Published online Jan 14, 2025. doi: 10.3748/wjg.v31.i2.100898
Revised: September 28, 2024
Accepted: November 20, 2024
Published online: January 14, 2025
Processing time: 110 Days and 19.1 Hours
Regulator of G protein signaling (RGS) proteins participate in tumor formation and metastasis by acting on the α-subunit of heterotrimeric G proteins. The speci
To explore the role and underlying mechanisms of action of RGS4 in GC develop
The prognostic significance of RGS4 in GC was analyzed using bioinformatics based public databases and verified by immunohistochemistry and quantitative polymerase chain reaction in 90 patients with GC. Function assays were employed to assess the carcinogenic impact of RGS4, and the mechanism of its possible influence was detected by western blot analysis. A nude mouse xenograft model was established to study the effects of RGS4 on GC growth in vitro.
RGS4 was highly expressed in GC tissues compared with matched adjacent normal tissues. Elevated RGS4 expression was correlated with increased tumor-node-metastasis stage, increased tumor grade as well as poorer overall survival in patients with GC. Cell experiments demonstrated that RGS4 knockdown suppressed GC cell proliferation, migration and invasion. Similarly, xenograft experiments confirmed that RGS4 silencing significantly inhibited tumor growth. Moreover, RGS4 knockdown resulted in reduced phosphorylation levels of focal adhesion kinase, phosphatidyl-inositol-3-kinase, and protein kinase B, decreased vimentin and N-cadherin, and elevated E-cadherin.
High RGS4 expression in GC indicates a worse prognosis and RGS4 is a prognostic marker. RGS4 influences tumor progression via the focal adhesion kinase/phosphatidyl-inositol-3-kinase/protein kinase B pathway and epithelial-mesenchymal transition.
Core Tip: This study explored the relevant role of regulator of G protein signaling 4 (RGS4) in gastric cancer (GC) and its possible mechanism. Through analysis of public databases, validation of clinical specimens, cell experiments and xenograft model, our study confirmed that RGS4 promotes GC progression through the focal adhesion kinase/phosphatidyl-inositol-3-kinase/protein kinase B pathway and epithelial-mesenchymal transition. RGS4 is a potential therapeutic target for GC and our study may further guide clinical practice.
- Citation: Chen PY, Wang PY, Liu B, Jia YP, Zhang ZX, Liu X, Wang DH, Yan YJ, Fu WH, Zhu F. RGS4 promotes the progression of gastric cancer through the focal adhesion kinase/phosphatidyl-inositol-3-kinase/protein kinase B pathway and epithelial-mesenchymal transition. World J Gastroenterol 2025; 31(2): 100898
- URL: https://www.wjgnet.com/1007-9327/full/v31/i2/100898.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i2.100898
Gastric cancer (GC) is a prevalent malignant tumor of the digestive tract, which is the fifth most common cancer and the fifth leading cause of cancer mortality worldwide[1]. Despite there has been significant progress in multidisciplinary treatment modalities, the overall survival (OS) rate remains low, particularly among patients in advanced stages of GC[2]. The poor prognosis of GC underscores the importance of identifying biomarkers related with cancer development. As one of the most widely known and ubiquitous membrane receptor families, G-protein-coupled receptors (GPCRs) are essential in a variety of physiological processes, such as cellular proliferation, motility, and gene expression[3]. GPCRs play an integral role in tumor’s malignant transformation[3,4]. The regulator of G protein signaling (RGS) protein family, comprising over 20 members, modulates cellular responses mediated by GPCRs through a conserved RGS domain with guanosine triphosphate (GTP)ase-activating protein activity[5], which interacts with GTP-bound Gα subunits of activated G-proteins and accelerates GTP hydrolysis, causing rapid G-protein deactivation and termination of GPCR signaling[6]. In cancer progression, RGS proteins act as pivotal gating switches that regulate the growth, division, differentiation, and mobility of tumor cells[7,8]. It is reasonable to speculate that the RGS gene family may be important molecular markers for GC diagnosis, treatment due to the significant role of GPCRs in this GC[9,10].
In our previous research, we found that RGS1 was associated with poor prognosis in GC[11], which has increased our interest in the RGS gene family. RGS4 belongs to the B/R4 RGS gene family and serves as a negative regulator of GPCRs, inhibiting the transmission of related signaling factors by hastening the hydrolysis of GTP molecules bound to the Gα subunit[12]. Interacting with various receptors, effectors, scaffolding proteins, and signaling entities, RGS4 forms intricate transduction assemblies that affect the localization, functionality, and stability of signals within the cells[5]. This protein exerts a crucial regulatory effect on tumor tissues or cells[13-16]. Given these biological functions, we speculate that RGS4 are essential in GC progression. Based on the public database and 90 patients with GC from Tianjin Medical University General Hospital, we aimed to investigate RGS4 expression in GC tissues and the relationship between RGS4 expression levels, clinicopathological parameters as well as OS of GC. Additionally, we examined the role of RGS4 in GC cell behavior and investigated its potential molecular mechanisms.
The normalized gene expression matrix, OS data, and relevant clinical and pathological parameters were obtained from The Cancer Genome Atlas Stomach Adenocarcinoma (TCGA-STAD) dataset (https://portal.gdc.cancer.gov/) and the Genotype-Tissue Expression dataset (https://xenabrowser.net/). Following data preprocessing steps such as probe annotation, normalization, and correction, the R software was used to analyze the gene expression differences of RGS4 between GC and normal tissues. Clinical and pathological data from 90 patients who underwent gastrectomy and were pathologically confirmed to have GC were collected at Tianjin Medical University General Hospital between 2017 and 2020. Each patient provided samples of GC and para-carcinoma normal tissues. All patients provided their consent.
The gene expression and OS data of RGS4 from TCGA-STAD database and clinical samples were organized. Whole group were categorized into RGS4 high-expression and RGS4 low-expression groups based on the median RGS4 expression value. The correlation between RGS4 expression and the prognosis of GC patients was analyzed using R software (survey package). Differences in gene expression were analyzed using Gene Set Enrichment Analysis (GSEA) (GSEA v3.0). Each analysis involved 1000 gene set permutations, with pathways deemed significantly enriched if they met the following criteria: Enrichment score exceeding 0.6 and P value < 0.05.
The human gastric mucosal cell line GES-1 and the human GC cell line MGC-803 were purchased from icellbioscience Co., Ltd. The human GC cell line NCI-N807 was purchased from ATCC Co., Ltd. AGS and HGC27 cell lines (two human GC cell lines) were acquired from Wuhan Prosa Life Science Co., Ltd. MGC-803 and NCI-N87, HGC27, and GES-1 cell lines were cultivated in RPMI 1640 medium (Gibco). AGS cells were grown in F-12 medium. All cells were cultivated with 50 mL/L CO2 at 37 °C.
The tumor and normal tissue specimens were incubated with the corresponding anti-RGS4 (Abcam, ab97037, 1:1000) overnight at 4 °C. Subsequently, the secondary antibody was used to incubate the specimens. The signals were detected using a DAB staining kit (Solarbio, China). The immunohistochemistry (IHC) scoring method was the same as that used by Chen et al[17].
Basic RNA extraction as well as real-time quantitative polymerase chain reaction (qPCR) methods were conducted as our previous research[11]. The primers used were shown below: RGS4 upstream-primer, 5’-ACATCGGCTAGGTTTC-3’, downstream-primer, 3’-GTTGTGGGAAGAATTGTGTTCAC-5’; GAPDH downstream-primer, 5’-GGTGAAGGTCGGAGTCAACG-3’, downstream-primer, 5’-CAAAGTTGTCATGGATGHACC-3’.
RIPA lysis buffer supplemented with phosphatase and protease inhibitors was used to isolate total protein. The proteins were transferred to polyvinylidene fluoride membranes following separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Next, primary antibodies [RGS4, ab308160, 1:1500, Abcam; focal adhesion kinase (FAK), ab40794, 1:2000, Abcam; p-FAK, ab81298, 1:1500, Abcam; phosphatidyl-inositol-3-kinase (PI3K), ab302958, 1:1500, Abcam; p-PI3K, ab182651, 1:200, Abcam; protein kinase B (AKT), ab8805, 1:500, Abcam; p-AKT, ab38449, 1:1500, Abcam; E-cadherin, ab40772, 1:1500, Abcam; N-cadherin, ab76011, 1:5000, Abcam; vimentin, ab20346, 1:1500, Abcam; β-actin, ab8227, 1:2000, Abcam] were used to nurture membranes. After incubation with the primary antibody, secondary antibodies were used to nurture membranes for 2 hours.
RGS4 short hairpin ribonucleic acid (shRNA) was synthesized by Tsingke Biotech Co. (Beijing, China). Supplementary Table 1 showed the top- and bottom-strand primer sequences of shRNA. ShRNA was cloned into the EcoRI/BamHI sites to generate recombinant plasmids. The recombinant plasmid was then transfected into cells to obtain MGC-803 cell line with RGS4 knockdown, and the vector plasmid was used as a blank control. To induce RGS4 overexpression, we integrated lentiviral expression vectors with DNA fragments of human RGS4 open reading frames to generate recombinant lentivirus, which was used to generate AGS cell lines overexpressing RGS4.
AGS and MGC-803 cells (2.0 × 103 cells/well) after transfection were cultivated in 96-well plates for 0, 12, 24, 48, 72, 96 hours with 50 mL/L CO2 at 37 °C. The cell counting kit-8 (CCK-8) reagent was then added at the endpoint time. Subse
Transfected AGS and MGC-803 cells were seeded in 6-well plates and hatched at 37 °C for 10 days to form single-cell colonies. Subsequently, the cells were fixed in 4% paraformaldehyde and then stained with 0.5% w/v crystal violet solution.
Transfected cells were seeded into 6-well plates (4 × 105 cells/well) and cultured in medium until 80% confluence. The tip of the 200 μL pipette was used to leave scratch. After 0, 24, and 48 hours post-scratch, wound closure was monitored.
The 24-well transwell chambers (Corning, United States) were coated overnight at 4 °C with or without 100 μL Matrigel matrix (Corning). A total of 1 × 104 cells in 150 μL of serum-free medium were placed in the upper chamber, with 500 μL of 10% foetal bovine serum culture medium added to the lower chamber. After 24 hours of incubation, cells that migrated to the lower chamber were stained with 0.5% crystal violet and counted.
BALB/c nude mice (4-6 weeks old) were randomly divided into four groups (n = 5 mice/group). The same amounts (1 × 107) of MGC-803 cells which were transfected with RGS4 (MGC803-shRGS4), blank control cells (MGC803-NC), AGS cells stably overexpressing RGS4 (AGS-RGS4), and blank control cells (AGS-NC) were injected into the axillary skin of nude mice. We measured the long diam and short diam of the tumor every two days. The tumor volume = (A × B2)/2 (A, long diam; B, short diam). After two weeks, the nude mice were executed. The removed tumor tissues were harvested for IHC as well as western blot analyses, and tumor sizes were analyzed, weighed, and photographed.
Data were analyzed and graphed using GraphPad Prism 10 and SPSS 25.0 software. Categorical variables were expressed as the frequency and percentages and continuous variables as means ± SD deviation. Kaplan-Meier method was used to plot survival curves. Significant differences in categorical variables were analyzed using the χ2 test and continuous variables were analyzed using Student’s t-test or one-way analysis of variance (ANOVA). Two tails P value < 0.05 was considered statistically significant.
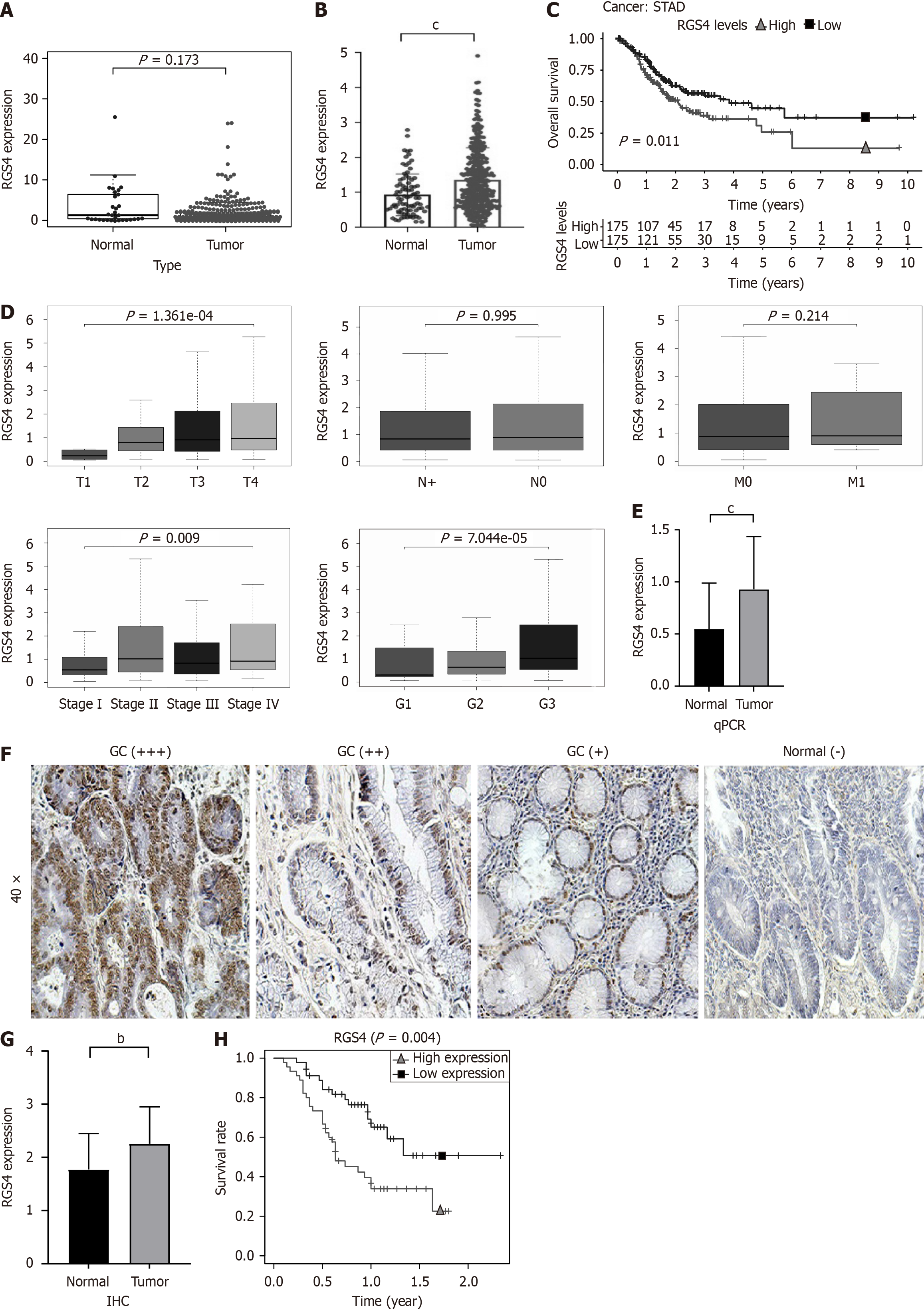
The results of bioinformatics showed that in the TCGA-STAD dataset, the expression level of RGS4 was not significantly different between the two groups - GC tissue (n = 375) and normal tissue (n = 32) (Figure 1A). To validate the expression of RGS4 at larger samples, the TCGA and Genotype-Tissue Expression datasets were integrated. The integrated analysis showed that compared to normal tissues (n = 88), the RGS4 was significantly increased in GC tissues (n = 375) (Figure 1B). From the results of the survival analysis, we can know that GC patients with high expression levels of RGS4 had a worse prognosis (P = 0.011, Figure 1C). Besides, the high expression of RGS4 was related with advanced T stage, tumor-node-metastasis (TNM) stage, and tumor grade (P < 0.05, Figure 1D).
We collected the clinical and pathological information of 90 patients with GC at our center, as well as their tissues including tumor and para-carcinoma. Subsequently, we performed qPCR and IHC assays on the tissue samples from each patient. As shown in Figure 1E, RGS4’s expression level in GC tissues was significantly higher than that in para-carcinoma normal tissues (0.9278 ± 0.5081 vs 0.5464 ± 0.4425, P < 0.05). IHC confirmed higher levels of RGS4 expression at protein level in GC tissues than in para-carcinoma tissues (2.254 ± 0.6992 vs 1.772 ± 0.6745, P < 0.01; Figure 1F and G). According to the expression level of the RGS4 gene, patients with GC (n = 90) were separated into an RGS4 high expression group (n = 45) and an RGS4 low expression group (n = 45). Survival curves displayed that the high expression group of RGS4 had a worse prognosis and shorter OS (P = 0.004, Figure 1H), which validated the results obtained in TCGA-STAD dataset (Figure 1C). The analysis of clinicopathological parameters of patients with GC in our center indicated that patients with high RGS4 expression had advanced T stage, N stage, tumor grade, and TNM stage (Table 1).
| Characteristics | RGS4 expression | P value | |
| Low group (n = 45) | High group (n = 45) | ||
| Age, median (IQR) | 64 (60, 68) | 66 (61, 70) | 0.199 |
| Gender, n (%) | 0.806 | ||
| Female | 12 (13.3) | 10 (11.1) | |
| Male | 33 (36.7) | 35 (38.9) | |
| T stage, n (%) | 0.011 | ||
| T1 | 9 (10) | 1 (1.1) | |
| T2 | 9 (10) | 4 (4.4) | |
| T3 | 7 (7.8) | 10 (11.1) | |
| T4 | 20 (22.2) | 30 (33.3) | |
| N stage, n (%) | 0.038 | ||
| N0 | 21 (23.3) | 11 (12.2) | |
| N1 | 10 (11.1) | 9 (10) | |
| N2 | 6 (6.7) | 5 (5.6) | |
| N3 | 8 (8.9) | 20 (22.2) | |
| M stage, n (%) | 1.000 | ||
| M0 | 43 (47.8) | 42 (46.7) | |
| M1 | 2 (2.2) | 3 (3.3) | |
| Grade, n (%) | 0.002 | ||
| G1 | 16 (17.8) | 6 (6.7) | |
| G2 | 18 (20) | 12 (13.3) | |
| G3 | 11 (12.2) | 27 (30) | |
| TNM stage, n (%) | 0.011 | ||
| Stage I | 15 (16.7) | 3 (3.3) | |
| Stage II | 11 (12.2) | 12 (13.3) | |
| Stage III | 17 (18.9) | 27 (30) | |
| Stage IV | 2 (2.2) | 3 (3.3) | |
| OS (month) | |||
| Median (range) | 28 (7-70) | 19 (3-56) | 0.004 |
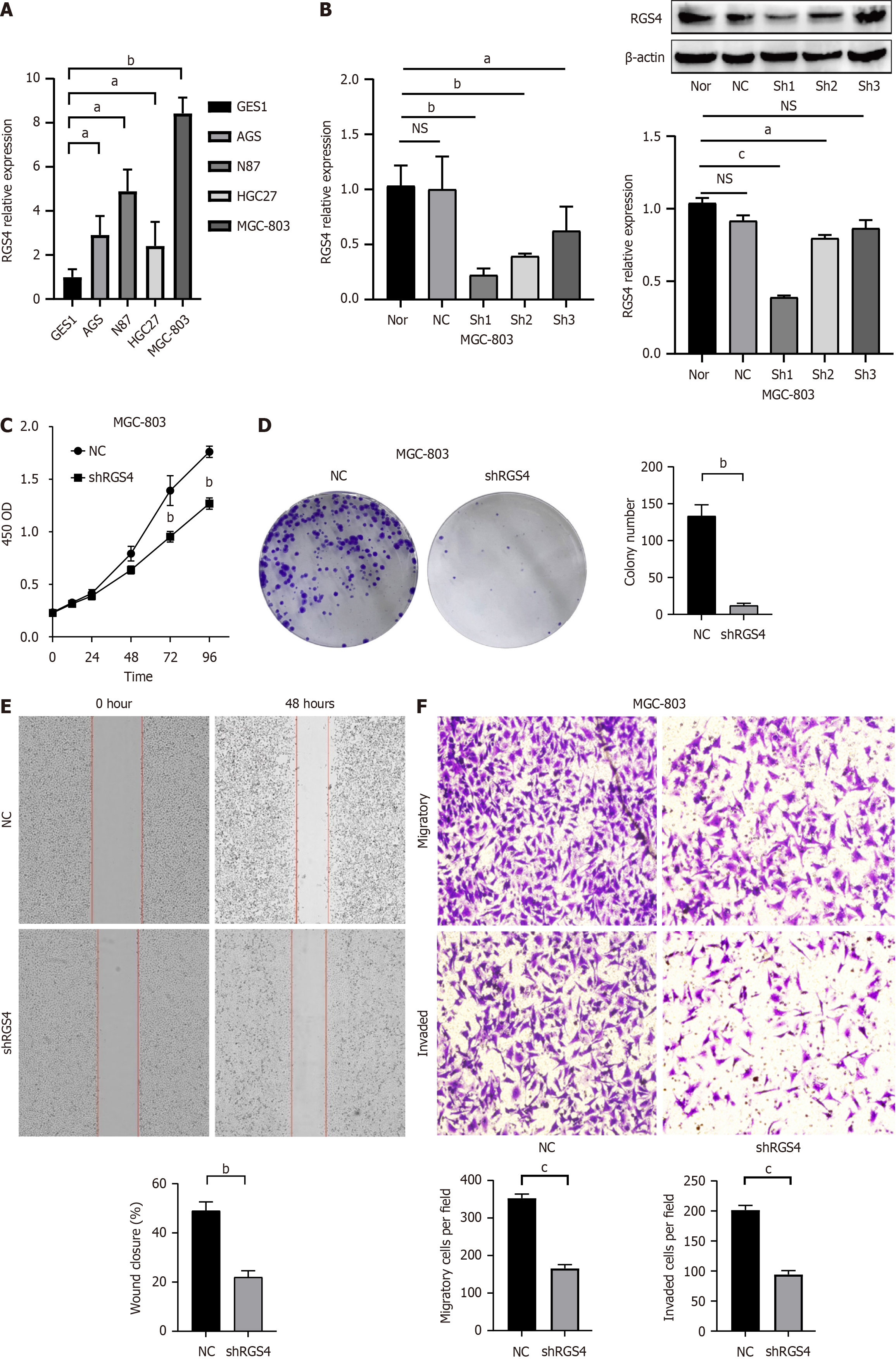
The expression levels of RGS4 in GES-1, NCI-N87, AGS, HGC27, and MGC-803 were assessed using qPCR. Figure 2A showed that the expression levels of RGS4 in NCI-N87, AGS, HGC27, and MGC803 GC cell lines were higher than those in the GES-1 cell line (P < 0.05). Due to MGC-803 cell lines showed the highest expression of RGS4, it was selected for subsequent experiments. MGC803 cells were transfected with shRNA-NC, RGS4 #1, or RGS4 #2. Transfection efficiency was shown in Figure 2B. RGS4 expression was significantly decreased following transfection with RGS4 #1 shRNA, leading to its selection for further experiments. CCK-8 proliferation assays revealed that RGS4 knockdown inhibited the proliferation of MGC-803 cells which were transfected compared with blank control cells (1.761 ± 0.054 vs 1.268 ± 0.054, P < 0.05; Figure 2C). Similarly, colony formation assays (Figure 2D) demonstrated that the number of cell colonies decreased significantly after RGS4 knockdown (133.3 ± 15.3 vs 12.0 ± 3.0, P < 0.05). Furthermore, wound healing (49% ± 3.61% vs 22% ± 2.65%, P < 0.05) and transwell assays (migration: 352.7 ± 11.2 vs 165.7 ± 10.1, P < 0.05; invasion: 201.7 ± 7.6 vs 94.3 ± 6.4, P < 0.05) indicated that RGS4 suppression significantly impaired cell migration and invasion (Figure 2E and F).
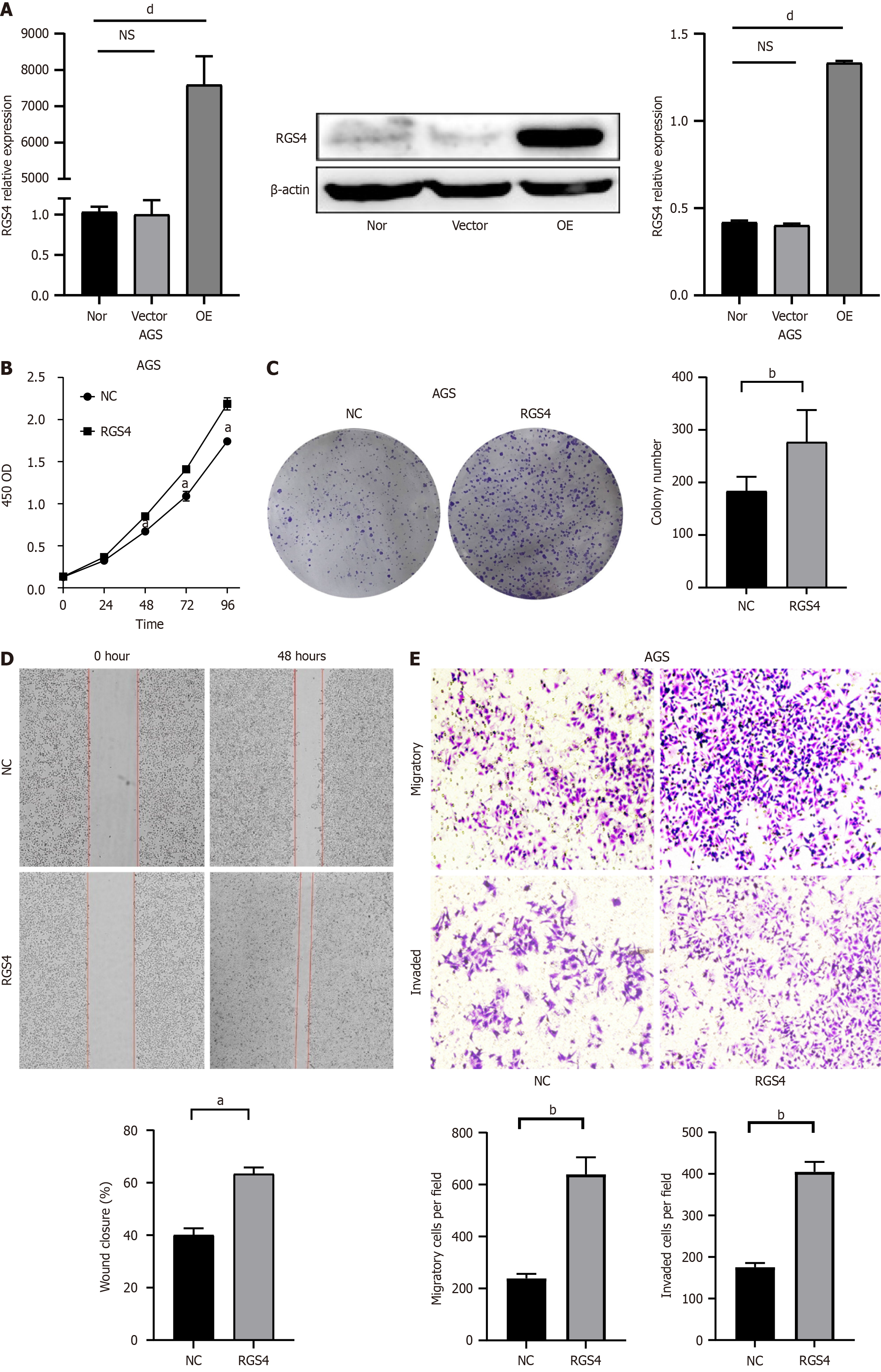
To further elucidate the effect of RGS4 overexpression on cell behavior, AGS cell lines stably overexpressing RGS4 were established and their transfection efficiency was verified. Compared with the vector and NC groups, RGS4 overexpression significantly increased protein and mRNA levels (Figure 3A). CCK-8 proliferation assays (Figure 3B) showed that AGS cells overexpressing RGS4 exhibited enhanced proliferative capabilities (1.741 ± 0.037 vs 2.187 ± 0.073, P < 0.05). Colony formation assays (Figure 3C) showed that RGS4 overexpression significantly promoted colony formation in AGS cells (183.3 ± 27.4 vs 276.7 ± 61.2, P < 0.05). Additionally, wound healing (40% ± 2.65% vs 63.33% ± 2.52%, P < 0.05) and transwell assays (migration: 238.7 ± 17.8 vs 639.7 ± 65.3, P < 0.05; invasion: 175.7 ± 10.1 vs 405.3 ± 23.4, P < 0.05) demonstrated that RGS4 overexpression significantly increased cell migration and invasion (Figure 3D and E).
We analyzed the potential mechanism of RGS4 affecting GC cells through GESA. The results of GSEA were showed in Figure 4A. Notably, extracellular matrix (ECM) receptor interactions and focal adhesion pathways exhibited the highest enrichment score. FAK was a key kinase of the focal adhesion pathway and FAK phosphorylation can promote cell proliferation by activating the PI3K/AKT pathway[18-20]. FAK also mediates various cellular functions, including epithelial-mesenchymal transition (EMT), which is necessary for tumor cells to migrate to and invade the ECM[20]. Additionally, the ECM receptor interaction pathway is closely related to EMT[21,22]. Accordingly, we hypothesized that RGS4 activated the FAK/PI3K/AKT pathway and EMT, thereby promoting GC progression. This hypothesis was corroborated. RGS4 knockdown reduced the ratios of p-FAK/FAK, p-PI3K/PI3K, and p-AKT/AKT (P < 0.05, Figure 4B), and up-regulated the level of epithelial marker E-cadherin (P < 0.05) and down-regulated mesenchymal markers N-cadherin and vimentin (P < 0.05, Figure 4C). In contrast, overexpressing RGS4 exhibited the opposite effect (Figure 4D and E). These findings supported the hypothesis that RGS4 enhances malignant behavior of GC cell by modulating the FAK/PI3K/AKT pathway and EMT.
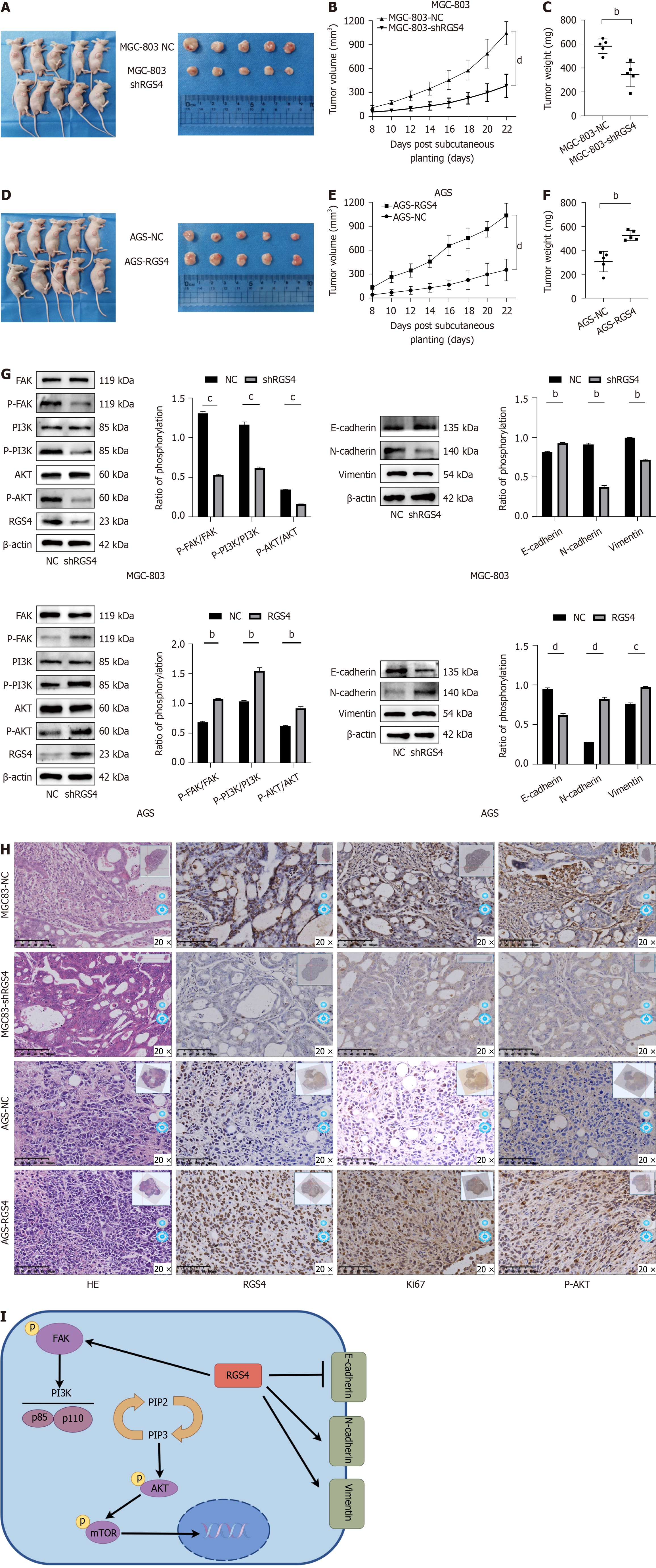
A subcutaneous tumorigenesis experiment was conducted using nude mice for investigating the effect of RGS4 on GC in vivo. The rate and volume of tumor growth significantly decreased in the MGC-803 cell line with RGS4 knockdown compared with the control group (1043.3 ± 146.1 vs 384.6 ± 145.1 mm3, P < 0.05, Figure 5A and B). Compared with the blank control group, the RGS4 knockdown group had a lower tumor weight at endpoint (582.0 ± 61.2 vs 344.8 ± 101.6 g, P < 0.05, Figure 5C). On the contrary (Figure 5D-F), compared with the control tumors, overexpressing RGS4 in AGS cells resulted in enhanced tumor growth in vivo, as evidenced by increased tumor volume (352.8 ± 137.6 vs 1035.3 ± 153.2 mm3, P < 0.05) and weight (305.6 ± 85.1 vs 523.4 ± 41.7 g, P < 0.05). Western blot analyses confirmed that RGS4 knockdown had a significant effect on signaling pathways, leading to decreased ratios of p-FAK/FAK, p-PI3K/PI3K, and p-AKT/AKT; up-regulated level of E-cadherin and down-regulated level of vimentin and N-cadherin (P < 0.05). Conversely, RGS4 overexpression notably increased these ratios and altered protein expression in a manner consistent with tumor progression (P < 0.05, Figure 5G). IHC staining further supported these findings, showing that RGS4 modulation influenced key markers of cell proliferation and signaling, with knockdown reduction and overexpression increasing the expression of Ki67 and phosphorylated AKT (Figure 5H).
GC is a highly malignant gastrointestinal tumor and main treatment method is still surgical resection. However, many patients have poor prognosis due to local recurrence or distant metastasis[23]. Therefore, we need to improve the pro
RGS has been proved to be related to the development of many tumors, including lung cancer, breast cancer, osteosarcoma and bladder cancer[25]. Previous research by our team revealed that RGS1 restrained the differentiation of GC cells, ultimately enhancing the malignancy of tumor. These findings indicate a potentially crucial role for the RGS gene in the advancement of GC. Therefore, we tried to investigate the effects of other RGS gene family members on GC. By mining public databases, we preliminarily identified that RGS4 was highly expressed in GC and contributed to worse prognosis in patients of GC. Subsequent patient validation at our center further confirmed that patients with higher RGS4 levels exhibited shorter OS and elevated T stage, N stage, TNM stage, and tumor grade. These results suggest that we can predict the prognosis of GC patients based on RGS4 expression. As a member of the G protein-coupled receptor family, PAR4 is associated with more aggressive forms of GC[26]. RGS4 formed a complex with the receptor and Gα in cells, inhibiting PAR4-mediated signaling[24]. Therefore, RGS4 may contribute to the malignant progression of GC by hindering PAR4-related signaling, leading to poor prognosis and advanced TNM stage in patients. In addition to their conventional role in GTPase-activating protein activity, RGS proteins can also influence other non-canonical functions in cancer. For instance, RGS5 and RGS16 have been shown to facilitate tumor invasion and metastasis through EMT[27,28]. Belonging to the B/R4 RGS gene family, RGS4, RGS5, and RGS16 exhibit similar patterns of cell growth and migration regulation[25]. It is plausible that RGS4 enhances metastasis in patients with GC by promoting EMT.
In this study, we conducted knockdown and overexpression experiments of the RGS4 gene to further elucidate its mechanism of action. Our results indicated that overexpression of RGS4 promoted malignant behaviors in GC cells, while knockdown of RGS4 reversed these effects. This suggests that RGS4 functions as a tumor-promoting factor in GC. Similarly, RGS4 also promotes the progression of glioblastoma[29], non-small cell lung cancer[30] and osteosarcoma[13]. GSEA results demonstrated that RGS4 expression was significantly associated with FAK signaling. Subsequent expe
In summary, this study found that RGS4 may promote the progression of GC via the FAK/PI3K/AKT pathway and EMT (Figure 5I). Inevitably, this study has several limitations. First, more clinical samples are necessary to confirm the prognostic significance of RGS4 in GC. Second, whether RGS4 affects FAK through GPCRs requires further clarification. Future investigations will involve additional experiments to better understand detailed mechanisms of RGS4-mediated oncogenesis in GC.
RGS4 is highly expressed in GC and can be regarded as a predictor to predict the prognosis of GC patients. RGS4 contributes to the progression of GC via the FAK/PI3K/AKT pathway and EMT and may be a potential therapeutic target for GC.
We thank Tianjin Medical University General Hospital and Jincheng People’s Hospital for their support.
| 1. | Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, Jemal A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5690] [Cited by in RCA: 8188] [Article Influence: 8188.0] [Reference Citation Analysis (2)] |
| 2. | Galletti G, Zhang C, Gjyrezi A, Cleveland K, Zhang J, Powell S, Thakkar PV, Betel D, Shah MA, Giannakakou P. Microtubule Engagement with Taxane Is Altered in Taxane-Resistant Gastric Cancer. Clin Cancer Res. 2020;26:3771-3783. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 3. | Cosín-Roger J, Ortiz-Masia D, Barrachina MD, Calatayud S. Metabolite Sensing GPCRs: Promising Therapeutic Targets for Cancer Treatment? Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 4. | Wu V, Yeerna H, Nohata N, Chiou J, Harismendy O, Raimondi F, Inoue A, Russell RB, Tamayo P, Gutkind JS. Illuminating the Onco-GPCRome: Novel G protein-coupled receptor-driven oncocrine networks and targets for cancer immunotherapy. J Biol Chem. 2019;294:11062-11086. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 145] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 5. | Yang C, Zhang X, Yang X, Lian F, Sun Z, Huang Y, Shen W. Function and regulation of RGS family members in solid tumours: a comprehensive review. Cell Commun Signal. 2023;21:316. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 6. | Roman DL, Traynor JR. Regulators of G protein signaling (RGS) proteins as drug targets: modulating G-protein-coupled receptor (GPCR) signal transduction. J Med Chem. 2011;54:7433-7440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 38] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 7. | Alqinyah M, Hooks SB. Regulating the regulators: Epigenetic, transcriptional, and post-translational regulation of RGS proteins. Cell Signal. 2018;42:77-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 8. | Sethakorn N, Dulin NO. RGS expression in cancer: oncomining the cancer microarray data. J Recept Signal Transduct Res. 2013;33:166-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 38] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 9. | Yan H, Zhang JL, Leung KT, Lo KW, Yu J, To KF, Kang W. An Update of G-Protein-Coupled Receptor Signaling and Its Deregulation in Gastric Carcinogenesis. Cancers (Basel). 2023;15. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 10. | Sriram K, Moyung K, Corriden R, Carter H, Insel PA. GPCRs show widespread differential mRNA expression and frequent mutation and copy number variation in solid tumors. PLoS Biol. 2019;17:e3000434. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 53] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 11. | Li S, Yang H, Li S, Zhao Z, Wang D, Fu W. High expression of regulator of G-protein signalling 1 is associated with the poor differentiation and prognosis of gastric cancer. Oncol Lett. 2021;21:322. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 12. | Watson N, Linder ME, Druey KM, Kehrl JH, Blumer KJ. RGS family members: GTPase-activating proteins for heterotrimeric G-protein alpha-subunits. Nature. 1996;383:172-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 440] [Cited by in RCA: 448] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 13. | Liu WG, Zhuo L, Lu Y, Wang L, Ji YX, Guo Q. miR-874-3p inhibits cell migration through targeting RGS4 in osteosarcoma. J Gene Med. 2020;22:e3213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 14. | Tang Y, Qing C, Wang J, Zeng Z. DNA Methylation-based Diagnostic and Prognostic Biomarkers for Glioblastoma. Cell Transplant. 2020;29:963689720933241. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 15. | Park HJ, Kim SH, Moon DO. Growth inhibition of human breast carcinoma cells by overexpression of regulator of G-protein signaling 4. Oncol Lett. 2017;13:4357-4363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 16. | Cheng C, Yue W, Li L, Li S, Gao C, Si L, Tian H. Regulator of G-protein signaling 4: A novel tumor suppressor with prognostic significance in non-small cell lung cancer. Biochem Biophys Res Commun. 2016;469:384-391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 17. | Chen X, Xie H, Wang X, Zheng Z, Jin S. CIRBP Knockdown Attenuates Tumourigenesis and Improves the Chemosensitivity of Pancreatic Cancer via the Downregulation of DYRK1B. Front Cell Dev Biol. 2021;9:667551. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 18. | Tan X, Yan Y, Song B, Zhu S, Mei Q, Wu K. Focal adhesion kinase: from biological functions to therapeutic strategies. Exp Hematol Oncol. 2023;12:83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 54] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 19. | Dawson JC, Serrels A, Stupack DG, Schlaepfer DD, Frame MC. Targeting FAK in anticancer combination therapies. Nat Rev Cancer. 2021;21:313-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 246] [Article Influence: 61.5] [Reference Citation Analysis (0)] |
| 20. | Lee BY, Timpson P, Horvath LG, Daly RJ. FAK signaling in human cancer as a target for therapeutics. Pharmacol Ther. 2015;146:132-149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 228] [Cited by in RCA: 324] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 21. | Malik R, Lelkes PI, Cukierman E. Biomechanical and biochemical remodeling of stromal extracellular matrix in cancer. Trends Biotechnol. 2015;33:230-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 254] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 22. | Ottone T, Silvestrini G, Piazza R, Travaglini S, Gurnari C, Marchesi F, Nardozza AM, Fabiani E, Attardi E, Guarnera L, Divona M, Ricci P, Irno Consalvo MA, Ienzi S, Arcese R, Biagi A, Fiori L, Novello M, Mauriello A, Venditti A, Anemona L, Voso MT. Expression profiling of extramedullary acute myeloid leukemia suggests involvement of epithelial-mesenchymal transition pathways. Leukemia. 2023;37:2383-2394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 23. | Malla RR, Nellipudi HR, Srilatha M, Nagaraju GP. HER-2 positive gastric cancer: Current targeted treatments. Int J Biol Macromol. 2024;274:133247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 24. | Kim Y, Ghil S. Regulators of G-protein signaling, RGS2 and RGS4, inhibit protease-activated receptor 4-mediated signaling by forming a complex with the receptor and Gα in live cells. Cell Commun Signal. 2020;18:86. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 25. | Li L, Xu Q, Tang C. RGS proteins and their roles in cancer: friend or foe? Cancer Cell Int. 2023;23:81. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 26. | Zhang Y, Yu G, Jiang P, Xiang Y, Li W, Lee W, Zhang Y. Decreased expression of protease-activated receptor 4 in human gastric cancer. Int J Biochem Cell Biol. 2011;43:1277-1283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 27. | Abe Y, Ogasawara S, Akiba J, Naito Y, Kondo R, Nakamura K, Kusukawa J, Yano H. Expression and role of regulator of G-protein signaling 5 in squamous cell carcinoma of the tongue. Clin Exp Dent Res. 2019;5:160-169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 28. | Huang R, Li G, Zhao Z, Zeng F, Zhang K, Liu Y, Wang K, Hu H. RGS16 promotes glioma progression and serves as a prognostic factor. CNS Neurosci Ther. 2020;26:791-803. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 29. | Guda MR, Velpula KK, Asuthkar S, Cain CP, Tsung AJ. Targeting RGS4 Ablates Glioblastoma Proliferation. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 30. | He Z, Yu L, Luo S, Li Q, Huang S, An Y. RGS4 Regulates Proliferation And Apoptosis Of NSCLC Cells Via microRNA-16 And Brain-Derived Neurotrophic Factor. Onco Targets Ther. 2019;12:8701-8714. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 31. | Ye Z, Xia Y, Li L, Li B, Chen W, Han S, Zhou X, Chen L, Yu W, Ruan Y, Cheng F. Effect of transmembrane protein 100 on prostate cancer progression by regulating SCNN1D through the FAK/PI3K/AKT pathway. Transl Oncol. 2023;27:101578. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 32. | Chen L, Niu W, Zang H, Qiu Y. DTX3L Accelerates Pancreatic cancer Progression via FAK/PI3K/AKT Axis. Biochem Genet. 2024;62:814-830. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 33. | Luo Y, Zhang D, Hou L, Lin N. Vernodalin Suppresses Tumor Proliferation and Increases Apoptosis of Gastric Cancer Cells Through Attenuation of FAK/PI3K/AKT/mTOR and MAPKs Signaling Pathways. Curr Pharm Biotechnol. 2023;24:708-717. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 34. | Wu YJ, Lin SH, Din ZH, Su JH, Liu CI. Sinulariolide Inhibits Gastric Cancer Cell Migration and Invasion through Downregulation of the EMT Process and Suppression of FAK/PI3K/AKT/mTOR and MAPKs Signaling Pathways. Mar Drugs. 2019;17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 35. | Huo FC, Zhu WT, Liu X, Zhou Y, Zhang LS, Mou J. Epidermal growth factor-like domain multiple 6 (EGFL6) promotes the migration and invasion of gastric cancer cells by inducing epithelial-mesenchymal transition. Invest New Drugs. 2021;39:304-316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 36. | Du WW, Li X, Li T, Li H, Khorshidi A, Liu F, Yang BB. The microRNA miR-17-3p inhibits mouse cardiac fibroblast senescence by targeting Par4. J Cell Sci. 2015;128:293-304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 41] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 37. | Li D, Xia L, Huang P, Wang Z, Guo Q, Huang C, Leng W, Qin S. Heterogeneity and plasticity of epithelial-mesenchymal transition (EMT) in cancer metastasis: Focusing on partial EMT and regulatory mechanisms. Cell Prolif. 2023;56:e13423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 75] [Reference Citation Analysis (0)] |