragments by digesting HBV-DNA with BamH I and Bgl II. These fragments were labelled with digoxigenin to get HBV-DNA and HBx-DNA probes. HBV-DNA was detected in HCC by dot blot and Southern blot hybridization with HBV-DNA probe, so the positive specimens in which HBV-DNA were integrated were selected. HBx-DNA was subsequently detected in the selected specimens with HBx-DNA probe.
RESULTS: HBV-DNA was detected in 75% HCC, among which integrated type, integrated + free type covered 63.6% and 36.4%. There was no free type. HBx-DNA was detected in 90.5% specimens of integrated type.
CONCLUSION: Hepatocarcinogenesis was highly related to HBV-DNA integration, and HBV-DNA mainly integrated into chromosome with incomplete virus DNA fragments among which HBx fragment was the predominant one.
- Citation: Gao FG, Sun WS, Cao YL, Zhang LN, Song J, Li HF, Yan SK. HBx-DNA probe preparation and its application in study of hepatocarcinogenesis. World J Gastroenterol 1998; 4(4): 320-322
- URL: https://www.wjgnet.com/1007-9327/full/v4/i4/320.htm
- DOI: https://dx.doi.org/10.3748/wjg.v4.i4.320
HBx gene, which is the smallest one of the four open reading frames (ORF) of HBV, has the transactivation function and is closely related to hepatocarcinogenesis[1,2]. In the study of hepatocellular carcinoma (HCC) formation, HBx-DNA probe is needed to investigate HBx gene integration and its mRNA transcription. We retrieved HBx-DNA fragment by digesting HBV-DNA and labeled it with digoxingenin. To study the HBx gene function in hepatocarcinogenesis, HBV-DNA was detected first in HCC by dot blot and Southern blot hybridization, and the positive specimens in which HBV-DNA were integrated were selected. Subsequently, integrated HBx-DNA was detected in the selected specimens with HBx-DNA probe. This study may provide some fundamental data for further research into hepatocarcinogenesis.
Subjects HCC tissues were obtained at surgical resection from Jinan Qianfeshan Hospital, which were confirmed later by pathology, and stored at -20 °C immediately after surgical resection for use.
Reagents Recombinant plasmid pBR322-2HBV is provided by Professor CB and DIG Hight Prime Labeling and Detection Starter Kit is product of Boehringer Mannheim Company, Germany. Restriction Endonucleases EcoR I, BamH I, Bgl II, Proteinase K are purchased from Huamei Company.
HBV-DNA retrieval Isolation of recombinant plasmid pBR322-2HBV and HBV-DNA retrieval were carried out as described by CAO et al[3]. pBR322-2HBV was transformed into the Escherichia coli strain HB101. A 500 mL bacterial culture was grown to OD600 = 1.0, chloramphenicol was added to a final concentration of 170 mg/L, and the culture was continued for another 4 h. The extracted pBR322-2HBV was digested with EcoR I, and electrophoresis was performed in 1.2% agarose gel. HBV-DNA 0.59 kb fragment (n1398-n1992) was retrieved according to the standard method[4].
DIG DNA labeling HBV and HBx-DNA labeling procedures were described before[3]. Template DNA (1.0 μg) and sterile redist water were added to a reaction vial. Denature the DNA and add DIG high prime and incubate for 20 h at 37 °C. Reaction was stopped by addtion of EDTA. Quantification of labeling efficiency was performed according to instructions of the manufacturer.
DNA extraction Genomic DNA from the tumors were isolated according to the standard methods[5].
HBV-DNA detection Denatured genomic DNA 5 μL, λDNA 10 ng, HBV-DNA 10 pg, and 1.0 pg were heated in a boiling water bath for 5 min-10 min, and chilled quickly in an ice/ethanol bath for 5 min. They were mixed with 20 × SSC and tipped to nitrocellulose filters. The filters were prehybridized for 8-12 h, hybridized for 24-36 h at 42 °C, washed and subjected to immunodetection according to the standard dot blot hybridization procedures.
Southern blot hybridization Positive specimens DNA which had been verified containing HBV-DNA by dot blot hybridization was then digested completely with specific restriction endonucleases, electrophoresed in agarose gels, denatured, and transferred to nitrocellulose filters. The filters were performed with standard Southern blot procedures.
HBx-DNA detection Selected positive specimens which had integ rated HBV-DNA only were confirmed by Southern blot hybridization and standard dot blot hybridization substituting HBV-DNA probe with HBx-DNA probe was performed.
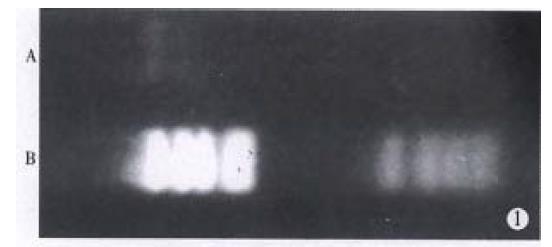
Retrieved HBx-DNA and marker pBR322/Hae III were subjected to electrophoresis in 1.5% agarose gels. HBx-DNA was 0.59 kb fragment and its concentration was 0.1 g/L (Figure 1).
Confirmed with homologous DNA by dot blot hybridization, HBV-DNA probe and HBx-DNA probe all had a high sensitivity of 1.0 pg, and had high specificity confirmed with heterologous DNA.
Thirty-three cases (75%) were confirmed to be infected by HBV previously with HBV-DNA probe by dot blot hybridization, and 11 cases had no evidences of HBV-DNA existence.
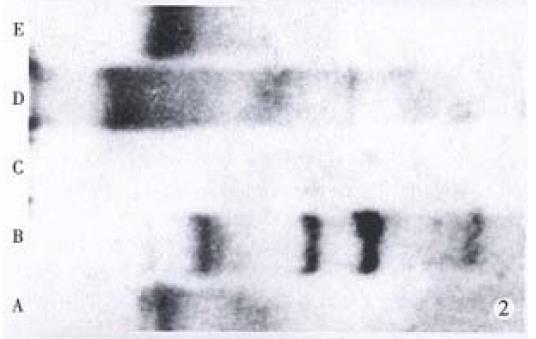
Based on HBV-DNA integration into chromosome or free existing in plasma, HBV-DNA positive HCC specimens can be divided into three types: integrated type which has integrated HBV-DNA only; free type which has free HBV-DNA in plasma;and integrated + free type which has HBV-DNA both in chromosome and in plasma. In this study, the integrated type and integrated + free type covered 63.6% and 36.4%. Free type was not found (Figure 2).
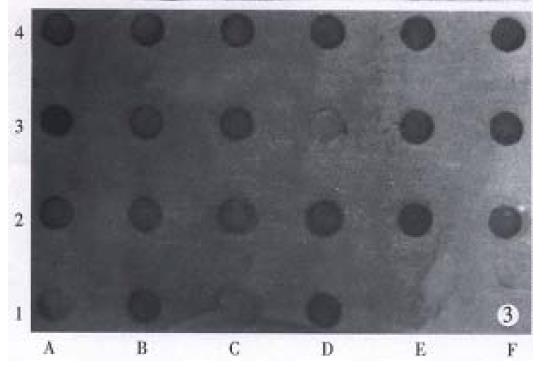
Confirmed with HBx-DNA probe by dot blot hybridization, 19 (90.5%) of 21 cases of integrated type HCC specimens, showed positive HBx-DNA (Figure 3).
Digoxigenin (DIG), a steroid hapten, is coupled to dUTP via an alkali-labile ester-bond. For DNA labeling, DIG-11-dUTP was labeled into HBx-DNA strands. DIG-labeled HBx-DNA probe was used for hybridization to membrane blotted nucleic acids and the hybridized probe was immunodetected with anti-digoxigenin-AP and then was visualized with the colorimetric substrates NBT/ BCIP. HBx-DNA probe prepared by us can detect 1.0 pg homologous DNA and had no reaction with heterologous λDNA which indicated a high sensitivity and specificity. The labeling process need no expensive equipment and without radioactive pollution. In this study, the HBx gene integration of integrated type HCC were detected using HBx-DNA probe, and HBx-DNA probe was used to observe the transcription ratio of integrated HBx-DNA. All these may provide some fundamental data for further research of hepatocarcinogenesis.
In our study, 75% HCC patients were found to be infected with HBV previously. Of all HBV-DNA positive HCC specimens, the integrated type and integrated + free type covered 63.6% and 36.4%, which indicated that HBV-DNA mainly integrated into cell chromosomes. Robinson et al[6] drew the similar conclusion. With PCR technique, Unsal H et al[7] studied the HBx existence in 80 HCC cases from Europe, Asia and Africa. The results showed that 78% cases had HBx-DNA including 28 cases from China. In this study, using intact HBV-DNA probe by Southern blot hybridization, integrated type HCC was selected first, and then integrated HBx-DNA was detected from the selected specimens. Excluding the possibility of pseudopositive results caused by free type HBV-DNA existing in plasma, our study can reflect HBx-DNA integration status exactly and efficiently. Of the integrated type HCC, 90.5% showed HBx-DNA integration, which suggest the important role of HBX gene in the development of HCC. On the other hand, the difference among HBV subgenic integration percentage may indicate that HBV-DNA integration was fragmental and always accompanied by virus DNA deletion.
It was reported that the integrated HBx gene can be transcribed as temple and hence guided HBx protein synthesis[8]. By now, more and more evidences have demonstrated that HBx protein has a transactivation function and integrated HBx-DNA or recombinated one with flank chromosome sequences has even stronger transactivation function than wild type one. Benn J et al [9] discovered that HBx protein deregulated checkpoint controls and consequently promoted the G0/G1 and G2/ M shift of cell cycle by activating ras or other unknown factors. Greenblatt MS et al[10] found that HBx protein can interact with p53 and suppress normal function of p53 protein. All these findings indicated that HBx function and the interaction among HBx protein, oncogene and tumor suppressor gene may play important roles in the development of HCC associated with HBV infection. Obviously, HBx mRNA transcription and its relationship with oncogene activation and tumor suppressor gene inactivation should be focused in further researches.
We would like to thank Professor Liu CB for providing us the recombinant plasmid pBR322-2HBV.
Project supported by the Natural Science Foundation of Shandong Province, No.931237222.
| 1. | Kekulé AS, Lauer U, Weiss L, Luber B, Hofschneider PH. Hepatitis B virus transactivator HBx uses a tumour promoter signalling pathway. Nature. 1993;361:742-745. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 277] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 2. | Twu JS, Lai MY, Chen DS, Robinson WS. Activation of protooncogene c-jun by the X protein of hepatitis B virus. Virology. 1993;192:346-350. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 89] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 3. | Cao YN, Sun WS, Song J, Wang DF, Zhang LN, Cao YL. The preparation of HBV-DNA probe labeled with digoxigenin. Acta Acad Med Shandong. 1993;31:279-282. |
| 4. | Wang SW. Gene diagnostic technique. Beijing: Beijing Medical University and PUMC Press. 1993;70-72. |
| 5. | Sambrook J. Molecular cloning. 2nd ed. New York: Cold Spring Harbor Labo-ratory Press. 1992;. |
| 6. | Robinson WS. The role of hepatitis B virus in development of primary hepa-tocellular carcinoma: Part II. J Gastro Hepat. 1993;8:96-105. |
| 7. | Unsal H, Yakicier C, Marçais C, Kew M, Volkmann M, Zentgraf H, Isselbacher KJ, Ozturk M. Genetic heterogeneity of hepatocellular carcinoma. Proc Natl Acad Sci USA. 1994;91:822-826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 109] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 8. | Wang WL, London WT, Feitelson MA. Hepatitis B x antigen in hepatitis B virus carrier patients with liver cancer. Cancer Res. 1991;51:4971-4977. [PubMed] |
| 9. | Benn J, Schneider RJ. Hepatitis B virus HBx protein deregulates cell cycle checkpoint controls. Proc Natl Acad Sci USA. 1995;92:11215-11219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 236] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 10. | Greenblatt MS, Bennett WP, Hollstein M, Harris CC. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54:4855-4878. [PubMed] |