Published online Feb 28, 2025. doi: 10.3748/wjg.v31.i8.101585
Revised: December 4, 2024
Accepted: January 6, 2025
Published online: February 28, 2025
Processing time: 125 Days and 13.2 Hours
Ras-related protein Rab24, which belongs to the small GTPase family, plays a crucial role in regulating intracellular protein trafficking. Dysregulation of Rab24 has been recently identified in hepatocellular carcinoma (HCC). However, its clinical significance and tumor related effects remain to be further clarified.
To explore the expression pattern of Rab24 and its role in HCC progression.
The expression profile of Rab24 was tested in HCC tissues together with adjacent tissues from transcriptional, mRNA, and protein levels. The prognostic role of Rab24 in HCC was assessed by univariate and multivariate analyses. Clinical outcomes were evaluated by the Kaplan-Meier analysis and log-rank test. The effect of Rab24 on cell proliferation was tested through cellular experiments and xenograft experiments.
Rab24 expression was elevated in HCC tissues compared to adjacent liver tissues. High expression of Rab24 was significantly associated with larger tumor size and advanced tumor stage. Moreover, HCC patients with high Rab24 expression showed poorer overall survival, and Rab24 was identified as an independent prognosis factor according to multivariate analysis. By using overexpression and shRNA knockdown strategies in HCC cell lines, we found that Rab24 can promote HCC proliferation. Finally, we validated that silencing Rab24 significantly attenuated xenograft growth in vivo.
Our study demonstrated that high expression of Rab24 was significantly correlated with poorer prognosis of HCC patients, indicating the potential of Rab24 as a novel clinical biomarker and therapeutic target.
Core Tip: This is a basic study to identify clinical significance and tumor related effects of ras-related protein Rab24 in hepatocellular carcinoma (HCC). High expression of Rab24 was significantly associated with larger tumor, advanced tumor stage and poorer overall survival. By using overexpression and shRNA knockdown strategies in HCC cell lines, we found that Rab24 can promote HCC proliferation. Finally, we validated that silencing Rab24 significantly attenuated xenograft growth in vivo. Our study firstly demonstrated that high expression of Rab24 was significantly correlated with poorer prognosis of HCC patients, indicating the potential of Rab24 as a novel clinical biomarker and therapeutic target.
- Citation: Ding H, Ding ZG, Liu S, Mao XN, Lu XS. Ras-related protein Rab24 plays a predictive role in hepatocellular carcinoma and enhanced tumor proliferation. World J Gastroenterol 2025; 31(8): 101585
- URL: https://www.wjgnet.com/1007-9327/full/v31/i8/101585.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i8.101585
Hepatobiliary cancer ranks the 5th in cancer-related death in male and the 7th in female within United States[1]. Globally, hepatocellular carcinoma (HCC) is the fourth most common cause of cancer-related death[2]. Great progress had been made during the past decades on understanding the risk factors for HCC, for example the chronic hepatitis B or hepatitis C infection, alcohol addiction, or exposure to toxins such as aflatoxins and aristolochic acid[2]. However, the high morbidity and mortality make HCC still a substantial burden worldwide. The treatment of HCC is largely depending on tumor stages. Surgical resection, interventional therapy, and liver transplantation are rational therapy for early stage HCC. However, treatment for advanced stage HCC remains unsatisfied. Molecular targeted therapy is a promising area and has made great progressions such as the kinase and immune targeted therapies. However, there are few The Food and Drug Administration approved molecular targeted drugs for the HCC treatment. Therefore, further identification of novel prognostic biomarkers will be invaluable on predicting HCC survival, identifying treatment targets, and deve
Small GTPases are a special type of G-protein, also known as small G-proteins. Small GTPase can bind to the nucleotide guanosine triphosphate (GTP) and hydrolyze it to guanosine diphosphate (GDP). Generally, a GTPase can be activated by binding GTP, while its GDP-binding status is inactive. Small GTPases function as molecular switches in various process of cellular signaling, including protein translation, protein activation, and protein translocation. Therefore, more and more attention has been focused on the relationship between small GTPases and diseases such as cardiovascular diseases[3], inflammation diseases[4], and malignancies[5,6]. Several small GTPases had been reported to be involved in HCC proliferation or metastasis including Rac1[7], Rab5[8], RalA[9], and Ras[10] etc.
Rab24 belongs to the Rab subfamily of small GTPases, which is an atypical member on its low GTPase activity due to an unusual serine-67 residue instead of a conserved glutamine in other Rab GTPases[11]. Dysregulated expression or mutation of Rab24 has been reported to be involved in the progression of several diseases including ataxia and car
Here we systematically explored the transcriptional level and protein expression level of Rab24 in HCC specimens from both TCGA dataset and our enrolled cohort. In addition to the expression data, we initially found that Rab24 expression can serve as a novel independent risk factor for clinical outcomes of HCC patients. Of note, we also conducted in vitro and in vivo experiments to validate the tumor-promoting role of Rab24 in HCC progression.
Here we took advantages of the Gene Expression Profiling Interactive Analysis (GEPIA) online server (http://gepia.cancer-pku.cn/index.html) to assess transcriptional level of Rab24 as well as its role in HCC survival[16]. The KM plotter website (http://kmplot.com/analysis/) was also used for stratification survival analyses[17].
The subjects of this study came from patients with HCC in the Department of General Surgery from January 2016 to December 2020 in Xinhua Hospital. This study enrolled two cohorts. The first cohort contains 30 HCC tissues and paired adjacent tissues that were fresh-resected in Xinhua Hospital, which was used for real-time PCR (RT-qPCR) analyses to test the mRNA level of Rab24.
The second cohort contains 147 HCC tissues from patients who had undergone R0 surgical resection at Xinhua Hospital. All samples were formalin-fixed paraffin-embedded and were used for immunohistochemistry (IHC) analyses. No patient received any adjuvant therapy before surgery. All patients in this cohort have intact follow-up information, and the median follow up time is 31 months (ranging 4-59 months). All patients in the two cohorts provided written informed consent. The study procedures were approved by the Ethics Committee of Xinhua Hospital.
Total RNA was extracted from HCC tissues with TRIzol reagent (Invitrogen, Carlsbad, CA, United States) and cDNA was synthesized using PrimeScript RT Reagent (Takara, Dalian, China) according to the manufacturer's instructions. As previously described[18], the qPCR was conducted by using the SYBR Mix (Hangzhou Bioer Technology Co. Ltd.) in an Applied Biosystems 7700 sequence detection system. The primers for RT-qPCR were as followings: Rab24, forward, 5’-CAGCTCTTTGAAACATCCAGCAAG-3’ and reverse, 5'- CCAGATCCACGCCCTTGTCC-3'; β-actin, forward, 5'- AACGAGCGGTTCCGATGCCCTGAG -3' and reverse, 5'- TGTCGCCTTCACCGTTCCAGTT-3'. The mRNA levels were evaluated by ΔΔCT method, and normalized to the β-actin housekeeping gene.
IHC experiments were conducted according to the standard procedures. Briefly, FFPE HCC tissue samples were cut into 6 μm sections, which were dried and deparaffinized. After antigen retrieval within citrate buffer, slide sections were then blocked for 5 minutes to prevent unspecific staining reaction, followed by incubating with rabbit anti-human polyclonal Rab24 antibody (Cat. #LS-C403469, LSBio) at 4 °C for 16 hours. Afterwards, slides were washed and incubated with secondary antibody. Then 3,3 Diaminobenzidine reaction buffer was used for the visualization. After counterstained, slides were dehydrated in an ascending series of alcohol and covered.
The IHC immunoreactive score (IRS) was assessed according to intensity and distribution percentage of positive stained cells. The scores of staining intensity (0 = negative, 1 = weak, 2 = moderate, and 3 = strong staining) and the percentage of positive stained cells (0 = no cells, 1 = 1%-25% positive cells, 2 = 25%-50% positive cells, 3 = 51%-75% positive cells, and 4 = > 75% positive cells) were multiplied to obtain IRS. According to the ROC curve, samples were regarded as low Rab24 expression with IRS 0-3, while as high Rab24 expression with IRS 4-12.
The L02 human hepatocytes and four human HCC cell lines (Huh7, HepG2, Hep3B, and MHCC-97H) were used in the present study. All cells were purchased from Shanghai Institute for Biological Science (China). All cell lines were cultured in DMEM medium supplemented with 10% fetal bovine serum (BSA) and maintained at 37 °C in a humidified incubator containing 5% CO2.
To overexpress Rab24, the full length Rab24 was cloned into pCDNA3.1 expression vector as described by others[19]. The dominant-negative mutants of Rab24, namely Rab24-D123I and Rab24-T120A, were generated using QuikChange site-directed mutagenesis strategy[20]. The pcDNA3.1 vector was used as control. The FuGENE® HD transfection reagent (Promega) was used for according to the manufacturer’s instructions. Briefly, cells were seeded on 6-well plates at 50%-60% confluence 24 hours before transfection, then 2 μg of pcDNA3.1-Rab24 plasmid DNA were mixed with FuGENE reagent and incubated in room temperature for 20 minutes. Afterwards, the plasmid mixture was added drop-wise to each well without removing DMEM. The cells were selected using increased concentration of neomycin to obtain stable cell line.
The lentivirus plasmids containing Rab24-shRNA were purchased from Santa Cruz (Cat. #sc-62918-SH). Meanwhile, we purchased the plasmids containing scramble-shRNA (Cat. #sc-108060) to use as control. The transfection of lentiviral vectors containing shRNAs was conducted as previously reported following the manufacturer's instructions[21]. Stable knockdown cells lines were selected using puromycin (3 μg/mL).
Cell counting kit-8 (CCK-8, Dojindo, Osaka, Japan) was used to measure the proliferative capacity of HCC cell lines. Briefly, stable transfected cells were seeded in 96-well plates. Each well was treated with 10 μL of CCK-8 reagent, followed by incubation at 37 °C for another 1 hour before the detection of absorbance at 450 nm by using a microplate reader. Cell viability was tested at different time points (day 1, 2, 3, 4, and 5).
This animal use was approved by the Animal Core Facility of Xinhua Hospital. All animal studies were conducted in accordance with Chinese animal welfare guideline. Five-week-old male Barcelona Clinic Liver Cancer (BCLC)/C nude mice obtained from Xinhua Hospital, which were randomly divided into two groups.
Huh7 cells (5 × 106) stably transfected with scramble-shRNA or Rab24-shRNA were subcutaneously injected into the mice flank. Tumor diameter was measured using caliper every five days and the tumor volume was calculated as (length × width × width)/2. Tumor xenografts were harvested after 30 days, which were subjected to IHC analysis targeting Rab24 and Ki-67.
Data are presented as the mean ± SD. Statistical analyses were conducted using SPSS 20.0 Software. Correlations between Rab24 expression and clinicopathological characteristics were conducted using χ2 test. Overall survival (OS) and disease-free survival (DFS) were assessed using the Kaplan-Meier method and significance was analyzed using the log-rank test. Multivariate cox regression analyses were carried out to identify independent prognostic factors. For the cellular and xenograft data, the difference between two groups was tested using Student’s t-test, while One-way ANOVA analysis was used compare data among more than two groups. A P value < 0.05 indicated a statistically significant result.
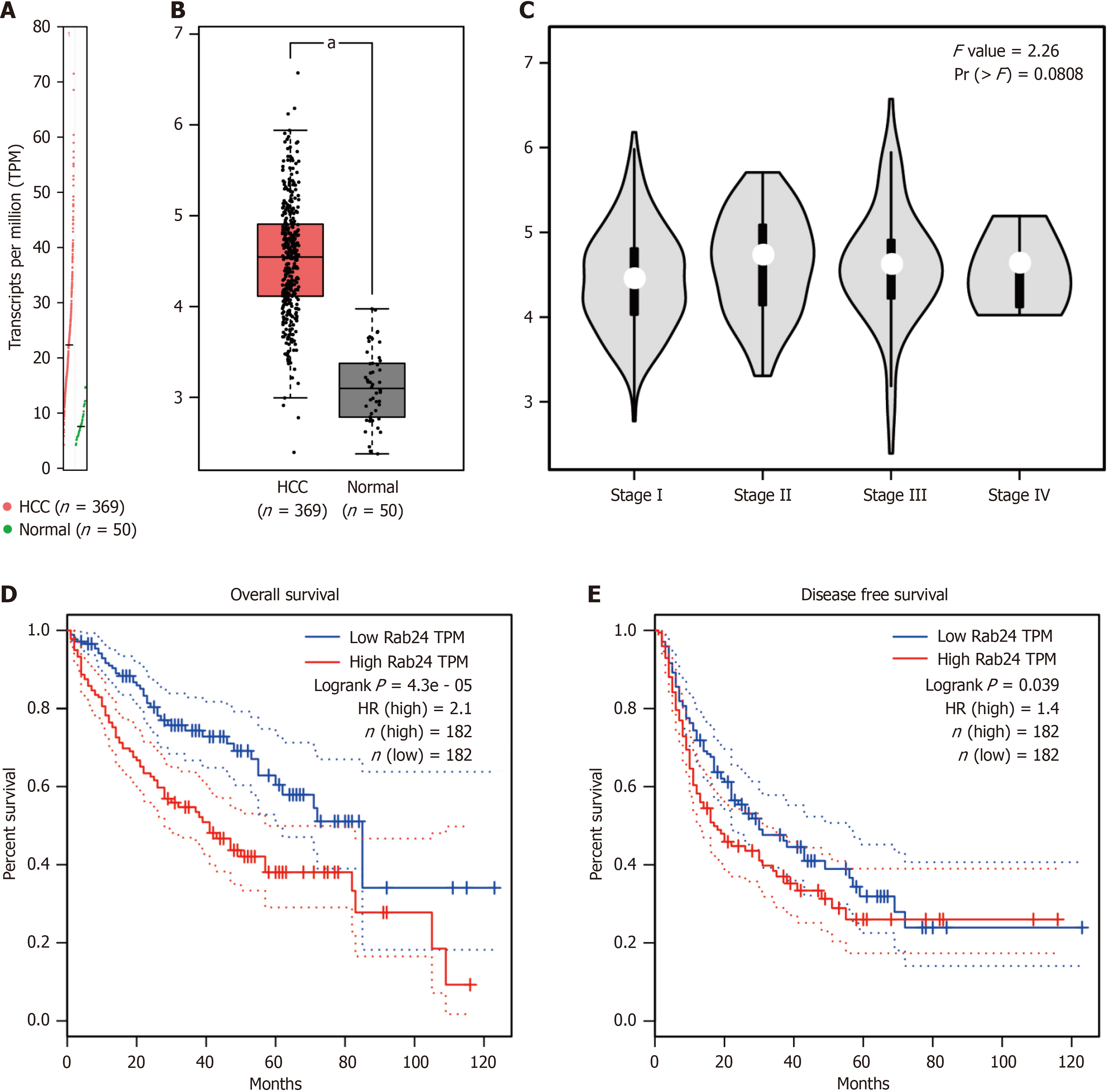
The RNA sequencing data in TCGA dataset was firstly used to analyze the transcriptional level of Rab24 via GEPIA online server. By analyzing 369 HCC tissues and 50 normal liver tissues, we observed that the transcripts per million level of Rab24 is significantly higher in HCC tissues than that in normal liver tissues (P < 0.05; Figure 1A and B). Additionally, higher Rab24 transcription level was observed in stage II-IV HCC specimens compared to that in stage I specimens, although the statistics did not reach statistical significance (P = 0.08; Figure 1C).
In addition, we evaluated the potential effect of Rab24 gene transcription on HCC prognosis in TCGA datasets. Accordingly, higher Rab24 transcripts is associated with unfavorable OS (P < 0.001; Figure 1D) and DFS (P = 0.039; Figure 1E) of HCC patients.
Since the TCGA datasets showed a potential clinical significance of Rab24 in HCC, we next set to collect retrospective cohorts from our hospital to further explore its mRNA and protein expression profiles.
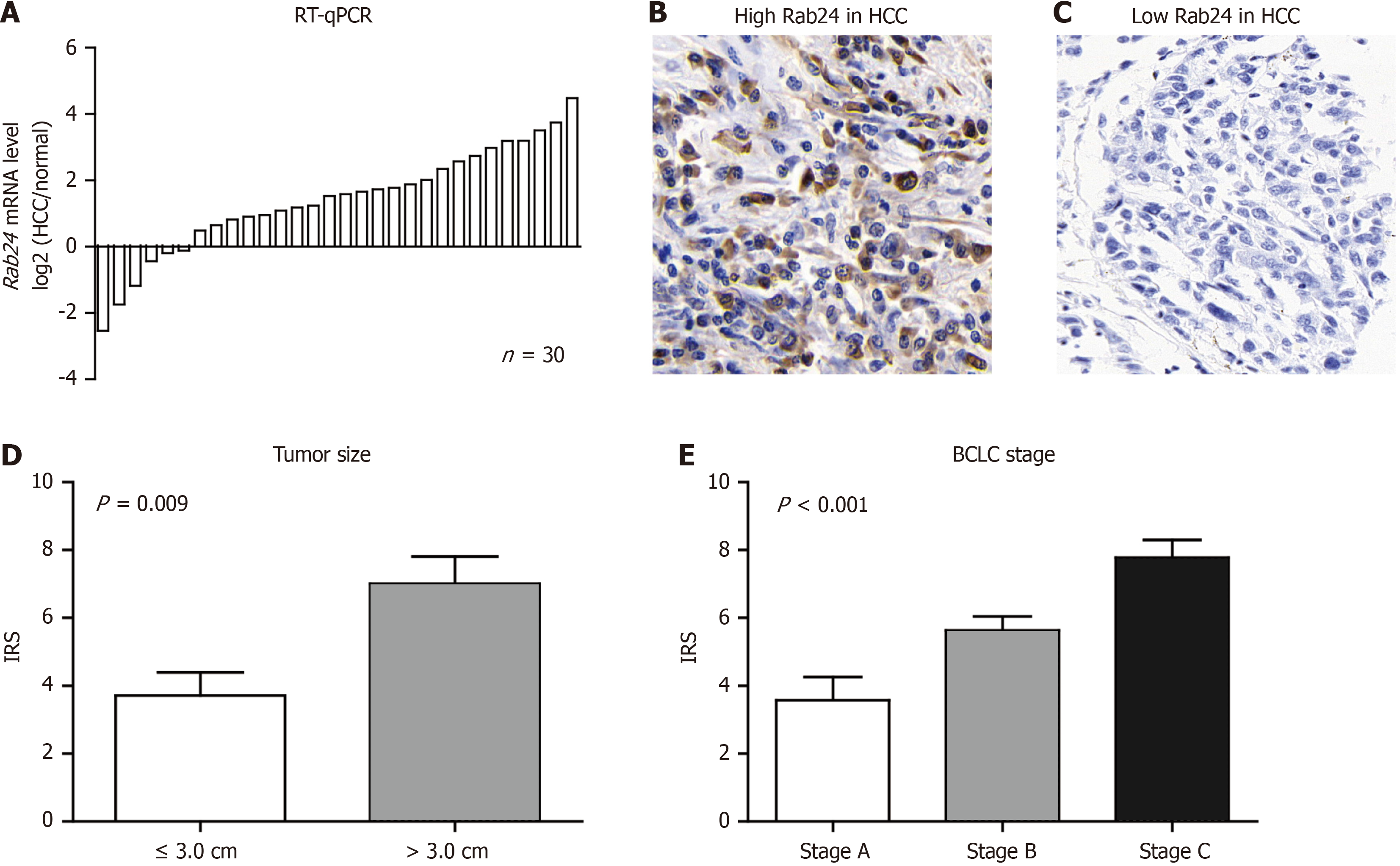
Basic information of the 30 patients, whose specimens were used for RT-qPCR analyses, were listed in Supplementary Table 1. RT-qPCR assay was then conducted to determine the Rab24 mRNA levels. As a result, most tumor samples (24/30 cases, 80%) expressed higher levels of Rab24 mRNA than that in adjacent tissues (Figure 2A), which is consistent with the transcriptional data from GEPIA.
As for the second cohort, which contains 147 cases for IHC test and survival analyses, the average age was 48.6 years old (ranging 27-79 years). Most of the cases were male (105/147, 71.4%) and most were characterized with positive hepatitis B Virus (HBV) infection history (104/147, 70.7%). Only 22.4% (33/147) patients were confirmed with multiple lesions while the other 77.6% (114/147) with single lesion. As for the tumor size, there were 51 cases (34.7%) with tumor size less or equal to 3.0 cm, and the other 96 cases (65.3%) with tumor size larger than 3.0 cm. We also retrieved the pathological information of all resected specimens in our cohort. Accordingly, there were 38.8% (57/147), 31.3% (46/147), and 29.9% (44/147) cases with BCLC stage A, B, C, respectively. As for the Child-Pugh class, 90 cases (61.2%) were identified as class A, 48 cases (32.7%) with class B, and the other 9 cases (6.1%) with class C. Besides, 40 of the 147 cases were identified as positive microvascular invasion, while the other 107 cases with no observed vascular invasion. As for the treatment, all patients underwent R0 resection with no positive resection margin. Among them, 89 cases (60.5%) underwent postoperative chemotherapy while the other 58 cases (39.5%) only underwent surgical resection.
IHC was next conducted to test protein level of Rab24 in enrolled HCC samples that mentioned above. As shown in Figure 2B and C, immunohistochemically staining reaction was mainly observed in the cytoplasm of HCC cells. Interes
To further investigate the correlations between Rab24 protein level and clinical parameters, we sub-grouped all patients into low-Rab24 group (n = 76) and high-Rab24 group (n = 71) as described in the Method section. χ2 test was next conducted to assess the clinical relevance of Rab24 HCC. Comparing to low-Rab24 group, patients in high-Rab24 group tend to exhibit multiple lesions, larger tumor size, advanced BCLC stage, or positive microvascular invasion (all P < 0.05; Table 1). In contrast, Rab24 expression level showed no statistically significant correlation with patients’ age, sex, HBV infection, serum alpha-fetoprotein (AFP), Child-Pugh class, surgical resection, nor therapy selection (all P > 0.05).
| Variables | Cases |
Rab24 expression | P value | |
| n = 147 | Low (n = 76) | High (n = 71) | ||
| Age, years | 0.824 | |||
| ≤ 50 | 78 | 41 | 37 | |
| > 50 | 69 | 35 | 34 | |
| Sex | 0.794 | |||
| Feale | 42 | 21 | 21 | |
| Male | 105 | 55 | 50 | |
| HBV history | 0.780 | |||
| Negative | 43 | 23 | 20 | |
| Positive | 104 | 53 | 51 | |
| Serum AFP, ng/mL | 0.181 | |||
| ≤ 400 | 60 | 35 | 25 | |
| > 400 | 87 | 41 | 46 | |
| Tumor number | 0.005 | |||
| 1 | 114 | 66 | 48 | |
| ≥ 2 | 33 | 10 | 23 | |
| Tumor size, cm | 0.003 | |||
| ≤ 3.0 | 51 | 35 | 16 | |
| > 3.0 | 96 | 41 | 55 | |
| Child-Pugh class | 0.406 | |||
| A | 90 | 50 | 40 | |
| B | 48 | 21 | 27 | |
| C | 9 | 5 | 4 | |
| BCLC stage | 0.001 | |||
| A | 57 | 40 | 17 | |
| B | 46 | 22 | 24 | |
| C | 44 | 14 | 30 | |
| Microvascular invasion | 0.035 | |||
| Absent | 107 | 61 | 46 | |
| Present | 40 | 15 | 25 | |
| Resection margin, cm | 0.959 | |||
| ≤ 1.0 | 68 | 35 | 33 | |
| > 1.0 | 79 | 41 | 38 | |
| Treatment | 0.739 | |||
| Surgery + chemotherapy | 89 | 47 | 42 | |
| Surgery only | 58 | 29 | 29 | |
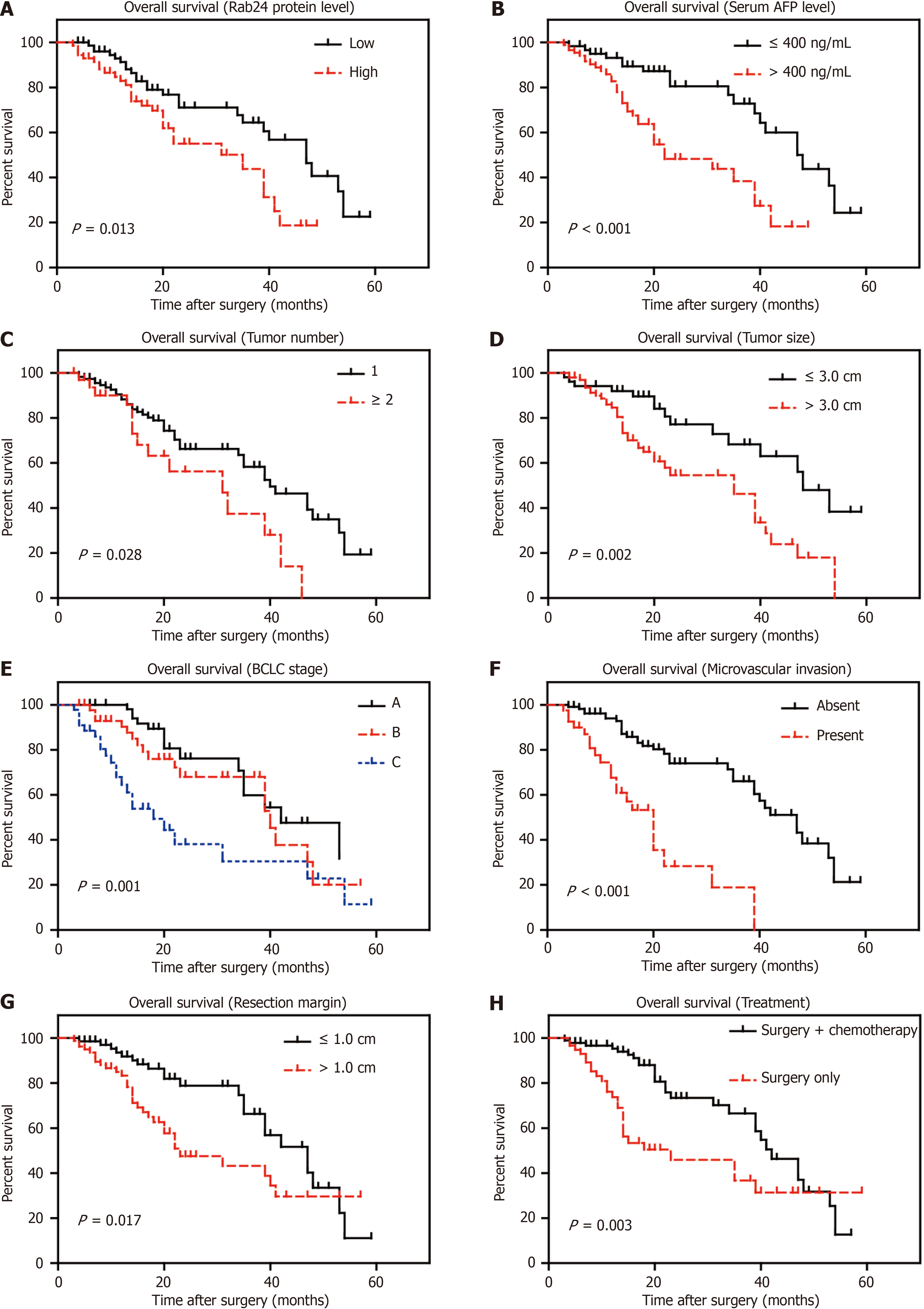
We next conducted Kaplan-Meier survival analyses to assess their prognostic effect of each clinicopathological parameter (Figure 3). Consistent with transcriptional levels in Figure 1D, HCC patients with higher Rab24 protein level showed poorer OS (mean 29.3 ± 2.3 months) compared to those with lower Rab24 expression (mean 40.3 ± 2.6 months; Table 2). Besides, univariate analyses demonstrated that patient’s OS time was negatively correlated with serum AFP (P < 0.001), tumor number (P = 0.028), tumor size (P = 0.002), BCLC stage (P = 0.001), microvascular invasion (P < 0.001), and resection margin (P = 0.017). As expected, patients underwent postoperative chemotherapy showed longer survival time compared to those without chemotherapy (39.3 ± 2.2 months vs 30.4 ± 3.6 months, P = 0.003).
| Variables | Cases | Overall survival months | P value | |
| n = 147 | Median | mean ± SD | ||
| Age, years | 0.153 | |||
| ≤ 50 | 78 | 39.0 | 38.0 ± 2.5 | |
| > 50 | 69 | 35.0 | 33.5 ± 3.1 | |
| Sex | 0.297 | |||
| Feale | 42 | 47.0 | 38.9 ± 3.1 | |
| Male | 105 | 34.0 | 34.3 ± 2.6 | |
| HBV history | 0.874 | |||
| Negative | 43 | 35.0 | 35.5 ± 3.3 | |
| Positive | 104 | 40.0 | 36.2 ± 2.5 | |
| Serum AFP, ng/mL | < 0.001 | |||
| ≤ 400 | 60 | 47.0 | 43.2 ± 2.6 | |
| > 400 | 87 | 22.0 | 27.7 ± 2.2 | |
| Tumor number | 0.028 | |||
| 1 | 114 | 40.0 | 37.8 ± 2.3 | |
| ≥ 2 | 33 | 31.0 | 27.5 ± 3.1 | |
| Tumor size, cm | 0.002 | |||
| ≤ 3.0 | 51 | 48.0 | 43.4 ± 3.1 | |
| > 3.0 | 96 | 35.0 | 30.8 ± 2.3 | |
| Child-Pugh class | 0.214 | |||
| A | 90 | 41.0 | 36.4 ± 2.2 | |
| B | 48 | 39.0 | 37.1 ± 4.4 | |
| C | 9 | 18.0 | 22.5 ± 5.6 | |
| BCLC stage | 0.001 | |||
| A | 57 | 42.0 | 40.2 ± 2.7 | |
| B | 46 | 40.0 | 36.6 ± 3.1 | |
| C | 44 | 18.0 | 26.3 ± 4.0 | |
| Microvascular invasion | < 0.001 | |||
| Absent | 107 | 47.0 | 40.6 ± 2.2 | |
| Present | 40 | 20.0 | 20.0 ± 2.4 | |
| Resection margin, cm | 0.017 | |||
| ≤ 1.0 | 68 | 47.0 | 40.5 ± 2.5 | |
| > 1.0 | 79 | 23.0 | 31.2 ± 3.0 | |
| Treatment | 0.003 | |||
| Surgery + chemotherapy | 89 | 42.0 | 39.3 ± 2.2 | |
| Surgery only | 58 | 23.0 | 30.4 ± 3.6 | |
| Rab24 protein level | 0.013 | |||
| Low | 76 | 47.0 | 40.3 ± 2.6 | |
| High | 71 | 35.0 | 29.3 ± 2.3 | |
Furthermore, we subjected all the significant parameters above into a Cox-regression model for multivariate analysis (Table 3). As a result, BCLC stage, microvascular invasion, resection margin, and chemotherapy all showed significance on predicting HCC OS. Of note, Rab24 was also validated as an independent prognosis predictor for HCC (HR = 2.115, 95%CI: 1.067-4.190, P = 0.032). Nevertheless, serum AFP, tumor number, and tumor size possessed no independent significant effect on HCC OS according to our cohort.
| Variables | HR | 95%CI | P value |
| Serum AFP (> 400 ng/mL vs ≤ 400 ng/mL) | 1.565 | 0.641-3.820 | 0.325 |
| Tumor number (≥ 2 vs 1) | 1.377 | 0.639-2.969 | 0.414 |
| Tumor size (> 3.0 cm vs ≤ 3.0 cm) | 1.048 | 0.558-1.967 | 0.884 |
| BCLC stage (B-C vs A) | 2.543 | 1.313-4.923 | 0.006 |
| Microvascular invasion (present vs absent) | 4.239 | 2.005-8.962 | < 0.001 |
| Resection margin (> 1.0 cm vs ≤ 1.0 cm) | 2.439 | 1.318-4.514 | 0.005 |
| Treatment (surgery vs surgery + chemotherapy) | 2.950 | 1.646-5.286 | < 0.001 |
| Rab24 protein level (high vs low) | 2.115 | 1.067-4.190 | 0.032 |
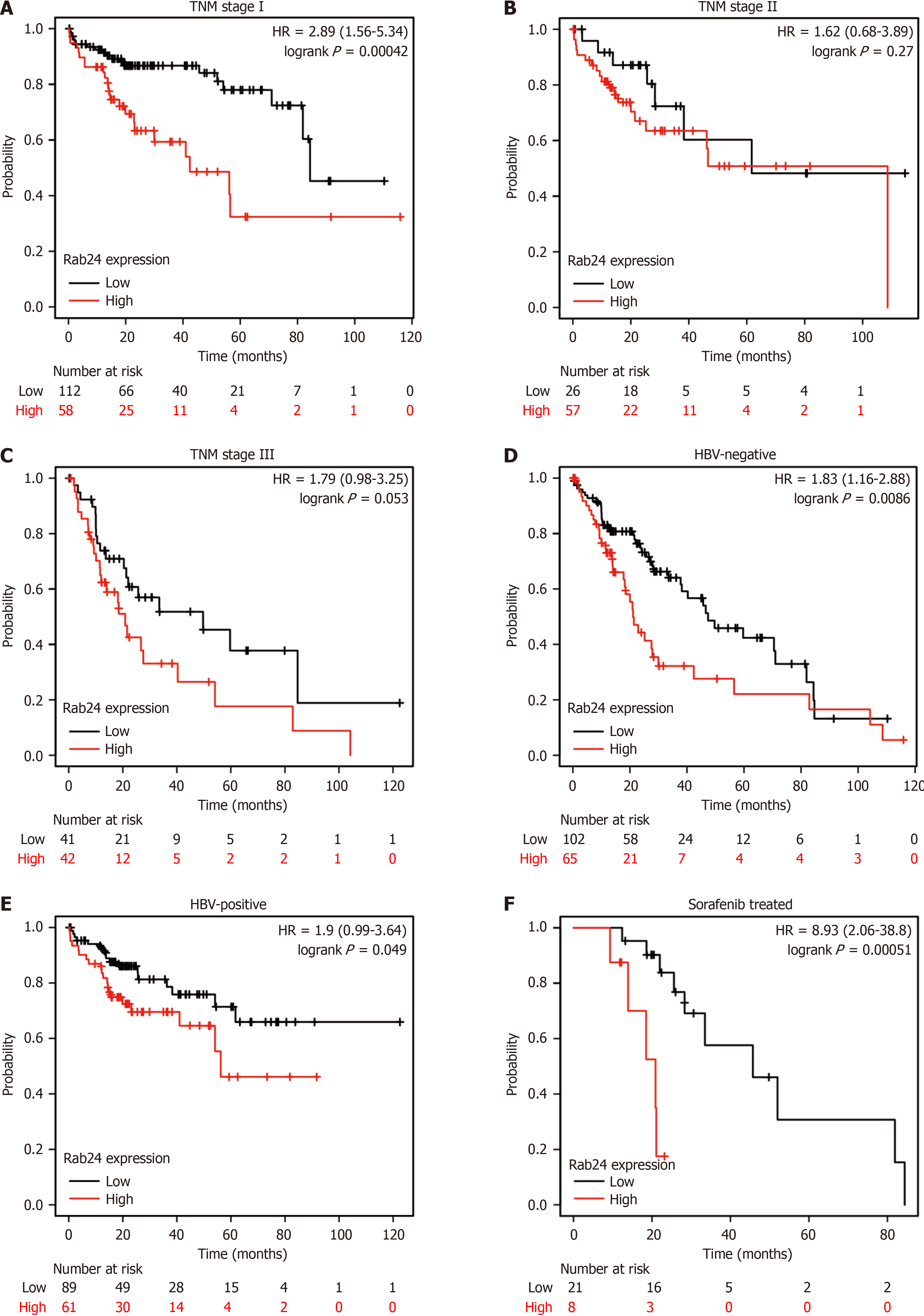
Since both univariate and multivariate analyses of our cohort demonstrated the role of Rab24 as a novel unfavorable prognostic predictor for HCC patients, we next retrieved the published data from other groups to cross-check our conclusion[17]. By independently testing the prognostic role of Rab24 in HCC patients with different tumor-node-metastasis (TNM) stages, we found that higher Rab24 is a remarkable unfavorable predictor for OS of stage I patients (P < 0.001; Figure 4A). Similarly, higher Rab24 seems to indicate poorer OS of stage II and stage III HCC patients, although the statistically differences were not significant (P = 0.27 and P = 0.053, respectively; Figure 4B and C). Besides TNM stage, we also conducted stratification analysis according to HBV infection history. Accordingly, higher Rab24 can help predict poorer OS in either HBV-negative or HBV-positive patients (P < 0.001 and P = 0.049, respectively; Figure 4D and E). In addition, we analyzed the prognostic effect of Rab24 on HCC patients who only underwent chemotherapy without surgical resection (Figure 4F), which obtained consistent results on that higher Rab24 is an unfavorable prognosis factor (P < 0.001).
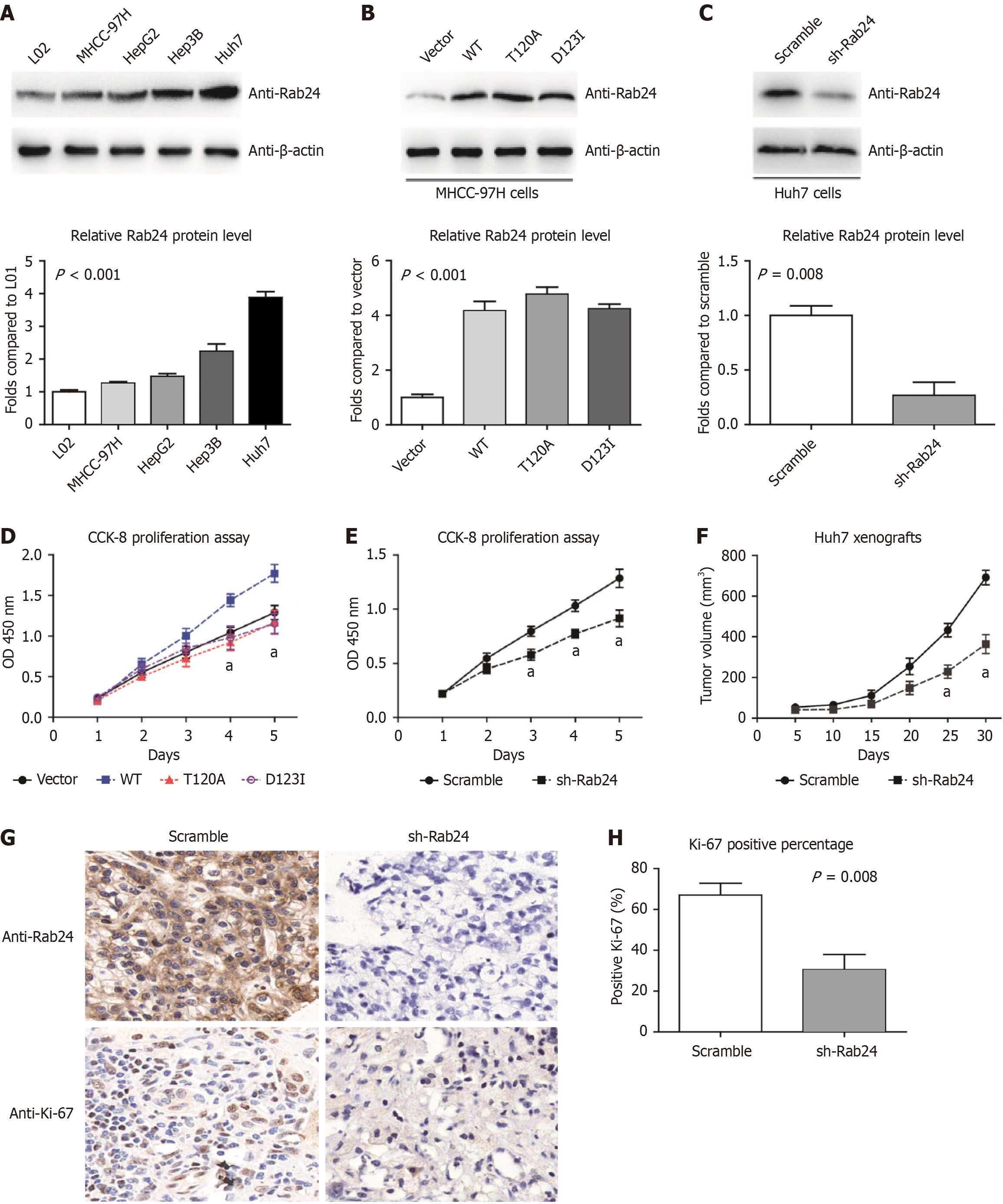
By comparing the expression level of SPRR2B in nontumorous L02 cells and several HCC cell lines (Huh7, HepG2, Hep3B, and MHCC-97H), we found that SPRR2B protein expression was higher in HCC cells than L02 cells (Figure 5A). Among the four HCC cell lines, MHCC-97H cells showed the lowest endogenous Rab24 Level, while Huh7 cells showed the highest expression. Therefore, we selected MHCC-97H cells for overexpression study (Figure 5B), while Huh7 cells for knockdown study (Figure 5C). By testing the proliferation capacities of transfected cells, we found that overexpressing wild-type Rab24 can remarkably promote MHCC-97H cell growth (Figure 5D). However, transfection of either Rab24-T120A or Rab24-D123I, two dominant-negative mutants of Rab24, exhibited no significant effect on HCC cell viability (Figure 5D). In opposite, silencing Rab24 by specific shRNA resulted in a significant decrease in Huh7 cell proliferation (Figure 5E).
To further explore the effect of Rab24 on tumor growth in vivo, xenograft in mice model was established using Huh7 cell lines. Mice were subcutaneously injected with shRNA-transfected cells, then the xenograft size was monitored every five days (Figure 5F). The xenograft growth curves showed that knockdown of Rab24 Led to a significant decrease in tumor growth compared to the scramble-shRNA group. Then isolated xenografts were subjected to IHC analyses targeting Rab24 and Ki-67. As expected, the sh-Rab24 group only exhibited slight immunoreactivity of Rab24 (Figure 5G). Furthermore, xenografts with Rab24-knockdown showed a decreased percentage of Ki-67 (Figure 5H), indicating the inhibition effect of Rab24-shRNA in HCC progression.
As a special member in Rab GTPase family, Rab24 is characterized with low intrinsic GTPase activity and inefficiently prenylation. Rab24 can play roles in growth, cellular differentiation, cell movement and lipid vesicle transport. For example, it modulates cell division process by facilitating meiotic apparatus assemblage and maturation progression[22,23]. Previous studies regarding Rab24 were predominantly focused on its involvement in autophagic pathway. For example, Rab24 can localize in autophagosomes and interact with protein LC3, a crucial protein for autophagy[19,24,25]. Besides promoting autophagosome maturation in starved or damaged cells[26,27], Rab24 was later reported to facilitate clearance of autophagic compartments under nutrient-rich conditions[28].
Nevertheless, Rab24 may exert distinct functions in different cell types. For instance, Rab24 is predominantly localized in the nuclei of COS-7 cells, which is different from its localization in endoplasmic reticulum/cis-Golgi system of other cell types[27]. Interestingly, the normal expression of Rab24 in liver help control blood glucose homeostasis via improving mitochondrial plasticity[29], however its dysregulated expression was identified in HCC[14]. Here we further investigate the transcription and expression profile of Rab24 in clinical HCC tissues. Accordingly, Rab24 is upregulated on both mRNA and protein levels in HCC, as reflected from TCGA dataset and our enrolled HCC cohort.
Based on retrospective cohorts, higher Rab24 was identified as a novel prognostic risk factor for the OS and DFS of HCC patients. Thus, we further conducted cellular experiments to explore its potential effect on HCC progression. By overexpressing or silencing Rab24 in different HCC cell lines, we demonstrated that Rab24 can positively regulate HCC cell proliferation. Besides the wild type Rab24, we also overexpressed two dominant-negative mutants (D123I and T120A), both have been reported to affect the physiological functions of Rab24. Of note, both mutants abolished the proliferation-promoting effect of Rab24 in HCC cells. Therefore, the natural conformation and activity of Rab24 may be critical for its tumor related role despite low intrinsic GTPase activity.
Our study has several limitations. Firstly, we only tested the protein level of Rab24 in limited HCC cases from our hospital, which may cause regional bias, limit the universal applicability of Rab24 and hinder the precise stratification of patients and the formulation of personalized treatment strategies. We tried to weaken this disadvantage by retrieving the RNA-sequencing data reported by Menyhárt et al[17] to cross-check our clinical findings. Secondly, due to lacking of specific antibodies, we didn’t test the phosphorylation pattern or GDP/GTP-binding status of Rab24 in our specimens, both had been reported to be closely correlated with its protein functions[11,30]. Without comprehensive data, the active state of Rab24 in HCC development may not be accurately assessed, affecting its accuracy as a prognostic marker and its potentiality as a therapeutic target. Thirdly, although we revealed a remarkable effect of Rab24 on enhancing HCC growth by in vitro and in vivo strategies, this study mainly focused on its clinical significance and did not fully dig into the underlying mechanisms. Literature search indicated that Rab24 can interact with cyclophilin A and gamma-aminobutyric acid type A receptor-associated protein in COS-7 cells[27], however our immunoprecipitation data didn’t identified corresponding interactions in HCC cells (data not shown). According to Chen et al’s report[15], the mechanism of Rab24 in liver cancer mainly involves the regulation of miRNAs. Specifically, the expression of miR-615-5p is reduced in liver cancer cells, and its downregulation is closely related to the increased methylation of the miR-615-5p promoter region caused by the absence of lysine demethylase 4B. Further studies focusing on illustrating its signaling pathway would be essential and invaluable to provide evidence for therapeutic development. In summary, the role of Rab24 in HCC is not only reflected in its potential as a prognostic predictor but also includes its potential as a therapeutic intervention target.
Overall, our results revealed Rab24 as a novel unfavorable prognosis indicator for HCC, which may function by enhancing HCC proliferation. Inhibiting the expression or activity of Rab24 would be a notable therapeutic direction for specific HCC patients.
The authors gratefully acknowledge all the students for skillful technical assistance.
| 1. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12667] [Cited by in RCA: 15305] [Article Influence: 3061.0] [Reference Citation Analysis (4)] |
| 2. | Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16:589-604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2184] [Cited by in RCA: 2893] [Article Influence: 482.2] [Reference Citation Analysis (17)] |
| 3. | Ferri N, Contini A, Bernini SK, Corsini A. Role of small GTPase protein Rac1 in cardiovascular diseases: development of new selective pharmacological inhibitors. J Cardiovasc Pharmacol. 2013;62:425-435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 4. | Seinen ML, van Nieuw Amerongen GP, de Boer NK, van Bodegraven AA. Rac Attack: Modulation of the Small GTPase Rac in Inflammatory Bowel Disease and Thiopurine Therapy. Mol Diagn Ther. 2016;20:551-557. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 5. | Vega FM, Ridley AJ. Rho GTPases in cancer cell biology. FEBS Lett. 2008;582:2093-2101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 568] [Cited by in RCA: 612] [Article Influence: 36.0] [Reference Citation Analysis (0)] |
| 6. | Karlsson R, Pedersen ED, Wang Z, Brakebusch C. Rho GTPase function in tumorigenesis. Biochim Biophys Acta. 2009;1796:91-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 187] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 7. | Cao X, Zhang L, Shi Y, Sun Y, Dai S, Guo C, Zhu F, Wang Q, Wang J, Wang X, Chen YH, Zhang L. Human tumor necrosis factor (TNF)-alpha-induced protein 8-like 2 suppresses hepatocellular carcinoma metastasis through inhibiting Rac1. Mol Cancer. 2013;12:149. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 69] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 8. | Qi S, Su L, Li J, Zhang C, Ma Z, Liu G, Zhang Q, Jia G, Piao Y, Zhang S. Arf6-driven endocytic recycling of CD147 determines HCC malignant phenotypes. J Exp Clin Cancer Res. 2019;38:471. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 9. | Ezzeldin M, Borrego-Diaz E, Taha M, Esfandyari T, Wise AL, Peng W, Rouyanian A, Asvadi Kermani A, Soleimani M, Patrad E, Lialyte K, Wang K, Williamson S, Abdulkarim B, Olyaee M, Farassati F. RalA signaling pathway as a therapeutic target in hepatocellular carcinoma (HCC). Mol Oncol. 2014;8:1043-1053. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 35] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 10. | Calvisi DF, Ladu S, Gorden A, Farina M, Conner EA, Lee JS, Factor VM, Thorgeirsson SS. Ubiquitous activation of Ras and Jak/Stat pathways in human HCC. Gastroenterology. 2006;130:1117-1128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 538] [Cited by in RCA: 552] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 11. | Erdman RA, Shellenberger KE, Overmeyer JH, Maltese WA. Rab24 is an atypical member of the Rab GTPase family. Deficient GTPase activity, GDP dissociation inhibitor interaction, and prenylation of Rab24 expressed in cultured cells. J Biol Chem. 2000;275:3848-3856. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 49] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 12. | Swaminathan B, Goikuria H, Vega R, Rodríguez-Antigüedad A, López Medina A, Freijo Mdel M, Vandenbroeck K, Alloza I. Autophagic marker MAP1LC3B expression levels are associated with carotid atherosclerosis symptomatology. PLoS One. 2014;9:e115176. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 38] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 13. | Agler C, Nielsen DM, Urkasemsin G, Singleton A, Tonomura N, Sigurdsson S, Tang R, Linder K, Arepalli S, Hernandez D, Lindblad-Toh K, van de Leemput J, Motsinger-Reif A, O'Brien DP, Bell J, Harris T, Steinberg S, Olby NJ. Canine hereditary ataxia in old english sheepdogs and gordon setters is associated with a defect in the autophagy gene encoding RAB24. PLoS Genet. 2014;10:e1003991. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 36] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 14. | He H, Dai F, Yu L, She X, Zhao Y, Jiang J, Chen X, Zhao S. Identification and characterization of nine novel human small GTPases showing variable expressions in liver cancer tissues. Gene Expr. 2002;10:231-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 105] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 15. | Chen Z, Wang X, Liu R, Chen L, Yi J, Qi B, Shuang Z, Liu M, Li X, Li S, Tang H. KDM4B-mediated epigenetic silencing of miRNA-615-5p augments RAB24 to facilitate malignancy of hepatoma cells. Oncotarget. 2017;8:17712-17725. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 16. | Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98-W102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5550] [Cited by in RCA: 7078] [Article Influence: 884.8] [Reference Citation Analysis (0)] |
| 17. | Menyhárt O, Nagy Á, Győrffy B. Determining consistent prognostic biomarkers of overall survival and vascular invasion in hepatocellular carcinoma. R Soc Open Sci. 2018;5:181006. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 242] [Cited by in RCA: 342] [Article Influence: 48.9] [Reference Citation Analysis (0)] |
| 18. | Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611-622. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9915] [Cited by in RCA: 11140] [Article Influence: 696.3] [Reference Citation Analysis (0)] |
| 19. | Munafó DB, Colombo MI. Induction of autophagy causes dramatic changes in the subcellular distribution of GFP-Rab24. Traffic. 2002;3:472-482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 154] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 20. | Maltese WA, Soule G, Gunning W, Calomeni E, Alexander B. Mutant Rab24 GTPase is targeted to nuclear inclusions. BMC Cell Biol. 2002;3:25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 21. | Ansorge S, Lanthier S, Transfiguracion J, Durocher Y, Henry O, Kamen A. Development of a scalable process for high-yield lentiviral vector production by transient transfection of HEK293 suspension cultures. J Gene Med. 2009;11:868-876. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 105] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 22. | Militello RD, Munafó DB, Berón W, López LA, Monier S, Goud B, Colombo MI. Rab24 is required for normal cell division. Traffic. 2013;14:502-518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 24] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 23. | Qiu D, Li S, Guo L, Yuan R, Ou X. Rab24 functions in meiotic apparatus assembly and maturational progression in mouse oocyte. Cell Cycle. 2019;18:2893-2901. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 24. | Gutierrez MG, Vázquez CL, Munafó DB, Zoppino FC, Berón W, Rabinovitch M, Colombo MI. Autophagy induction favours the generation and maturation of the Coxiella-replicative vacuoles. Cell Microbiol. 2005;7:981-993. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 207] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 25. | Ao X, Zou L, Wu Y. Regulation of autophagy by the Rab GTPase network. Cell Death Differ. 2014;21:348-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 245] [Cited by in RCA: 322] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 26. | Egami Y, Kiryu-Seo S, Yoshimori T, Kiyama H. Induced expressions of Rab24 GTPase and LC3 in nerve-injured motor neurons. Biochem Biophys Res Commun. 2005;337:1206-1213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 27. | Wu M, Yin G, Zhao X, Ji C, Gu S, Tang R, Dong H, Xie Y, Mao Y. Human RAB24, interestingly and predominantly distributed in the nuclei of COS-7 cells, is colocalized with cyclophilin A and GABARAP. Int J Mol Med. 2006;17:749-754. [PubMed] |
| 28. | Ylä-Anttila P, Mikkonen E, Happonen KE, Holland P, Ueno T, Simonsen A, Eskelinen EL. RAB24 facilitates clearance of autophagic compartments during basal conditions. Autophagy. 2015;11:1833-1848. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 41] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 29. | Seitz S, Kwon Y, Hartleben G, Jülg J, Sekar R, Krahmer N, Najafi B, Loft A, Gancheva S, Stemmer K, Feuchtinger A, Hrabe de Angelis M, Müller TD, Mann M, Blüher M, Roden M, Berriel Diaz M, Behrends C, Gilleron J, Herzig S, Zeigerer A. Hepatic Rab24 controls blood glucose homeostasis via improving mitochondrial plasticity. Nat Metab. 2019;1:1009-1026. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 30. | Ding J, Soule G, Overmeyer JH, Maltese WA. Tyrosine phosphorylation of the Rab24 GTPase in cultured mammalian cells. Biochem Biophys Res Commun. 2003;312:670-675. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 77] [Article Influence: 3.7] [Reference Citation Analysis (0)] |