Published online May 14, 2025. doi: 10.3748/wjg.v31.i18.105729
Revised: March 27, 2025
Accepted: April 14, 2025
Published online: May 14, 2025
Processing time: 97 Days and 18.1 Hours
As a member of the chaperonin-containing tailless complex polypeptide 1 (TCP1) complex, which plays a pivotal role in ensuring the accurate folding of numerous proteins, chaperonin-containing TCP1 subunit 6A (CCT6A) participates in various physiological and pathological processes. However, its effects on cell death and cancer therapy and the underlying mechanisms need further exploration in colorectal cancer (CRC) cells.
To explore the effects of CCT6A on cell death and cancer therapy and the under
Cell proliferation was evaluated using the MTS assay, EdU staining, and colony growth assays. The expression of CCT6A was monitored by immunoblotting and quantitative PCR. CCT6A was knocked out by CRISPR-Cas9, and overexpressed by transfecting plasmids. Autophagy was examined by immunoblotting and the mCherry-GFP-LC3 assay. To monitor apoptosis and necroptosis, immunoblotting, co-immunoprecipitation, and flow cytometry were employed.
Cisplatin (DDP) exerted cytotoxic effects on CRC cells while simultaneously downregulating the expression of CCT6A. Depletion of CCT6A amplified the cytotoxic effects of DDP, whereas overexpression of CCT6A attenuated these adverse effects. CCT6A suppressed autophagy, apoptosis, and necroptosis under both basal and DDP-treated conditions. Autophagy inhibitors significantly enhanced the cytotoxic effects of DDP, whereas a necroptosis inhibitor partially reversed the cell viability loss induced by DDP. Furthermore, inhibiting autophagy enhanced both apoptosis and necroptosis induced by DDP.
CCT6A negatively modulates autophagy, apoptosis, and necroptosis, and CCT6A confers resistance to DDP therapy in CRC, suggesting its potential as a therapeutic target.
Core Tip: This study evaluated the role of chaperonin-containing tailless complex polypeptide 1 subunit 6A (CCT6A) in colorectal cancer (CRC) therapy. CCT6A expression was downregulated by cisplatin (DDP) in CRC cells, enhancing its cytotoxicity. Depletion of CCT6A potentiated DDP's effects by promoting autophagy, apoptosis, and necroptosis, whereas its overexpression provided resistance. Immunoblotting and autophagy confirmed CCT6A's negative regulatory role in autophagy. Markers of apoptosis and necroptosis were upregulated in CCT6A-depleted cells, indicating increased cell death. Autophagy inhibition exacerbated cell death, compromising viability and proliferation. Thus, CCT6A is a potential therapeutic target in CRC, modulating key cellular processes and enhancing DDP efficacy.
- Citation: Ma JX, Li XJ, Li YL, Liu MC, Du RH, Cheng Y, Li LJ, Ai ZY, Jiang JT, Yan SY. Chaperonin-containing tailless complex polypeptide 1 subunit 6A negatively regulates autophagy and protects colorectal cancer cells from cisplatin-induced cytotoxicity. World J Gastroenterol 2025; 31(18): 105729
- URL: https://www.wjgnet.com/1007-9327/full/v31/i18/105729.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i18.105729
Although the incidence of colorectal cancer (CRC) has decreased in average-age onset patients in high-income nations, it continues to rank as the world's third most frequently diagnosed cancer, with rising rates observed in developing economies[1,2]. Despite advances in comprehensive treatment strategies include surgery, chemotherapy, radiation therapy, targeted therapy, and immunotherapy, CRC is notorious for its multidrug resistance[3]. Thus, early detection and prompt intervention are of paramount importance in enhancing survival outcomes.
Chaperonin-containing tailless complex polypeptide 1 (TCP1) subunit 6A (CCT6A), also known as the chaperonin containing TCP1 subunit 6A, encodes a vital protein that constitutes the TCP1 complex (alternatively named TRiC)[4,5]. This chaperonin complex, indispensable for protein folding, comprises two intricately stacked rings, each harboring eight distinct proteins. By harnessing ATP energy, the TCP1 complex orchestrates the folding of unfolded polypeptide chains into functional proteins, including essential components like actin and tubulin, which serve as the foundational pillars of cellular architecture and motility[6]. The ubiquitous expression of CCT6A across diverse tissues underscores its central importance in intracellular protein folding, a fundamental biological process that is crucial for cellular growth, differentiation, and the maintenance of homeostasis[7,8]. Its high degree of conservation and multifaceted functions make CCT6A and its associated complex cornerstone components of cellular life. As such, CCT6A has garnered significant attention in biomedical research and disease diagnosis, serving as a crucial target for investigating protein folding disorders and related pathologies[9,10]. Yan et al[5] reported that CCT6A exerts beneficial effects on alleviating pulmonary fibrosis by effectively inhibiting the hypoxia-inducible factor 1 alpha-mediated lactate production pathway. Moreover, a recent study from our group and others indicated that CCT6A plays a pro-cancer role in various types of cancer, including CRC and lung adenocarcinoma[5,9,11].
While autophagy protects against stress, prolonged activation can lead to autophagic cell death, a phenomenon termed autophagic cell death. Apoptosis, autophagic cell death, and necroptosis represent three intricate and interconnected forms of programmed cell death, each playing a pivotal role in the landscape of cancer therapy[12]. Generally, inhibition of autophagy can sensitize cells to apoptosis induction by chemotherapy and radiotherapy[13-15]. Numerous proteins and intricate signaling pathways are intricately involved in regulating both autophagy and apoptosis. Notably, Beclin-1 protein, a pivotal autophagy regulator, engages in a complex interplay with anti-apoptotic proteins such as B-cell lymphoma 2 (Bcl-2), thereby exerting dual influence on both of these vital cellular processes[16]. Necroptosis serves as a backup mechanism when apoptosis is blocked[17]. Shared signaling molecules, such as receptor-interacting protein kinases (RIPs), can switch the cell's fate from apoptosis to necroptosis under certain conditions. Necroptosis involves the activation of specific kinases (RIP1 and RIP3) that can phosphorylate and inactivate anti-apoptotic proteins, thereby promoting cell death[18]. For apoptosis-resistant cancer cells, targeting necroptosis emerges as a promising strategy, as it can trigger tumor cell death and stimulate antitumor immune responses. The delicate balance between these processes is critical, and strategies that selectively target their interactions in cancer cells hold significant potential for enhancing the efficacy of anticancer treatments.
In the present investigation, we demonstrated that cisplatin (DDP) significantly diminished both the viability and proliferation of CRC cells, concurrently suppressing the expression level of CCT6A. Notably, the overexpression (OE) of CCT6A mitigated the cytotoxic effects of DDP, whereas its depletion intensified these impacts. Employing rigorous detection methodologies and pertinent indicators, we confirmed that CCT6A negatively regulates autophagy, apoptosis, and necroptosis in CRC. Interestingly, the introduction of autophagy inhibitors not only augmented the apoptosis and necroptosis elicited by DDP but also alleviated the associated loss of cell viability, indicating that autophagy serves as a protective mechanism during these biological processes.
DDP, HY-17394, and 3-Methyladenine (3-MA, HY-19312) were obtained from MedChemExpress (Monmouth Junction, NJ, United States). Chloroquine diphosphate salt (CQ, C6628), necrostatin-1 (Nec-1, 4311-88-0), and polyclonal antibodies against light chain 3 (LC3, L7543) were obtained from Sigma-Aldrich (St. Louis, MO, United States). Primary antibodies against CCT6A (19793-1-AP), tubulin (66240-1-Ig), phosphorylated Unc-51-like autophagy activating kinase 1 (phospho-Ulk1) (Ser556, 80218-1-RR), Beclin-1 (11306-1-AP), and p62 (66184-1-Ig) were purchased from Proteintech Group (Wuhan, China). Primary antibodies against phosphorylated mammalian target of rapamycin (phosphor-mTOR) (S2448, 5536S), total-mTOR (2983S), phospho-p70 (T389, 9234S), total-p70 (34475S), total-mixed lineage kinase domain-like (MLKL, 14993S), total-RIP3 (13526S), phospho-RIP1 (S166, 65746S), phospho-RIP3 (S227, 93654S), Bcl-extra large (Bcl-xL) (2764S), poly(ADP-ribose) polymerase 1 (PARP-1, 9542S), and cleaved caspase-3 (9661S) were purchased from Cell Signaling Technology (Danvers, MA, United States). Total-RIP1 (610458) and phospho-MLKL (S358, 187091) were purchased from BD Transduction Laboratories (Franklin Lakes, NJ, United States) and Abcam (Cambridge, United Kingdom), respectively. A Cell Apoptosis Detection Kit (KGA1102-50) was obtained from KeyGene Biotech (Wageningen, Netherlands).
Human SW480 and HT29 CRC cell lines were obtained from American Type Culture Collection (Manassas, VA, United States) and cultured in Dulbecco’s Modified Eagle Medium (C11995500BT; Gibco, Waltham, MA, United States) supplemented with 10% fetal bovine serum (10099-141; Gibco) as well as 1% antibiotics (UB89609; BIOODIN, Shandong, China) at 37 °C, 5% CO2. The Attractive Transfection Reagent (301005; QIAGEN, Hilden, Germany) was utilized to perform transfections when cells grew to 70%-80% confluency, according to the manufacturer’s instructions. The OE-control (Ctrl) and OE-CCT6A plasmids were purchased from Fenghui Biotechnology (Hunan, China), and the knockout (KO)-CCT6A (YKO-RP003-hCCT6A-gRNA1: 5’-CGGTCAACATCAGCGCAGCGCGG-3’, YKO-RP003-hCCT6A-gRNA2: 5’-GGTCAACTTCAGCGCAGCGCGGG-3’) plasmids were obtained from Genai Biotechnology (Shanghai, China). To generate KO-CCT6A cells, transfected and subsequently cultured monoclonal cells were employed to rigorously screen and confirm the successful elimination of CCT6A expression.
The whole protein lysates were extracted from the indicated cells with RIPA lysis buffer (89901; Thermo Fisher Scientific, Waltham, MA, United States) and denatured at 100 °C for 15 minutes. The samples were separated by SDS-PAGE (Gel42012; Elife Technology, Shanghai, China), electrotransferred to polyvinylidene fluoride membranes (88520, Invitrogen, Carlsbad, CA, United States), and blocked in 5% skim milk. Subsequently, the membranes were incubated at 4 °C overnight with the corresponding primary antibody, and then at room temperature for 1 hour with the secondary antibody. Then the bands were visualized using ECL Luminescent Solution (P0018S; Beyotime Biotech, Beijing, China), and the grayscale values were analyzed by ImageJ software (National Institutes of Health, Bethesda, MD, United States).
The cells were lysed using co-immunoprecipitation (Co-IP) lysis buffer (P0013) purchased from Beyotime Biotech (Beijing, China), followed by the addition of specific primary antibodies to the lysates. Then these lysates were incubated at 4 °C overnight. Subsequently, Protein A/G magnetic beads (HY-K0202; MedChemExpress) were utilized with a hopper magnet to precipitate the proteins. The proteins were separated through heating, and the resulting samples underwent immunoblot analysis.
Using the RNA Easy Fast Tissue/Cell Kit (DP451; TIANGEN, Beijing, China), total cellular RNA was extracted according to the manufacturer's protocol. Reverse transcription was performed using 1 µg RNA at 37 °C for 15 minutes with the 5 × EasyQuick RT MasterMix (CW2634M; CoWin Biosciences, Jiangsu, China), followed by heat inactivation at 85 °C for 5 seconds. After a 10-minute denaturation step, quantitative PCR (qPCR) was conducted using a cycling program of 95 °C (15 seconds), 60 °C (45 seconds), and 72 °C (1 minutes) for up to 40 cycles. The data were calculated based on the internal control of β-actin, and the sequences of primers (Table 1).
| Gene | Primer | Nucleotides |
| CCT6A | Forward (5′ → 3′) | CTGAAACAGGCGGATCTCTACA |
| Reverse (5′ → 3′) | CCCTGTCCATCTCTCTGCTTAC | |
| β-Actin | Forward (5′ → 3′) | GCCTGACGGCCAGGTCATCAC |
| Reverse (5′ → 3′) | CGGATGTCCACGTCACACTTC |
A total of 7500 cells were seeded into each well of 96-well plates and cultured overnight. Following the indicated treatment for the appropriate period, 10 µL MTS/PMS (20:1) was added to each well, and continuously cultivated for 1-2 hours. Subsequently, cell viability was detected at 492 nm using a microplate reader. PMS (P9625) was obtained from Sigma-Aldrich and MTS (G111) was purchased from Promega Corporation (Fitchburg, WI, United States).
For the colony growth assay, cells were cultured in a 12-well plate at a consistent density of 100 cells per well for approximately 14 days. Following this incubation period, the cell colonies were fixed in 4% paraformaldehyde (PFA) at room temperature for 15 minutes. Subsequently, the colonies were stained with Giemsa dye. After staining, images were captured, and the colonies were counted using ImageJ software.
The EdU Cell Proliferation Detection Kit with Alexa Fluor594 (C0078S) was purchased from Beyotime. Cells were divided into a 24-well plate and incubated overnight. Subsequent to specific treatments, staining procedures were conducted according to the manufacturer's instructions. Thereafter, the pictures were taken using fluorescence microscopy (Nikon Co., Tokyo, Japan), and the data were analyzed using Adobe Photoshop and ImageJ software.
The indicated treatments were carried out after the cells were plated on cover slips. Cells were fixed in freshly prepared 4% PFA at room temperature for 15 minutes, followed by washing with phosphate-buffered saline (PBS). Subsequently, the cells were permeabilized with 0.1% Triton X-100, blocked in 0.5% bovine serum albumin at room temperature for 10 minutes, and incubated with the indicated primary antibody at 4 °C overnight. After three washes, the cells were incubated with secondary antibody for 1 hour at room temperature. After another three washes, VECTASHIELD with DAPI (H1200; Newark, CA, United States) was used to stain the nuclei, and images were acquired by fluorescence microscopy (Nikon).
The mCherry-GFP-LC3B adenovirus (Vigene Biosciences,Shandong, China) was utilized to infect the indicated cells in a 6-well plate. Subsequently, the specified treatments were carried out. Then cells were fixed in freshly prepared 4% PFA, and then they were washed three times with PBS. The images were acquired by fluorescence microscopy following stained with VECTASHIELD with DAPI.
After undergoing the appropriate treatment, cells were trypsinized (excluding EGTA/EDTA), harvested while retaining all floating cells, and subsequently washed with PBS. These cells were then stained with fluorescein isothiocyanate-labeled annexin V (FITC) and propidium iodide, following the guidelines provided by the Annexin-V-FITC Apoptosis Detection Kit (K101-100; BioVision Inc., Milpitas, CA, United States). Finally, the stained samples were analyzed using flow cytometry.
The normally distributed data are presented as the mean ± SD, reflecting at least three independent experiments. Statistical significance for comparisons between two groups was analyzed using the two-sided Student's t-test, whereas for comparisons among multiple groups, one-way analysis of variance with the Student-Newman-Keuls post hoc test was employed. P < 0.05 was considered statistically significant.
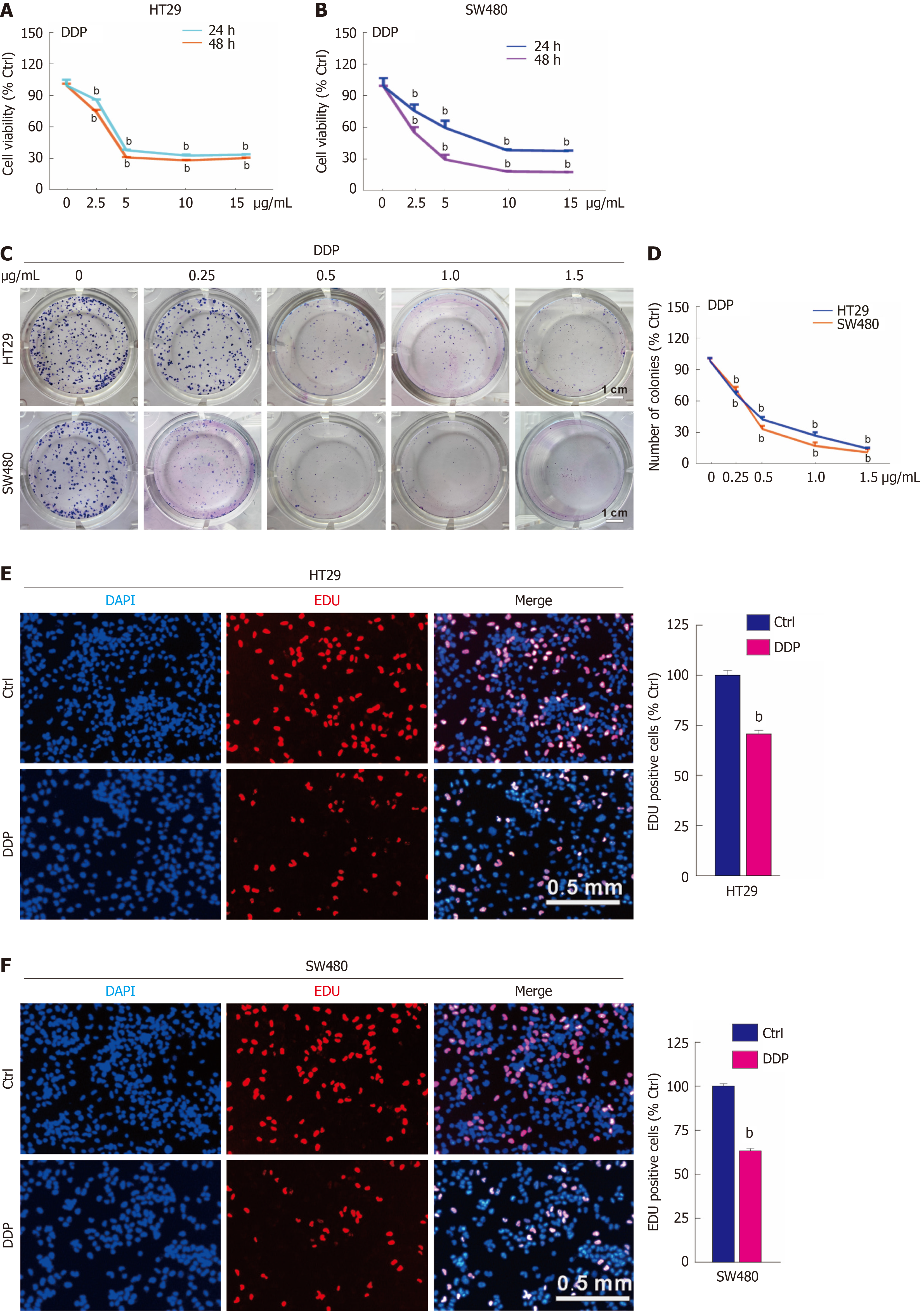
As DDP is a widely used antitumor drug, its cytotoxic effects were monitored by a variety of methods. In particular, the MTS assay prominently demonstrated that DDP elicited a dose-responsive decline in the viability of HT29 and SW480 cells (Figure 1A and B). Notably, at the 24-hour time point, a concentration of 10 μg/mL DDP effectively diminished cellular activity to less than 50% in both HT29 and SW480 cells. By contrast, achieving a comparable reduction at 48 hours necessitated a lower concentration of 5 μg/mL (Figure 1A and B). Consequently, unless otherwise specified, a concentration of 5 μg/mL DDP was selected as the standard for the majority of subsequent experiments. In the colony growth assay, due to the initial seeding of a limited number of cells, administering standard drug concentrations would fully suppress their proliferation into monoclonal colonies. Therefore, lower concentrations of the drugs were employed for treatment, ensuring that a discernible effect on colony formation could be observed and accurately assessed. As shown, DDP significantly reduced the number and size of clones, which might be attributed to decreased cell proliferation and increased cell death (Figure 1C and D). In the EdU staining assay, which visualizes newly synthesized DNA by direct integration, DDP significantly reduced the percentage of EdU-positive cells in both HT29 and SW480 cells (Figure 1E and F). These findings underscore the ability of DDP to inhibit the proliferation of CRC cells.
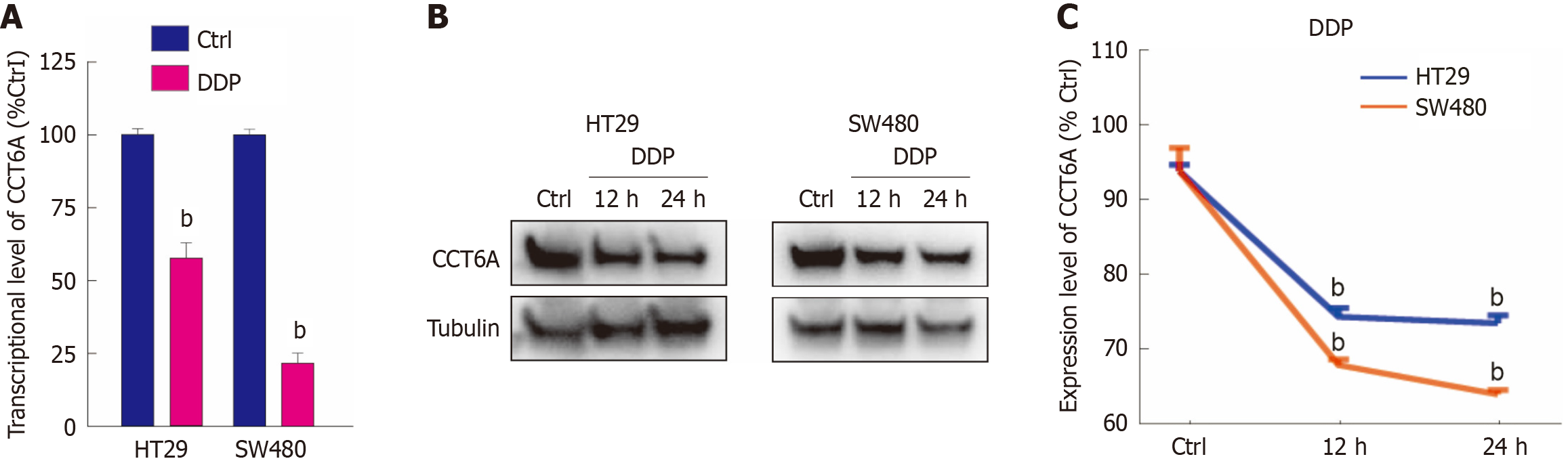
CCT6A is an essential subunit of TCP1 complex. Our previous study verified that it is significantly overexpressed in CRC tumors compared to normal tissues, and a highly elevated level of CCT6A expression correlates with worse prognoses in CRC[11]. In our previous study, we observed that platinum-based drugs affect its level. Upon analyzing the transcriptional level, we observed the pronounced downregulation of CCT6A expression in both HT29 and SW480 cells following DDP treatment (Figure 2A). Furthermore, DDP also decreased the protein level of CCT6A in HT29 cells in a time-dependent manner (Figure 2B and C). A similar pattern was observed in SW480 cells, confirming the consistency of this phenomenon across cell lines (Figure 2B and C). Given that prior studies established the role of CCT6A in promoting cell proliferation[5,9], and our findings revealed an association between CCT6A levels and DDP treatment, we subsequently investigated whether CCT6A could potentially modulate DDP's regulatory effects on cell proliferation.
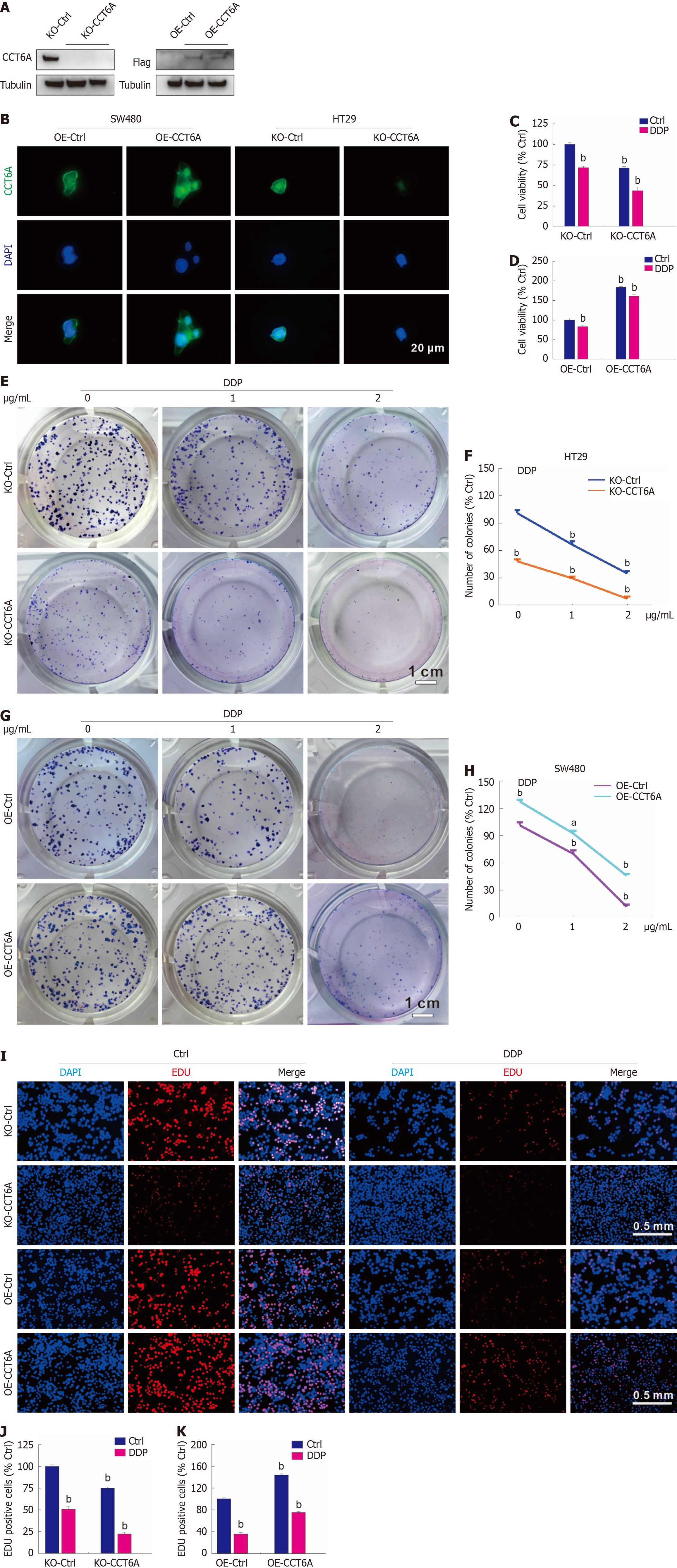
Our former investigation verified that CCT6A is highly expressed in HT29 cells, and rarely expressed in SW480 cells[11]. Therefore, we knocked out CCT6A in HT29 cells, and overexpressed CCT6A in SW480 cells. As shown in Figure 3A, no CCT6A bands were monitored in the KO-CCT6A cells. By contrast, Flag-specific bands were observed in the OE-CCT6A cells (Figure 3A). Furthermore, we conducted an immunofluorescence assay utilizing a CCT6A-specific antibody, which revealed a marked increase in fluorescence intensity in OE-CCT6A cells compared to OE-Ctrl cells (Figure 3B). Conversely, KO-CCT6A cells exhibited virtually no fluorescence, underscoring the specificity and efficacy of our approach (Figure 3B). Employing these engineered cell lines, we discovered that the KO of CCT6A led to a marked decrease in both basal cell viability and further reduced the viability already compromised by DDP treatment. Conversely, OE of CCT6A had the opposite effect, demonstrating an improvement in both basal and DDP-challenged cell survival (Figure 3C and D). Furthermore, we observed that knocking out CCT6A exacerbated the reduction in colony number induced by DDP treatment, whereas overexpressing CCT6A partially mitigated the suppressive effect of DDP on colony formation (Figure 3E-H). In the EdU staining assay, similarly phenotypes were obtained, echoing the findings from previous experiments (Figure 3I-K). The aforementioned results collectively indicate that the level of CCT6A is inversely correlated with the cytotoxicity of DDP in CRC cells, suggesting a pivotal role of CCT6A in modulating the sensitivity of these cells to DDP treatment.
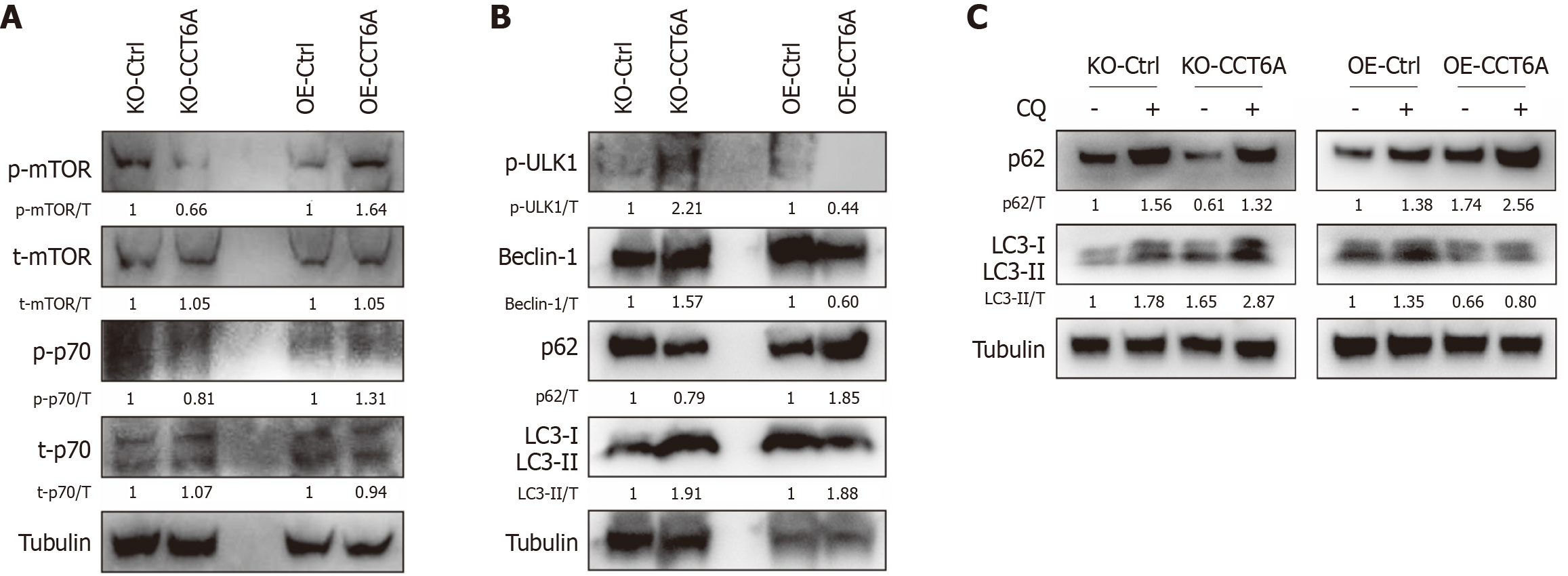
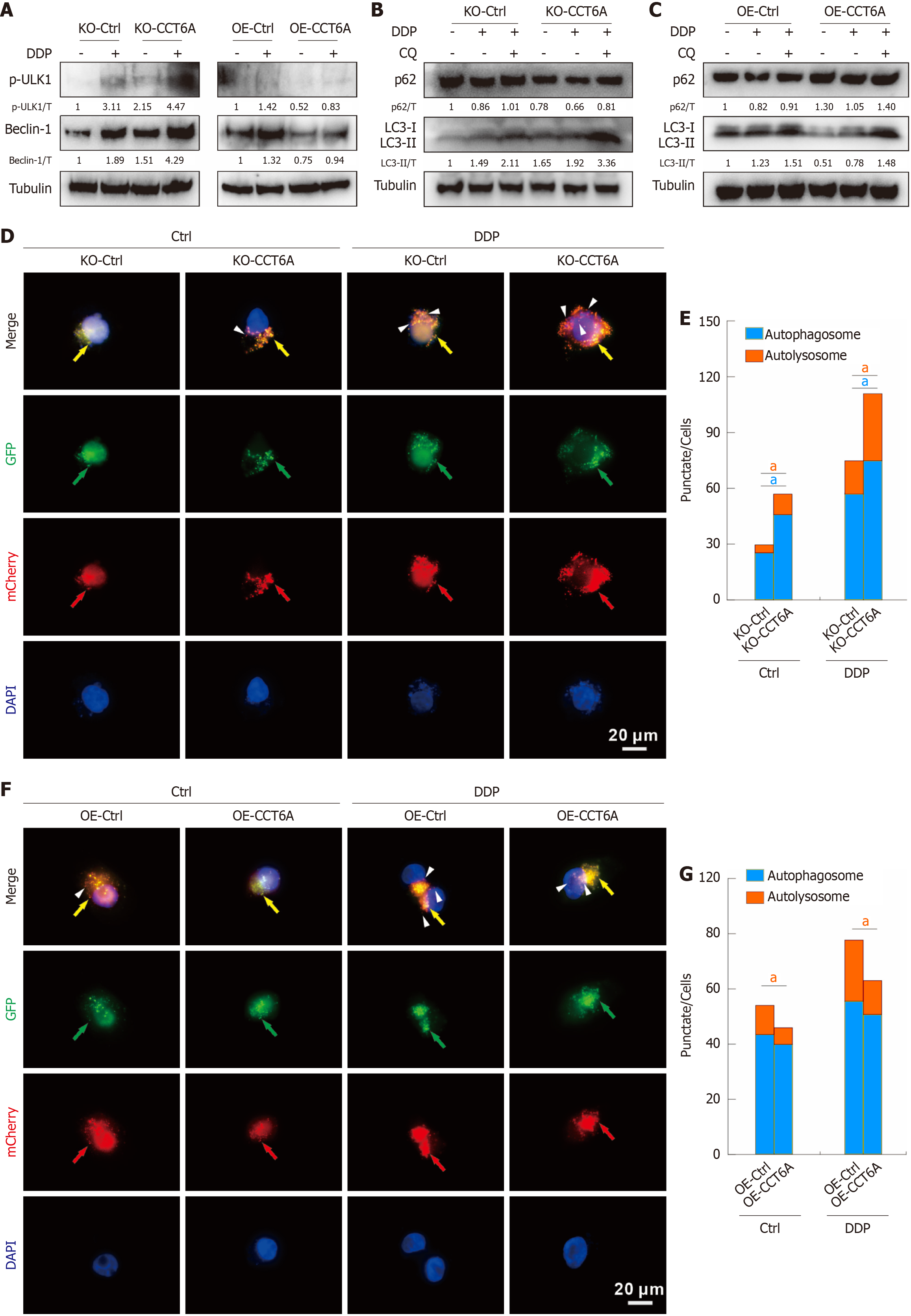
As CCT6A positively regulates cell proliferation, we determined whether it could impact the activity of mTOR signaling, which plays essential roles in modulating transcription and translation[19]. As anticipated, OE of CCT6A led to a notable increase in the phosphorylation level of mTOR at the serine 2448 site, which serves as a hallmark for its kinase activity (Figure 4A). Furthermore, p70 S6 kinase, a crucial downstream effector of mTOR signaling, known for its essential role in promoting cell growth and facilitating G1 phase progression of the cell cycle[19], exhibited increased phosphorylation levels in OE-CCT6A cells compared to OE-Ctrl cells (Figure 4A). This finding provides compelling evidence that OE of CCT6A stimulates the overall activity of the mTOR signaling cascade. Conversely, ablation of CCT6A resulted in a decrease in its activity, further substantiating the positive regulatory role of CCT6A on mTOR signaling (Figure 4A). Apart from its role in proliferation, the mTOR signaling pathway is widely known for its function as a negative regulator of autophagy, a crucial conservative degradation mechanism operative within eukaryotic cells[20]. p-Ulk1 (S555) and Beclin-1 are two essential proteins for autophagy initiation, and their levels were negatively correlated with the level of CCT6A (Figure 4B). Moreover, KO-CCT6A facilitated an increase in the level of the autophagosome marker LC3-II, while concurrently decreasing the level of the autophagic substrate p62 (Figure 4B). Conversely, OE-CCT6A exhibited the reverse effect, demonstrating an opposing trend to that observed with KO-CCT6A (Figure 4B). Moreover, inclusion of the autophagic flux inhibitor CQ effectively halted the degradation of p62 and conspicuously elevated the abundance of LC3-II (Figure 4C). These cumulative findings underscore the pivotal role of CCT6A in negatively modulating autophagy in CRC cells.
Chemotherapy and radiotherapy commonly act as cellular stressors, serving as potent inducers of autophagy[20]. We further investigated whether CCT6A affects the impact of DDP on autophagy. In both KO-Ctrl and OE-Ctrl cells, DDP treatment alone increased the levels of p-Ulk1 and Beclin-1 (Figure 5A). Meanwhile, the ablation of CCT6A further augmented their levels, whereas its OE had the opposite effect (Figure 5A). Moreover, DDP promoted the accumulation of LC3-II and facilitated the degradation of p62 (Figure 5B and C). In DDP-treated cells, CCT6A KO enhanced the degradation of p62 and its expression reduced p62 degradation. Meanwhile, KO-CCT6A increased LC3-II level in DDP-treated cells, whereas OE-CCT6A presented opposite phenotype (Figure 5B and C). Importantly, the inclusion of CQ enhanced LC3-II accumulation and abrogated p62 degradation induced by DDP (Figure 5B and C), confirming that CCT6A indeed influences DDP-induced autophagic flux. To gain further insights, we employed the mCherry-GFP-LC3 assay, leveraging the differential pH stability of mCherry and GFP fluorescent proteins[20]. In detail, during the initial stage of autophagosome formation, LC3 embeds into the autophagosome membrane, resulting in the appearance of yellow punctate structures (GFP+/mCherry+). Subsequently, upon fusion of the autophagosome and lysosome, the acidic milieu within the lysosome extinguishes the GFP fluorescence, with only the red punctate structures remaining (GFP-/mCherry+). Our results demonstrated that both DDP treatment and CCT6A KO enhanced the formation of autophagosome dots (yellow) and autolysosome dots (sole red), indicative of accelerated complete autophagic flux (Figure 5D and E). Meanwhile, KO of CCT6A further enhanced the punctate number of autophagosome dots and autolysosome dots in the DDP-treated cells (Figure 5D and E). By contrast, OE of CCT6A diminished both basal and DDP-induced autolysosome dots (Figure 5F and G). These findings underscore the pivotal role of CCT6A in negatively regulating the autophagy modulated by DDP.
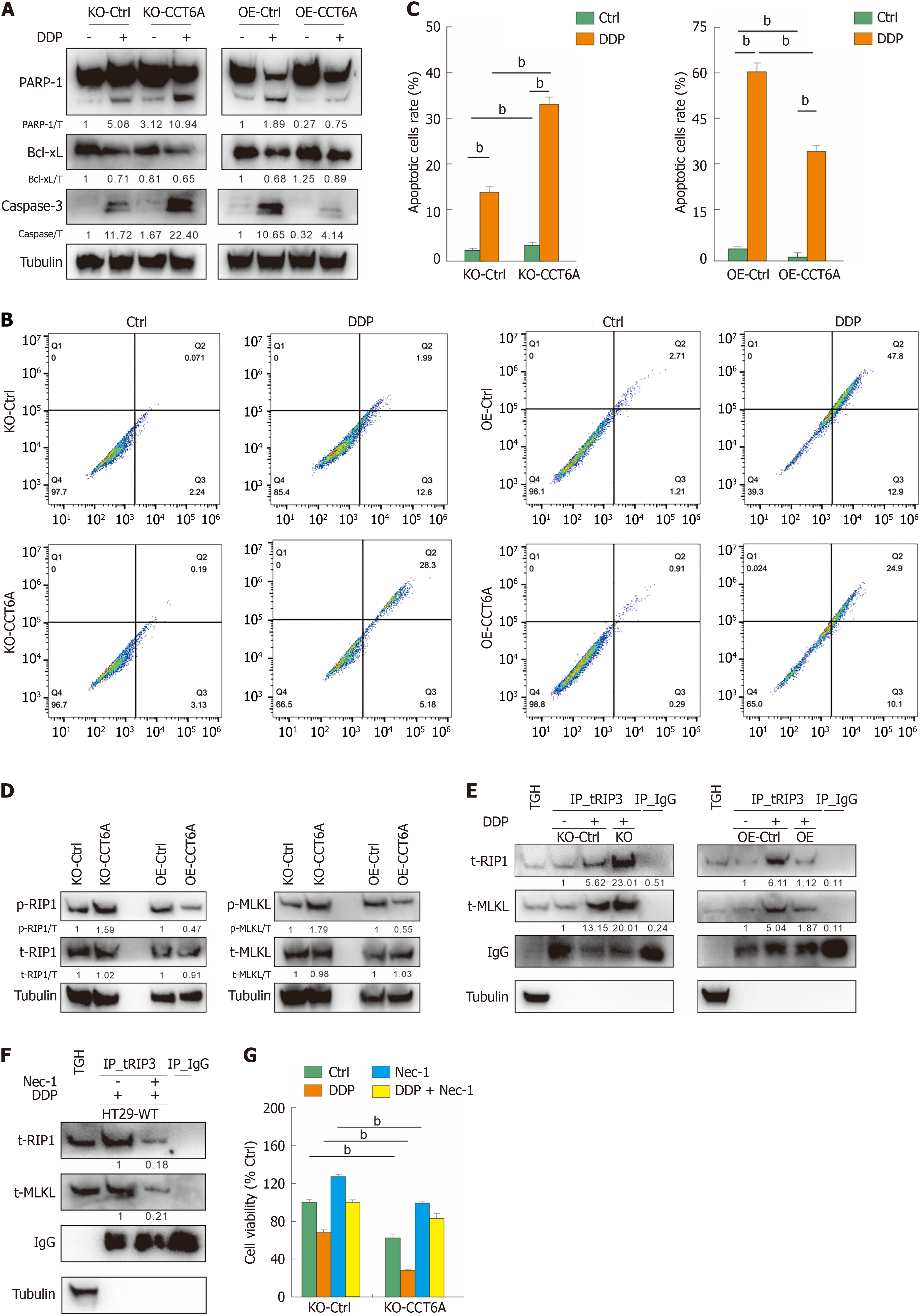
As a commonly used chemotherapy agent, DDP has the ability to efficiently trigger apoptosis in diverse cellular types. DDP alone exhibited the potent ability to elicit PARP-1 and caspase-3 cleavage, two widely recognized hallmarks of apoptosis, while concurrently downregulating the expression of the apoptosis-inhibitory protein Bcl-xL[21] (Figure 6A). CCT6A KO enhanced the apoptotic cell death induced by DDP, while OE-CCT6A presented the opposite trend (Figure 6A). Similar results were also observed in flow cytometry experiments (Figure 6B and C), indicating that CCT6A negatively regulated the apoptosis induced by DDP. Apart from apoptosis, necroptosis is another extensively researched form of programmed cell death, characterized by activation of the RIP1-RIP3-MLKL complex[18]. In DDP-treated cells, ablation of CCT6A led to an elevation in both phospho-RIP1 and phospho-MLKL levels, whereas OE of CCT6A resulted in a decrease in their respective levels (Figure 6D). Subsequently, we conducted a Co-IP assay utilizing a RIP3-specific antibody, revealing that DDP significantly enhanced the formation of the RIP1-RIP3-MLKL complex in both HT29 and SW480 cells (Figure 6E). This observation confirmed that DDP has the capacity to induce necroptosis in CRC cells. Furthermore, upon exposure to DDP, CCT6A KO further augmented the accumulation of the RIP1-RIP3-MLKL complex, whereas OE-CCT6A led to its decreased formation (Figure 6E). Nec-1 is a widely used necroptosis inhibitor, and its effect was verified by the observation that it clearly reduced the RIP1-RIP3-MLKL complex formation induced by DDP (Figure 6F). In the MTS assay, it rescued the cell viability increased by DDP in both KO-Ctrl and KO-CCT6A cells (Figure 6G). These findings strongly suggest that CCT6A exerts negative regulatory effects on the process of necroptosis.
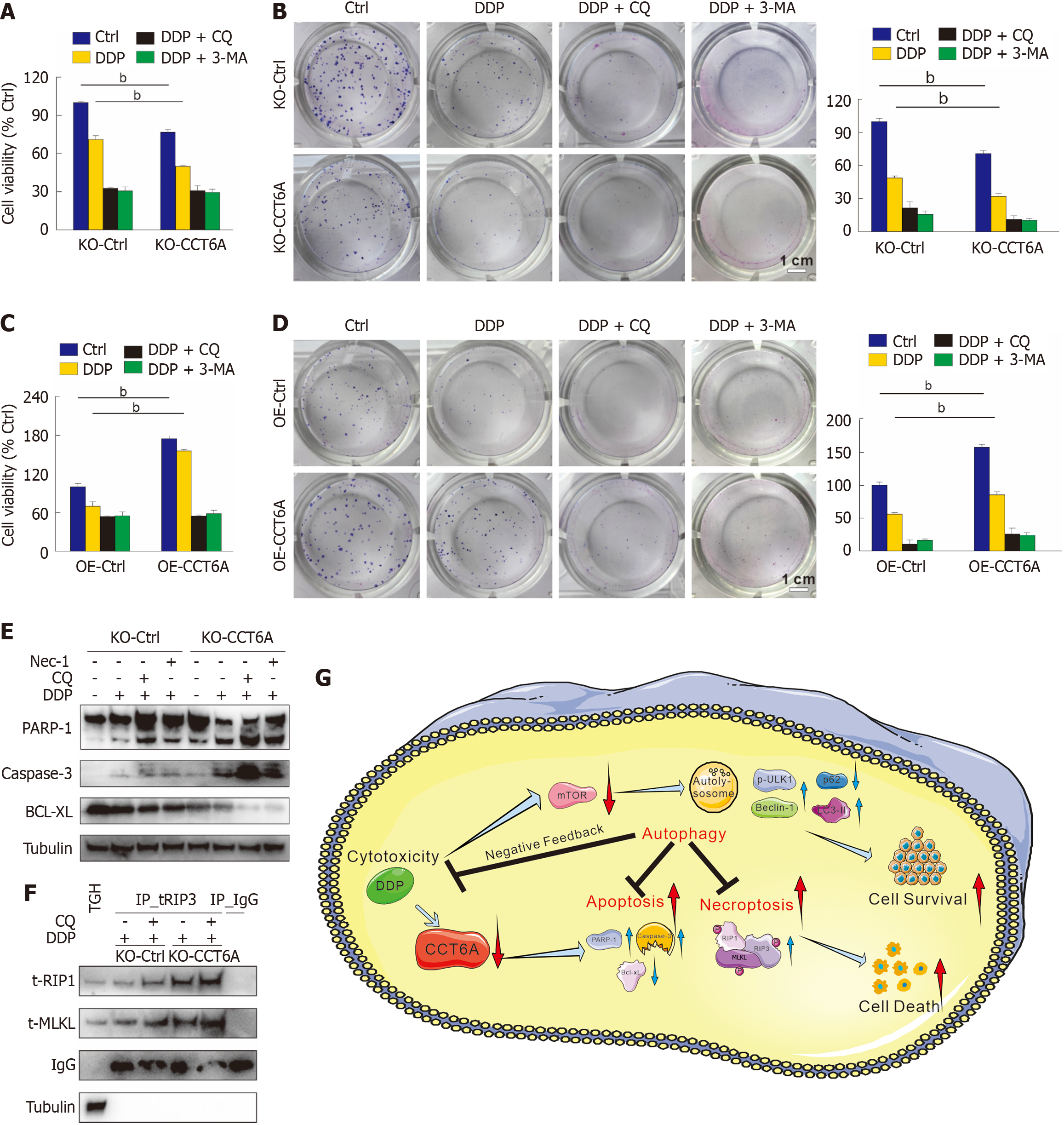
Autophagy plays a dual and intricate role in cancer treatment. On the one hand, it can facilitate cancer cell survival under stressful conditions, potentially leading to treatment resistance. On the other hand, excessive autophagy leads to autophagic cell death, thereby enhancing the therapeutic efficacy[14,20]. Here, we evaluated whether inhibiting autophagy could modulate the cytotoxicity of DDP that is regulated by CCT6A. In the MTS assay, the administration of CQ further diminished cell viability in both KO-Ctrl and KO-CCT6A cells upon treatment with DDP (Figure 7A). Notably, CQ mitigated the disparities in DDP-induced cytotoxicity observed between KO-Ctrl and KO-CCT6A cells, suggesting a modulatory role of autophagy in this context (Figure 7A). Similarly, the inclusion of CQ not only further suppressed cell proliferation but also attenuated the distinction in proliferation rates between the KO-CCT6A and KO-Ctrl cells in the colony growth assay (Figure 7B). Although the previous results indicated that OE-CCT6A inhibited basal and DDP-induced autophagy, we found that CQ still had the ability to inhibit cell activity (Figure 7C). Additionally, CQ effectively diminished colony growth in both OE-Ctrl and OE-CCT6A cells when combined with DDP treatment (Figure 7D). 3-MA, a widely employed inhibitor of autophagy initiation, demonstrated a comparable influence on cellular activity to that of CQ (Figure 7A-D), thereby reinforcing the pivotal role of autophagy in the observed phenomena. The addition of CQ promoted PARP-1 and caspase-3 cleavage, and reduced Bcl-xL levels in both KO-Ctrl and KO-CCT6A cells (Figure 7E). Moreover, CQ augmented the formation of the necrosome in both KO-Ctrl and KO-CCT6A cell lines, suggesting that autophagy suppression facilitated the progression of necroptosis (Figure 7F). Furthermore, while Nec-1 partially mitigated the cell viability decline induced by DDP in both KO-Ctrl and KO-CCT6A cells (Figure 6G), its administration was observed to exacerbate PARP-1 and caspase-3 cleavage, accompanied by reduced Bcl-xL levels (Figure 7E), thereby promoting apoptosis. These results suggest an intrinsic connection among autophagy, apoptosis, and necroptosis in CRC cells.
This study establishes CCT6A as a central regulator of DDP response in CRC, operating through bidirectional control of cell survival and death pathways. The observed reciprocal relationship where DDP suppresses CCT6A expression, while elevated CCT6A levels counteract DDP cytotoxicity, highlights its dual role as both a chemotherapy target and resistance mediator. Furthermore, CCT6A exerts a negative regulatory role in both basal and DDP-triggered autophagy, and suppression of autophagy significantly enhances DDP's cytotoxicity. Additionally, CCT6A participates in the modulation of DDP-induced apoptosis and necroptosis, where inhibiting either autophagy or necroptosis pathways potentiates DDP's apoptotic capabilities, indicating the intricate interplay among these three processes.
As an evolutionarily conserved process involves the turnover of intracellular materials in eukaryotes, autophagy degrades long-lived/damaged proteins and organelles to maintain internal homeostasis[20]. During chemotherapy and radiotherapy, cancer cells often utilize autophagy as a survival mechanism to evade the death fate[14,22]. As a result, chemotherapy-induced autophagy frequently triggers a negative feedback loop that diminishes the cytotoxic effects of the drugs, posing challenges for effective cancer treatment. Here, autophagy inhibitors can potentiate the cytotoxic effects of DDP, suggesting their potential as adjuvant therapies to enhance the efficacy of chemotherapy. We also found that DDP reduced the expression of CCT6A at both the transcription and translation levels, which is involved in modulating cell proliferation, migration, and invasion in cancer cells[7,8]. Deprivation of CCT6A promoted DDP cytotoxicity as determined by monitoring the cell viability and colony growth, whereas its OE showed the opposite effect. Meanwhile, we showed that the level of CCT6A was negatively correlated with the mTOR activity, which is a negative regulator of autophagy[20,23]. Through immunoblotting and the mCherry-GFP-LC3 assay, we verified that CCT6A negatively regulates both basal and DDP-induced autophagic flux. It is interesting to note that KO of CCT6A not only promoted the DDP-induced autophagy but also enhanced its cytotoxicity, which may be due to the fact that CCT6A can impact other signaling pathways. In addition, deprivation of autophagy in KO-CCT6A cells further promoted the cell viability loss and apoptosis induced by DDP. On the other hand, although CCT6A OE attenuated the DDP-induced autophagy, CQ/3-MA could still enhance the cytotoxicity effects of DDP. These observations suggest that CCT6A negatively regulates autophagy, and autophagy deprivation enhances the chemotherapy efficacy of DDP.
Generally, necroptosis is a regulated form of cell death that occurs when apoptosis is blocked. Some inducers of apoptosis, such as tumor necrosis factor alpha, can increase necroptosis in the presence of Z-VAD-FMK, a widely used pan-apoptosis inhibitor[24]. Several reports have shown that RIP1 plays a switching role among apoptosis, necroptosis, and survival mechanisms[17,25]. However, research has also demonstrated that the same therapeutic intervention can concurrently elicit both apoptotic and necrotic apoptotic cell death pathways[26]. Upon the induction of necroptosis, RIP1 and RIP3 undergo phosphorylation and form a complex that triggers the phosphorylation of MLKL. Subsequently, phospho-MLKL translocates to the cell membrane, where it forms pore complexes that compromise membrane integrity, ultimately culminating in cell death[18,27]. Herein, we determined that DDP elicits apoptosis and necroptosis in CRC cells, whereas CCT6A exerts a negative regulatory role in these cell death pathways. Interestingly, the employment of the necroptosis inhibitor Nec-1 notably diminished the cytotoxic effects of DDP on cells, yet paradoxically, it subtly augmented the apoptotic response elicited by DDP. Thus, it is plausible that additional mechanisms of cell death are concurrently operative and contributing to the observed phenomena. Conversely, the autophagy inhibitor CQ clearly potentiated the cytotoxic effects of DDP, corroborating the protective role autophagy plays in tumor chemotherapy[13-15]. Meanwhile, the inhibition of autophagy enhances the capability of DDP to elicit apoptosis and necroptosis, highlighting its pivotal role in modulating cellular death pathways during chemotherapy. Therefore, these findings reveal CCT6A as a linchpin coordinating multimodal death pathway interactions, offering a strategic entry point for combinatorial regimens targeting autophagy-apoptosis-necroptosis networks in chemotherapy-resistant malignancies.
CCT6A positively regulates cell proliferation and negatively modulates autophagy, apoptosis, and necroptosis. The concurrent inhibition of CCT6A and autophagy synergistically potentiates the cytotoxic effects of DDP, suggesting a potential therapeutic strategy to enhance the efficacy of this chemotherapy agent.
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [PubMed] [DOI] [Full Text] |
| 2. | Eng C, Yoshino T, Ruíz-García E, Mostafa N, Cann CG, O'Brian B, Benny A, Perez RO, Cremolini C. Colorectal cancer. Lancet. 2024;404:294-310. [PubMed] [DOI] [Full Text] |
| 3. | Biller LH, Schrag D. Diagnosis and Treatment of Metastatic Colorectal Cancer: A Review. JAMA. 2021;325:669-685. [PubMed] [DOI] [Full Text] |
| 4. | Yu SK, Yu T, Wang YM, Sun A, Liu J, Lu KH. CCT6A facilitates lung adenocarcinoma progression and glycolysis via STAT1/HK2 axis. J Transl Med. 2024;22:460. [PubMed] [DOI] [Full Text] |
| 5. | Yan P, Yang K, Xu M, Zhu M, Duan Y, Li W, Liu L, Liang C, Li Z, Pan X, Wang L, Yu G. CCT6A alleviates pulmonary fibrosis by inhibiting HIF-1α-mediated lactate production. J Mol Cell Biol. 2024;16:mjae021. [PubMed] [DOI] [Full Text] |
| 6. | Frydman J, Nimmesgern E, Erdjument-Bromage H, Wall JS, Tempst P, Hartl FU. Function in protein folding of TRiC, a cytosolic ring complex containing TCP-1 and structurally related subunits. EMBO J. 1992;11:4767-4778. [PubMed] [DOI] [Full Text] |
| 7. | Yang X, Tong Y, Ye W, Chen L. HOXB2 increases the proliferation and invasiveness of colon cancer cells through the upregulation of CCT6A. Mol Med Rep. 2022;25:174. [PubMed] [DOI] [Full Text] |
| 8. | Ying Z, Tian H, Li Y, Lian R, Li W, Wu S, Zhang HZ, Wu J, Liu L, Song J, Guan H, Cai J, Zhu X, Li J, Li M. CCT6A suppresses SMAD2 and promotes prometastatic TGF-β signaling. J Clin Invest. 2017;127:1725-1740. [PubMed] [DOI] [Full Text] |
| 9. | Zhang H, Zheng T, Qin C, Zhang X, Lin H, Huang X, Liu Q, Chang S, Zhang L, Guo J, Zhang Y, Bian C, Liu H. CCT6A promotes cell proliferation in colon cancer by targeting BIRC5 associated with p53 status. Cancer Gene Ther. 2024;31:1151-1163. [PubMed] [DOI] [Full Text] |
| 10. | Yu T, Zhang Q, Yu SK, Nie FQ, Zhang ML, Wang Q, Lu KH. THOC3 interacts with YBX1 to promote lung squamous cell carcinoma progression through PFKFB4 mRNA modification. Cell Death Dis. 2023;14:475. [PubMed] [DOI] [Full Text] |
| 11. | Ma J, Wei Q, Zhang L, Sun F, Li W, Du R, Liu M, Yan S, Wang C. CCT6A functions as promising diagnostic biomarker and promotes cell proliferation in colorectal cancer. J Cancer. 2024;15:5897-5909. [PubMed] [DOI] [Full Text] |
| 12. | Galluzzi L, Vitale I, Aaronson SA, Abrams JM, Adam D, Agostinis P, Alnemri ES, Altucci L, Amelio I, Andrews DW, Annicchiarico-Petruzzelli M, Antonov AV, Arama E, Baehrecke EH, Barlev NA, Bazan NG, Bernassola F, Bertrand MJM, Bianchi K, Blagosklonny MV, Blomgren K, Borner C, Boya P, Brenner C, Campanella M, Candi E, Carmona-Gutierrez D, Cecconi F, Chan FK, Chandel NS, Cheng EH, Chipuk JE, Cidlowski JA, Ciechanover A, Cohen GM, Conrad M, Cubillos-Ruiz JR, Czabotar PE, D'Angiolella V, Dawson TM, Dawson VL, De Laurenzi V, De Maria R, Debatin KM, DeBerardinis RJ, Deshmukh M, Di Daniele N, Di Virgilio F, Dixit VM, Dixon SJ, Duckett CS, Dynlacht BD, El-Deiry WS, Elrod JW, Fimia GM, Fulda S, García-Sáez AJ, Garg AD, Garrido C, Gavathiotis E, Golstein P, Gottlieb E, Green DR, Greene LA, Gronemeyer H, Gross A, Hajnoczky G, Hardwick JM, Harris IS, Hengartner MO, Hetz C, Ichijo H, Jäättelä M, Joseph B, Jost PJ, Juin PP, Kaiser WJ, Karin M, Kaufmann T, Kepp O, Kimchi A, Kitsis RN, Klionsky DJ, Knight RA, Kumar S, Lee SW, Lemasters JJ, Levine B, Linkermann A, Lipton SA, Lockshin RA, López-Otín C, Lowe SW, Luedde T, Lugli E, MacFarlane M, Madeo F, Malewicz M, Malorni W, Manic G, Marine JC, Martin SJ, Martinou JC, Medema JP, Mehlen P, Meier P, Melino S, Miao EA, Molkentin JD, Moll UM, Muñoz-Pinedo C, Nagata S, Nuñez G, Oberst A, Oren M, Overholtzer M, Pagano M, Panaretakis T, Pasparakis M, Penninger JM, Pereira DM, Pervaiz S, Peter ME, Piacentini M, Pinton P, Prehn JHM, Puthalakath H, Rabinovich GA, Rehm M, Rizzuto R, Rodrigues CMP, Rubinsztein DC, Rudel T, Ryan KM, Sayan E, Scorrano L, Shao F, Shi Y, Silke J, Simon HU, Sistigu A, Stockwell BR, Strasser A, Szabadkai G, Tait SWG, Tang D, Tavernarakis N, Thorburn A, Tsujimoto Y, Turk B, Vanden Berghe T, Vandenabeele P, Vander Heiden MG, Villunger A, Virgin HW, Vousden KH, Vucic D, Wagner EF, Walczak H, Wallach D, Wang Y, Wells JA, Wood W, Yuan J, Zakeri Z, Zhivotovsky B, Zitvogel L, Melino G, Kroemer G. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018;25:486-541. [PubMed] [DOI] [Full Text] |
| 13. | Li J, Hou N, Faried A, Tsutsumi S, Kuwano H. Inhibition of autophagy augments 5-fluorouracil chemotherapy in human colon cancer in vitro and in vivo model. Eur J Cancer. 2010;46:1900-1909. [PubMed] [DOI] [Full Text] |
| 14. | Kimura T, Takabatake Y, Takahashi A, Isaka Y. Chloroquine in cancer therapy: a double-edged sword of autophagy. Cancer Res. 2013;73:3-7. [PubMed] [DOI] [Full Text] |
| 15. | Li Q, Ma J, Zhang Y, Sun F, Li W, Shen W, Ai Z, Li C, Wang S, Wei X, Yan S. PFKFB3 deprivation attenuates the cisplatin resistance via blocking its autophagic elimination in colorectal cancer cells. Front Pharmacol. 2024;15:1433137. [PubMed] [DOI] [Full Text] |
| 16. | Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, Herman B, Levine B. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol. 1998;72:8586-8596. [PubMed] [DOI] [Full Text] |
| 17. | Dillon CP, Weinlich R, Rodriguez DA, Cripps JG, Quarato G, Gurung P, Verbist KC, Brewer TL, Llambi F, Gong YN, Janke LJ, Kelliher MA, Kanneganti TD, Green DR. RIPK1 blocks early postnatal lethality mediated by caspase-8 and RIPK3. Cell. 2014;157:1189-1202. [PubMed] [DOI] [Full Text] |
| 18. | Li J, McQuade T, Siemer AB, Napetschnig J, Moriwaki K, Hsiao YS, Damko E, Moquin D, Walz T, McDermott A, Chan FK, Wu H. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell. 2012;150:339-350. [PubMed] [DOI] [Full Text] |
| 19. | Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274-293. [PubMed] [DOI] [Full Text] |
| 20. | Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, Adhihetty PJ, Adler SG, Agam G, Agarwal R, Aghi MK, Agnello M, Agostinis P, Aguilar PV, Aguirre-Ghiso J, Airoldi EM, Ait-Si-Ali S, Akematsu T, Akporiaye ET, Al-Rubeai M, Albaiceta GM, Albanese C, Albani D, Albert ML, Aldudo J, Algül H, Alirezaei M, Alloza I, Almasan A, Almonte-Beceril M, Alnemri ES, Alonso C, Altan-Bonnet N, Altieri DC, Alvarez S, Alvarez-Erviti L, Alves S, Amadoro G, Amano A, Amantini C, Ambrosio S, Amelio I, Amer AO, Amessou M, Amon A, An Z, Anania FA, Andersen SU, Andley UP, Andreadi CK, Andrieu-Abadie N, Anel A, Ann DK, Anoopkumar-Dukie S, Antonioli M, Aoki H, Apostolova N, Aquila S, Aquilano K, Araki K, Arama E, Aranda A, Araya J, Arcaro A, Arias E, Arimoto H, Ariosa AR, Armstrong JL, Arnould T, Arsov I, Asanuma K, Askanas V, Asselin E, Atarashi R, Atherton SS, Atkin JD, Attardi LD, Auberger P, Auburger G, Aurelian L, Autelli R, Avagliano L, Avantaggiati ML, Avrahami L, Awale S, Azad N, Bachetti T, Backer JM, Bae DH, Bae JS, Bae ON, Bae SH, Baehrecke EH, Baek SH, Baghdiguian S, Bagniewska-Zadworna A, Bai H, Bai J, Bai XY, Bailly Y, Balaji KN, Balduini W, Ballabio A, Balzan R, Banerjee R, Bánhegyi G, Bao H, Barbeau B, Barrachina MD, Barreiro E, Bartel B, Bartolomé A, Bassham DC, Bassi MT, Bast RC Jr, Basu A, Batista MT, Batoko H, Battino M, Bauckman K, Baumgarner BL, Bayer KU, Beale R, Beaulieu JF, Beck GR Jr, Becker C, Beckham JD, Bédard PA, Bednarski PJ, Begley TJ, Behl C, Behrends C, Behrens GM, Behrns KE, Bejarano E, Belaid A, Belleudi F, Bénard G, Berchem G, Bergamaschi D, Bergami M, Berkhout B, Berliocchi L, Bernard A, Bernard M, Bernassola F, Bertolotti A, Bess AS, Besteiro S, Bettuzzi S, Bhalla S, Bhattacharyya S, Bhutia SK, Biagosch C, Bianchi MW, Biard-Piechaczyk M, Billes V, Bincoletto C, Bingol B, Bird SW, Bitoun M, Bjedov I, Blackstone C, Blanc L, Blanco GA, Blomhoff HK, Boada-Romero E, Böckler S, Boes M, Boesze-Battaglia K, Boise LH, Bolino A, Boman A, Bonaldo P, Bordi M, Bosch J, Botana LM, Botti J, Bou G, Bouché M, Bouchecareilh M, Boucher MJ, Boulton ME, Bouret SG, Boya P, Boyer-Guittaut M, Bozhkov PV, Brady N, Braga VM, Brancolini C, Braus GH, Bravo-San Pedro JM, Brennan LA, Bresnick EH, Brest P, Bridges D, Bringer MA, Brini M, Brito GC, Brodin B, Brookes PS, Brown EJ, Brown K, Broxmeyer HE, Bruhat A, Brum PC, Brumell JH, Brunetti-Pierri N, Bryson-Richardson RJ, Buch S, Buchan AM, Budak H, Bulavin DV, Bultman SJ, Bultynck G, Bumbasirevic V, Burelle Y, Burke RE, Burmeister M, Bütikofer P, Caberlotto L, Cadwell K, Cahova M, Cai D, Cai J, Cai Q, Calatayud S, Camougrand N, Campanella M, Campbell GR, Campbell M, Campello S, Candau R, Caniggia I, Cantoni L, Cao L, Caplan AB, Caraglia M, Cardinali C, Cardoso SM, Carew JS, Carleton LA, Carlin CR, Carloni S, Carlsson SR, Carmona-Gutierrez D, Carneiro LA, Carnevali O, Carra S, Carrier A, Carroll B, Casas C, Casas J, Cassinelli G, Castets P, Castro-Obregon S, Cavallini G, Ceccherini I, Cecconi F, Cederbaum AI, Ceña V, Cenci S, Cerella C, Cervia D, Cetrullo S, Chaachouay H, Chae HJ, Chagin AS, Chai CY, Chakrabarti G, Chamilos G, Chan EY, Chan MT, Chandra D, Chandra P, Chang CP, Chang RC, Chang TY, Chatham JC, Chatterjee S, Chauhan S, Che Y, Cheetham ME, Cheluvappa R, Chen CJ, Chen G, Chen GC, Chen G, Chen H, Chen JW, Chen JK, Chen M, Chen M, Chen P, Chen Q, Chen Q, Chen SD, Chen S, Chen SS, Chen W, Chen WJ, Chen WQ, Chen W, Chen X, Chen YH, Chen YG, Chen Y, Chen Y, Chen Y, Chen YJ, Chen YQ, Chen Y, Chen Z, Chen Z, Cheng A, Cheng CH, Cheng H, Cheong H, Cherry S, Chesney J, Cheung CH, Chevet E, Chi HC, Chi SG, Chiacchiera F, Chiang HL, Chiarelli R, Chiariello M, Chieppa M, Chin LS, Chiong M, Chiu GN, Cho DH, Cho SG, Cho WC, Cho YY, Cho YS, Choi AM, Choi EJ, Choi EK, Choi J, Choi ME, Choi SI, Chou TF, Chouaib S, Choubey D, Choubey V, Chow KC, Chowdhury K, Chu CT, Chuang TH, Chun T, Chung H, Chung T, Chung YL, Chwae YJ, Cianfanelli V, Ciarcia R, Ciechomska IA, Ciriolo MR, Cirone M, Claerhout S, Clague MJ, Clària J, Clarke PG, Clarke R, Clementi E, Cleyrat C, Cnop M, Coccia EM, Cocco T, Codogno P, Coers J, Cohen EE, Colecchia D, Coletto L, Coll NS, Colucci-Guyon E, Comincini S, Condello M, Cook KL, Coombs GH, Cooper CD, Cooper JM, Coppens I, Corasaniti MT, Corazzari M, Corbalan R, Corcelle-Termeau E, Cordero MD, Corral-Ramos C, Corti O, Cossarizza A, Costelli P, Costes S, Cotman SL, Coto-Montes A, Cottet S, Couve E, Covey LR, Cowart LA, Cox JS, Coxon FP, Coyne CB, Cragg MS, Craven RJ, Crepaldi T, Crespo JL, Criollo A, Crippa V, Cruz MT, Cuervo AM, Cuezva JM, Cui T, Cutillas PR, Czaja MJ, Czyzyk-Krzeska MF, Dagda RK, Dahmen U, Dai C, Dai W, Dai Y, Dalby KN, Dalla Valle L, Dalmasso G, D'Amelio M, Damme M, Darfeuille-Michaud A, Dargemont C, Darley-Usmar VM, Dasarathy S, Dasgupta B, Dash S, Dass CR, Davey HM, Davids LM, Dávila D, Davis RJ, Dawson TM, Dawson VL, Daza P, de Belleroche J, de Figueiredo P, de Figueiredo RC, de la Fuente J, De Martino L, De Matteis A, De Meyer GR, De Milito A, De Santi M, de Souza W, De Tata V, De Zio D, Debnath J, Dechant R, Decuypere JP, Deegan S, Dehay B, Del Bello B, Del Re DP, Delage-Mourroux R, Delbridge LM, Deldicque L, Delorme-Axford E, Deng Y, Dengjel J, Denizot M, Dent P, Der CJ, Deretic V, Derrien B, Deutsch E, Devarenne TP, Devenish RJ, Di Bartolomeo S, Di Daniele N, Di Domenico F, Di Nardo A, Di Paola S, Di Pietro A, Di Renzo L, DiAntonio A, Díaz-Araya G, Díaz-Laviada I, Diaz-Meco MT, Diaz-Nido J, Dickey CA, Dickson RC, Diederich M, Digard P, Dikic I, Dinesh-Kumar SP, Ding C, Ding WX, Ding Z, Dini L, Distler JH, Diwan A, Djavaheri-Mergny M, Dmytruk K, Dobson RC, Doetsch V, Dokladny K, Dokudovskaya S, Donadelli M, Dong XC, Dong X, Dong Z, Donohue TM Jr, Doran KS, D'Orazi G, Dorn GW 2nd, Dosenko V, Dridi S, Drucker L, Du J, Du LL, Du L, du Toit A, Dua P, Duan L, Duann P, Dubey VK, Duchen MR, Duchosal MA, Duez H, Dugail I, Dumit VI, Duncan MC, Dunlop EA, Dunn WA Jr, Dupont N, Dupuis L, Durán RV, Durcan TM, Duvezin-Caubet S, Duvvuri U, Eapen V, Ebrahimi-Fakhari D, Echard A, Eckhart L, Edelstein CL, Edinger AL, Eichinger L, Eisenberg T, Eisenberg-Lerner A, Eissa NT, El-Deiry WS, El-Khoury V, Elazar Z, Eldar-Finkelman H, Elliott CJ, Emanuele E, Emmenegger U, Engedal N, Engelbrecht AM, Engelender S, Enserink JM, Erdmann R, Erenpreisa J, Eri R, Eriksen JL, Erman A, Escalante R, Eskelinen EL, Espert L, Esteban-Martínez L, Evans TJ, Fabri M, Fabrias G, Fabrizi C, Facchiano A, Færgeman NJ, Faggioni A, Fairlie WD, Fan C, Fan D, Fan J, Fang S, Fanto M, Fanzani A, Farkas T, Faure M, Favier FB, Fearnhead H, Federici M, Fei E, Felizardo TC, Feng H, Feng Y, Feng Y, Ferguson TA, Fernández ÁF, Fernandez-Barrena MG, Fernandez-Checa JC, Fernández-López A, Fernandez-Zapico ME, Feron O, Ferraro E, Ferreira-Halder CV, Fesus L, Feuer R, Fiesel FC, Filippi-Chiela EC, Filomeni G, Fimia GM, Fingert JH, Finkbeiner S, Finkel T, Fiorito F, Fisher PB, Flajolet M, Flamigni F, Florey O, Florio S, Floto RA, Folini M, Follo C, Fon EA, Fornai F, Fortunato F, Fraldi A, Franco R, Francois A, François A, Frankel LB, Fraser ID, Frey N, Freyssenet DG, Frezza C, Friedman SL, Frigo DE, Fu D, Fuentes JM, Fueyo J, Fujitani Y, Fujiwara Y, Fujiya M, Fukuda M, Fulda S, Fusco C, Gabryel B, Gaestel M, Gailly P, Gajewska M, Galadari S, Galili G, Galindo I, Galindo MF, Galliciotti G, Galluzzi L, Galluzzi L, Galy V, Gammoh N, Gandy S, Ganesan AK, Ganesan S, Ganley IG, Gannagé M, Gao FB, Gao F, Gao JX, García Nannig L, García Véscovi E, Garcia-Macía M, Garcia-Ruiz C, Garg AD, Garg PK, Gargini R, Gassen NC, Gatica D, Gatti E, Gavard J, Gavathiotis E, Ge L, Ge P, Ge S, Gean PW, Gelmetti V, Genazzani AA, Geng J, Genschik P, Gerner L, Gestwicki JE, Gewirtz DA, Ghavami S, Ghigo E, Ghosh D, Giammarioli AM, Giampieri F, Giampietri C, Giatromanolaki A, Gibbings DJ, Gibellini L, Gibson SB, Ginet V, Giordano A, Giorgini F, Giovannetti E, Girardin SE, Gispert S, Giuliano S, Gladson CL, Glavic A, Gleave M, Godefroy N, Gogal RM Jr, Gokulan K, Goldman GH, Goletti D, Goligorsky MS, Gomes AV, Gomes LC, Gomez H, Gomez-Manzano C, Gómez-Sánchez R, Gonçalves DA, Goncu E, Gong Q, Gongora C, Gonzalez CB, Gonzalez-Alegre P, Gonzalez-Cabo P, González-Polo RA, Goping IS, Gorbea C, Gorbunov NV, Goring DR, Gorman AM, Gorski SM, Goruppi S, Goto-Yamada S, Gotor C, Gottlieb RA, Gozes I, Gozuacik D, Graba Y, Graef M, Granato GE, Grant GD, Grant S, Gravina GL, Green DR, Greenhough A, Greenwood MT, Grimaldi B, Gros F, Grose C, Groulx JF, Gruber F, Grumati P, Grune T, Guan JL, Guan KL, Guerra B, Guillen C, Gulshan K, Gunst J, Guo C, Guo L, Guo M, Guo W, Guo XG, Gust AA, Gustafsson ÅB, Gutierrez E, Gutierrez MG, Gwak HS, Haas A, Haber JE, Hadano S, Hagedorn M, Hahn DR, Halayko AJ, Hamacher-Brady A, Hamada K, Hamai A, Hamann A, Hamasaki M, Hamer I, Hamid Q, Hammond EM, Han F, Han W, Handa JT, Hanover JA, Hansen M, Harada M, Harhaji-Trajkovic L, Harper JW, Harrath AH, Harris AL, Harris J, Hasler U, Hasselblatt P, Hasui K, Hawley RG, Hawley TS, He C, He CY, He F, He G, He RR, He XH, He YW, He YY, Heath JK, Hébert MJ, Heinzen RA, Helgason GV, Hensel M, Henske EP, Her C, Herman PK, Hernández A, Hernandez C, Hernández-Tiedra S, Hetz C, Hiesinger PR, Higaki K, Hilfiker S, Hill BG, Hill JA, Hill WD, Hino K, Hofius D, Hofman P, Höglinger GU, Höhfeld J, Holz MK, Hong Y, Hood DA, Hoozemans JJ, Hoppe T, Hsu C, Hsu CY, Hsu LC, Hu D, Hu G, Hu HM, Hu H, Hu MC, Hu YC, Hu ZW, Hua F, Hua Y, Huang C, Huang HL, Huang KH, Huang KY, Huang S, Huang S, Huang WP, Huang YR, Huang Y, Huang Y, Huber TB, Huebbe P, Huh WK, Hulmi JJ, Hur GM, Hurley JH, Husak Z, Hussain SN, Hussain S, Hwang JJ, Hwang S, Hwang TI, Ichihara A, Imai Y, Imbriano C, Inomata M, Into T, Iovane V, Iovanna JL, Iozzo RV, Ip NY, Irazoqui JE, Iribarren P, Isaka Y, Isakovic AJ, Ischiropoulos H, Isenberg JS, Ishaq M, Ishida H, Ishii I, Ishmael JE, Isidoro C, Isobe K, Isono E, Issazadeh-Navikas S, Itahana K, Itakura E, Ivanov AI, Iyer AK, Izquierdo JM, Izumi Y, Izzo V, Jäättelä M, Jaber N, Jackson DJ, Jackson WT, Jacob TG, Jacques TS, Jagannath C, Jain A, Jana NR, Jang BK, Jani A, Janji B, Jannig PR, Jansson PJ, Jean S, Jendrach M, Jeon JH, Jessen N, Jeung EB, Jia K, Jia L, Jiang H, Jiang H, Jiang L, Jiang T, Jiang X, Jiang X, Jiang X, Jiang Y, Jiang Y, Jiménez A, Jin C, Jin H, Jin L, Jin M, Jin S, Jinwal UK, Jo EK, Johansen T, Johnson DE, Johnson GV, Johnson JD, Jonasch E, Jones C, Joosten LA, Jordan J, Joseph AM, Joseph B, Joubert AM, Ju D, Ju J, Juan HF, Juenemann K, Juhász G, Jung HS, Jung JU, Jung YK, Jungbluth H, Justice MJ, Jutten B, Kaakoush NO, Kaarniranta K, Kaasik A, Kabuta T, Kaeffer B, Kågedal K, Kahana A, Kajimura S, Kakhlon O, Kalia M, Kalvakolanu DV, Kamada Y, Kambas K, Kaminskyy VO, Kampinga HH, Kandouz M, Kang C, Kang R, Kang TC, Kanki T, Kanneganti TD, Kanno H, Kanthasamy AG, Kantorow M, Kaparakis-Liaskos M, Kapuy O, Karantza V, Karim MR, Karmakar P, Kaser A, Kaushik S, Kawula T, Kaynar AM, Ke PY, Ke ZJ, Kehrl JH, Keller KE, Kemper JK, Kenworthy AK, Kepp O, Kern A, Kesari S, Kessel D, Ketteler R, Kettelhut Ido C, Khambu B, Khan MM, Khandelwal VK, Khare S, Kiang JG, Kiger AA, Kihara A, Kim AL, Kim CH, Kim DR, Kim DH, Kim EK, Kim HY, Kim HR, Kim JS, Kim JH, Kim JC, Kim JH, Kim KW, Kim MD, Kim MM, Kim PK, Kim SW, Kim SY, Kim YS, Kim Y, Kimchi A, Kimmelman AC, Kimura T, King JS, Kirkegaard K, Kirkin V, Kirshenbaum LA, Kishi S, Kitajima Y, Kitamoto K, Kitaoka Y, Kitazato K, Kley RA, Klimecki WT, Klinkenberg M, Klucken J, Knævelsrud H, Knecht E, Knuppertz L, Ko JL, Kobayashi S, Koch JC, Koechlin-Ramonatxo C, Koenig U, Koh YH, Köhler K, Kohlwein SD, Koike M, Komatsu M, Kominami E, Kong D, Kong HJ, Konstantakou EG, Kopp BT, Korcsmaros T, Korhonen L, Korolchuk VI, Koshkina NV, Kou Y, Koukourakis MI, Koumenis C, Kovács AL, Kovács T, Kovacs WJ, Koya D, Kraft C, Krainc D, Kramer H, Kravic-Stevovic T, Krek W, Kretz-Remy C, Krick R, Krishnamurthy M, Kriston-Vizi J, Kroemer G, Kruer MC, Kruger R, Ktistakis NT, Kuchitsu K, Kuhn C, Kumar AP, Kumar A, Kumar A, Kumar D, Kumar D, Kumar R, Kumar S, Kundu M, Kung HJ, Kuno A, Kuo SH, Kuret J, Kurz T, Kwok T, Kwon TK, Kwon YT, Kyrmizi I, La Spada AR, Lafont F, Lahm T, Lakkaraju A, Lam T, Lamark T, Lancel S, Landowski TH, Lane DJ, Lane JD, Lanzi C, Lapaquette P, Lapierre LR, Laporte J, Laukkarinen J, Laurie GW, Lavandero S, Lavie L, LaVoie MJ, Law BY, Law HK, Law KB, Layfield R, Lazo PA, Le Cam L, Le Roch KG, Le Stunff H, Leardkamolkarn V, Lecuit M, Lee BH, Lee CH, Lee EF, Lee GM, Lee HJ, Lee H, Lee JK, Lee J, Lee JH, Lee JH, Lee M, Lee MS, Lee PJ, Lee SW, Lee SJ, Lee SJ, Lee SY, Lee SH, Lee SS, Lee SJ, Lee S, Lee YR, Lee YJ, Lee YH, Leeuwenburgh C, Lefort S, Legouis R, Lei J, Lei QY, Leib DA, Leibowitz G, Lekli I, Lemaire SD, Lemasters JJ, Lemberg MK, Lemoine A, Leng S, Lenz G, Lenzi P, Lerman LO, Lettieri Barbato D, Leu JI, Leung HY, Levine B, Lewis PA, Lezoualc'h F, Li C, Li F, Li FJ, Li J, Li K, Li L, Li M, Li M, Li Q, Li R, Li S, Li W, Li W, Li X, Li Y, Lian J, Liang C, Liang Q, Liao Y, Liberal J, Liberski PP, Lie P, Lieberman AP, Lim HJ, Lim KL, Lim K, Lima RT, Lin CS, Lin CF, Lin F, Lin F, Lin FC, Lin K, Lin KH, Lin PH, Lin T, Lin WW, Lin YS, Lin Y, Linden R, Lindholm D, Lindqvist LM, Lingor P, Linkermann A, Liotta LA, Lipinski MM, Lira VA, Lisanti MP, Liton PB, Liu B, Liu C, Liu CF, Liu F, Liu HJ, Liu J, Liu JJ, Liu JL, Liu K, Liu L, Liu L, Liu Q, Liu RY, Liu S, Liu S, Liu W, Liu XD, Liu X, Liu XH, Liu X, Liu X, Liu X, Liu Y, Liu Y, Liu Z, Liu Z, Liuzzi JP, Lizard G, Ljujic M, Lodhi IJ, Logue SE, Lokeshwar BL, Long YC, Lonial S, Loos B, López-Otín C, López-Vicario C, Lorente M, Lorenzi PL, Lõrincz P, Los M, Lotze MT, Lovat PE, Lu B, Lu B, Lu J, Lu Q, Lu SM, Lu S, Lu Y, Luciano F, Luckhart S, Lucocq JM, Ludovico P, Lugea A, Lukacs NW, Lum JJ, Lund AH, Luo H, Luo J, Luo S, Luparello C, Lyons T, Ma J, Ma Y, Ma Y, Ma Z, Machado J, Machado-Santelli GM, Macian F, MacIntosh GC, MacKeigan JP, Macleod KF, MacMicking JD, MacMillan-Crow LA, Madeo F, Madesh M, Madrigal-Matute J, Maeda A, Maeda T, Maegawa G, Maellaro E, Maes H, Magariños M, Maiese K, Maiti TK, Maiuri L, Maiuri MC, Maki CG, Malli R, Malorni W, Maloyan A, Mami-Chouaib F, Man N, Mancias JD, Mandelkow EM, Mandell MA, Manfredi AA, Manié SN, Manzoni C, Mao K, Mao Z, Mao ZW, Marambaud P, Marconi AM, Marelja Z, Marfe G, Margeta M, Margittai E, Mari M, Mariani FV, Marin C, Marinelli S, Mariño G, Markovic I, Marquez R, Martelli AM, Martens S, Martin KR, Martin SJ, Martin S, Martin-Acebes MA, Martín-Sanz P, Martinand-Mari C, Martinet W, Martinez J, Martinez-Lopez N, Martinez-Outschoorn U, Martínez-Velázquez M, Martinez-Vicente M, Martins WK, Mashima H, Mastrianni JA, Matarese G, Matarrese P, Mateo R, Matoba S, Matsumoto N, Matsushita T, Matsuura A, Matsuzawa T, Mattson MP, Matus S, Maugeri N, Mauvezin C, Mayer A, Maysinger D, Mazzolini GD, McBrayer MK, McCall K, McCormick C, McInerney GM, McIver SC, McKenna S, McMahon JJ, McNeish IA, Mechta-Grigoriou F, Medema JP, Medina DL, Megyeri K, Mehrpour M, Mehta JL, Mei Y, Meier UC, Meijer AJ, Meléndez A, Melino G, Melino S, de Melo EJ, Mena MA, Meneghini MD, Menendez JA, Menezes R, Meng L, Meng LH, Meng S, Menghini R, Menko AS, Menna-Barreto RF, Menon MB, Meraz-Ríos MA, Merla G, Merlini L, Merlot AM, Meryk A, Meschini S, Meyer JN, Mi MT, Miao CY, Micale L, Michaeli S, Michiels C, Migliaccio AR, Mihailidou AS, Mijaljica D, Mikoshiba K, Milan E, Miller-Fleming L, Mills GB, Mills IG, Minakaki G, Minassian BA, Ming XF, Minibayeva F, Minina EA, Mintern JD, Minucci S, Miranda-Vizuete A, Mitchell CH, Miyamoto S, Miyazawa K, Mizushima N, Mnich K, Mograbi B, Mohseni S, Moita LF, Molinari M, Molinari M, Møller AB, Mollereau B, Mollinedo F, Mongillo M, Monick MM, Montagnaro S, Montell C, Moore DJ, Moore MN, Mora-Rodriguez R, Moreira PI, Morel E, Morelli MB, Moreno S, Morgan MJ, Moris A, Moriyasu Y, Morrison JL, Morrison LA, Morselli E, Moscat J, Moseley PL, Mostowy S, Motori E, Mottet D, Mottram JC, Moussa CE, Mpakou VE, Mukhtar H, Mulcahy Levy JM, Muller S, Muñoz-Moreno R, Muñoz-Pinedo C, Münz C, Murphy ME, Murray JT, Murthy A, Mysorekar IU, Nabi IR, Nabissi M, Nader GA, Nagahara Y, Nagai Y, Nagata K, Nagelkerke A, Nagy P, Naidu SR, Nair S, Nakano H, Nakatogawa H, Nanjundan M, Napolitano G, Naqvi NI, Nardacci R, Narendra DP, Narita M, Nascimbeni AC, Natarajan R, Navegantes LC, Nawrocki ST, Nazarko TY, Nazarko VY, Neill T, Neri LM, Netea MG, Netea-Maier RT, Neves BM, Ney PA, Nezis IP, Nguyen HT, Nguyen HP, Nicot AS, Nilsen H, Nilsson P, Nishimura M, Nishino I, Niso-Santano M, Niu H, Nixon RA, Njar VC, Noda T, Noegel AA, Nolte EM, Norberg E, Norga KK, Noureini SK, Notomi S, Notterpek L, Nowikovsky K, Nukina N, Nürnberger T, O'Donnell VB, O'Donovan T, O'Dwyer PJ, Oehme I, Oeste CL, Ogawa M, Ogretmen B, Ogura Y, Oh YJ, Ohmuraya M, Ohshima T, Ojha R, Okamoto K, Okazaki T, Oliver FJ, Ollinger K, Olsson S, Orban DP, Ordonez P, Orhon I, Orosz L, O'Rourke EJ, Orozco H, Ortega AL, Ortona E, Osellame LD, Oshima J, Oshima S, Osiewacz HD, Otomo T, Otsu K, Ou JH, Outeiro TF, Ouyang DY, Ouyang H, Overholtzer M, Ozbun MA, Ozdinler PH, Ozpolat B, Pacelli C, Paganetti P, Page G, Pages G, Pagnini U, Pajak B, Pak SC, Pakos-Zebrucka K, Pakpour N, Palková Z, Palladino F, Pallauf K, Pallet N, Palmieri M, Paludan SR, Palumbo C, Palumbo S, Pampliega O, Pan H, Pan W, Panaretakis T, Pandey A, Pantazopoulou A, Papackova Z, Papademetrio DL, Papassideri I, Papini A, Parajuli N, Pardo J, Parekh VV, Parenti G, Park JI, Park J, Park OK, Parker R, Parlato R, Parys JB, Parzych KR, Pasquet JM, Pasquier B, Pasumarthi KB, Patschan D, Patterson C, Pattingre S, Pattison S, Pause A, Pavenstädt H, Pavone F, Pedrozo Z, Peña FJ, Peñalva MA, Pende M, Peng J, Penna F, Penninger JM, Pensalfini A, Pepe S, Pereira GJ, Pereira PC, Pérez-de la Cruz V, Pérez-Pérez ME, Pérez-Rodríguez D, Pérez-Sala D, Perier C, Perl A, Perlmutter DH, Perrotta I, Pervaiz S, Pesonen M, Pessin JE, Peters GJ, Petersen M, Petrache I, Petrof BJ, Petrovski G, Phang JM, Piacentini M, Pierdominici M, Pierre P, Pierrefite-Carle V, Pietrocola F, Pimentel-Muiños FX, Pinar M, Pineda B, Pinkas-Kramarski R, Pinti M, Pinton P, Piperdi B, Piret JM, Platanias LC, Platta HW, Plowey ED, Pöggeler S, Poirot M, Polčic P, Poletti A, Poon AH, Popelka H, Popova B, Poprawa I, Poulose SM, Poulton J, Powers SK, Powers T, Pozuelo-Rubio M, Prak K, Prange R, Prescott M, Priault M, Prince S, Proia RL, Proikas-Cezanne T, Prokisch H, Promponas VJ, Przyklenk K, Puertollano R, Pugazhenthi S, Puglielli L, Pujol A, Puyal J, Pyeon D, Qi X, Qian WB, Qin ZH, Qiu Y, Qu Z, Quadrilatero J, Quinn F, Raben N, Rabinowich H, Radogna F, Ragusa MJ, Rahmani M, Raina K, Ramanadham S, Ramesh R, Rami A, Randall-Demllo S, Randow F, Rao H, Rao VA, Rasmussen BB, Rasse TM, Ratovitski EA, Rautou PE, Ray SK, Razani B, Reed BH, Reggiori F, Rehm M, Reichert AS, Rein T, Reiner DJ, Reits E, Ren J, Ren X, Renna M, Reusch JE, Revuelta JL, Reyes L, Rezaie AR, Richards RI, Richardson DR, Richetta C, Riehle MA, Rihn BH, Rikihisa Y, Riley BE, Rimbach G, Rippo MR, Ritis K, Rizzi F, Rizzo E, Roach PJ, Robbins J, Roberge M, Roca G, Roccheri MC, Rocha S, Rodrigues CMP, Rodríguez CI, de Cordoba SR, Rodriguez-Muela N, Roelofs J, Rogov VV, Rohn TT, Rohrer B, Romanelli D, Romani L, Romano PS, Roncero MI, Rosa JL, Rosello A, Rosen KV, Rosenstiel P, Rost-Roszkowska M, Roth KA, Roué G, Rouis M, Rouschop KM, Ruan DT, Ruano D, Rubinsztein DC, Rucker EB 3rd, Rudich A, Rudolf E, Rudolf R, Ruegg MA, Ruiz-Roldan C, Ruparelia AA, Rusmini P, Russ DW, Russo GL, Russo G, Russo R, Rusten TE, Ryabovol V, Ryan KM, Ryter SW, Sabatini DM, Sacher M, Sachse C, Sack MN, Sadoshima J, Saftig P, Sagi-Eisenberg R, Sahni S, Saikumar P, Saito T, Saitoh T, Sakakura K, Sakoh-Nakatogawa M, Sakuraba Y, Salazar-Roa M, Salomoni P, Saluja AK, Salvaterra PM, Salvioli R, Samali A, Sanchez AM, Sánchez-Alcázar JA, Sanchez-Prieto R, Sandri M, Sanjuan MA, Santaguida S, Santambrogio L, Santoni G, Dos Santos CN, Saran S, Sardiello M, Sargent G, Sarkar P, Sarkar S, Sarrias MR, Sarwal MM, Sasakawa C, Sasaki M, Sass M, Sato K, Sato M, Satriano J, Savaraj N, Saveljeva S, Schaefer L, Schaible UE, Scharl M, Schatzl HM, Schekman R, Scheper W, Schiavi A, Schipper HM, Schmeisser H, Schmidt J, Schmitz I, Schneider BE, Schneider EM, Schneider JL, Schon EA, Schönenberger MJ, Schönthal AH, Schorderet DF, Schröder B, Schuck S, Schulze RJ, Schwarten M, Schwarz TL, Sciarretta S, Scotto K, Scovassi AI, Screaton RA, Screen M, Seca H, Sedej S, Segatori L, Segev N, Seglen PO, Seguí-Simarro JM, Segura-Aguilar J, Seki E, Sell C, Seiliez I, Semenkovich CF, Semenza GL, Sen U, Serra AL, Serrano-Puebla A, Sesaki H, Setoguchi T, Settembre C, Shacka JJ, Shajahan-Haq AN, Shapiro IM, Sharma S, She H, Shen CK, Shen CC, Shen HM, Shen S, Shen W, Sheng R, Sheng X, Sheng ZH, Shepherd TG, Shi J, Shi Q, Shi Q, Shi Y, Shibutani S, Shibuya K, Shidoji Y, Shieh JJ, Shih CM, Shimada Y, Shimizu S, Shin DW, Shinohara ML, Shintani M, Shintani T, Shioi T, Shirabe K, Shiri-Sverdlov R, Shirihai O, Shore GC, Shu CW, Shukla D, Sibirny AA, Sica V, Sigurdson CJ, Sigurdsson EM, Sijwali PS, Sikorska B, Silveira WA, Silvente-Poirot S, Silverman GA, Simak J, Simmet T, Simon AK, Simon HU, Simone C, Simons M, Simonsen A, Singh R, Singh SV, Singh SK, Sinha D, Sinha S, Sinicrope FA, Sirko A, Sirohi K, Sishi BJ, Sittler A, Siu PM, Sivridis E, Skwarska A, Slack R, Slaninová I, Slavov N, Smaili SS, Smalley KS, Smith DR, Soenen SJ, Soleimanpour SA, Solhaug A, Somasundaram K, Son JH, Sonawane A, Song C, Song F, Song HK, Song JX, Song W, Soo KY, Sood AK, Soong TW, Soontornniyomkij V, Sorice M, Sotgia F, Soto-Pantoja DR, Sotthibundhu A, Sousa MJ, Spaink HP, Span PN, Spang A, Sparks JD, Speck PG, Spector SA, Spies CD, Springer W, Clair DS, Stacchiotti A, Staels B, Stang MT, Starczynowski DT, Starokadomskyy P, Steegborn C, Steele JW, Stefanis L, Steffan J, Stellrecht CM, Stenmark H, Stepkowski TM, Stern ST, Stevens C, Stockwell BR, Stoka V, Storchova Z, Stork B, Stratoulias V, Stravopodis DJ, Strnad P, Strohecker AM, Ström AL, Stromhaug P, Stulik J, Su YX, Su Z, Subauste CS, Subramaniam S, Sue CM, Suh SW, Sui X, Sukseree S, Sulzer D, Sun FL, Sun J, Sun J, Sun SY, Sun Y, Sun Y, Sun Y, Sundaramoorthy V, Sung J, Suzuki H, Suzuki K, Suzuki N, Suzuki T, Suzuki YJ, Swanson MS, Swanton C, Swärd K, Swarup G, Sweeney ST, Sylvester PW, Szatmari Z, Szegezdi E, Szlosarek PW, Taegtmeyer H, Tafani M, Taillebourg E, Tait SW, Takacs-Vellai K, Takahashi Y, Takáts S, Takemura G, Takigawa N, Talbot NJ, Tamagno E, Tamburini J, Tan CP, Tan L, Tan ML, Tan M, Tan YJ, Tanaka K, Tanaka M, Tang D, Tang D, Tang G, Tanida I, Tanji K, Tannous BA, Tapia JA, Tasset-Cuevas I, Tatar M, Tavassoly I, Tavernarakis N, Taylor A, Taylor GS, Taylor GA, Taylor JP, Taylor MJ, Tchetina EV, Tee AR, Teixeira-Clerc F, Telang S, Tencomnao T, Teng BB, Teng RJ, Terro F, Tettamanti G, Theiss AL, Theron AE, Thomas KJ, Thomé MP, Thomes PG, Thorburn A, Thorner J, Thum T, Thumm M, Thurston TL, Tian L, Till A, Ting JP, Titorenko VI, Toker L, Toldo S, Tooze SA, Topisirovic I, Torgersen ML, Torosantucci L, Torriglia A, Torrisi MR, Tournier C, Towns R, Trajkovic V, Travassos LH, Triola G, Tripathi DN, Trisciuoglio D, Troncoso R, Trougakos IP, Truttmann AC, Tsai KJ, Tschan MP, Tseng YH, Tsukuba T, Tsung A, Tsvetkov AS, Tu S, Tuan HY, Tucci M, Tumbarello DA, Turk B, Turk V, Turner RF, Tveita AA, Tyagi SC, Ubukata M, Uchiyama Y, Udelnow A, Ueno T, Umekawa M, Umemiya-Shirafuji R, Underwood BR, Ungermann C, Ureshino RP, Ushioda R, Uversky VN, Uzcátegui NL, Vaccari T, Vaccaro MI, Váchová L, Vakifahmetoglu-Norberg H, Valdor R, Valente EM, Vallette F, Valverde AM, Van den Berghe G, Van Den Bosch L, van den Brink GR, van der Goot FG, van der Klei IJ, van der Laan LJ, van Doorn WG, van Egmond M, van Golen KL, Van Kaer L, van Lookeren Campagne M, Vandenabeele P, Vandenberghe W, Vanhorebeek I, Varela-Nieto I, Vasconcelos MH, Vasko R, Vavvas DG, Vega-Naredo I, Velasco G, Velentzas AD, Velentzas PD, Vellai T, Vellenga E, Vendelbo MH, Venkatachalam K, Ventura N, Ventura S, Veras PS, Verdier M, Vertessy BG, Viale A, Vidal M, Vieira HL, Vierstra RD, Vigneswaran N, Vij N, Vila M, Villar M, Villar VH, Villarroya J, Vindis C, Viola G, Viscomi MT, Vitale G, Vogl DT, Voitsekhovskaja OV, von Haefen C, von Schwarzenberg K, Voth DE, Vouret-Craviari V, Vuori K, Vyas JM, Waeber C, Walker CL, Walker MJ, Walter J, Wan L, Wan X, Wang B, Wang C, Wang CY, Wang C, Wang C, Wang C, Wang D, Wang F, Wang F, Wang G, Wang HJ, Wang H, Wang HG, Wang H, Wang HD, Wang J, Wang J, Wang M, Wang MQ, Wang PY, Wang P, Wang RC, Wang S, Wang TF, Wang X, Wang XJ, Wang XW, Wang X, Wang X, Wang Y, Wang Y, Wang Y, Wang YJ, Wang Y, Wang Y, Wang YT, Wang Y, Wang ZN, Wappner P, Ward C, Ward DM, Warnes G, Watada H, Watanabe Y, Watase K, Weaver TE, Weekes CD, Wei J, Weide T, Weihl CC, Weindl G, Weis SN, Wen L, Wen X, Wen Y, Westermann B, Weyand CM, White AR, White E, Whitton JL, Whitworth AJ, Wiels J, Wild F, Wildenberg ME, Wileman T, Wilkinson DS, Wilkinson S, Willbold D, Williams C, Williams K, Williamson PR, Winklhofer KF, Witkin SS, Wohlgemuth SE, Wollert T, Wolvetang EJ, Wong E, Wong GW, Wong RW, Wong VK, Woodcock EA, Wright KL, Wu C, Wu D, Wu GS, Wu J, Wu J, Wu M, Wu M, Wu S, Wu WK, Wu Y, Wu Z, Xavier CP, Xavier RJ, Xia GX, Xia T, Xia W, Xia Y, Xiao H, Xiao J, Xiao S, Xiao W, Xie CM, Xie Z, Xie Z, Xilouri M, Xiong Y, Xu C, Xu C, Xu F, Xu H, Xu H, Xu J, Xu J, Xu J, Xu L, Xu X, Xu Y, Xu Y, Xu ZX, Xu Z, Xue Y, Yamada T, Yamamoto A, Yamanaka K, Yamashina S, Yamashiro S, Yan B, Yan B, Yan X, Yan Z, Yanagi Y, Yang DS, Yang JM, Yang L, Yang M, Yang PM, Yang P, Yang Q, Yang W, Yang WY, Yang X, Yang Y, Yang Y, Yang Z, Yang Z, Yao MC, Yao PJ, Yao X, Yao Z, Yao Z, Yasui LS, Ye M, Yedvobnick B, Yeganeh B, Yeh ES, Yeyati PL, Yi F, Yi L, Yin XM, Yip CK, Yoo YM, Yoo YH, Yoon SY, Yoshida K, Yoshimori T, Young KH, Yu H, Yu JJ, Yu JT, Yu J, Yu L, Yu WH, Yu XF, Yu Z, Yuan J, Yuan ZM, Yue BY, Yue J, Yue Z, Zacks DN, Zacksenhaus E, Zaffaroni N, Zaglia T, Zakeri Z, Zecchini V, Zeng J, Zeng M, Zeng Q, Zervos AS, Zhang DD, Zhang F, Zhang G, Zhang GC, Zhang H, Zhang H, Zhang H, Zhang H, Zhang J, Zhang J, Zhang J, Zhang J, Zhang JP, Zhang L, Zhang L, Zhang L, Zhang L, Zhang MY, Zhang X, Zhang XD, Zhang Y, Zhang Y, Zhang Y, Zhang Y, Zhang Y, Zhao M, Zhao WL, Zhao X, Zhao YG, Zhao Y, Zhao Y, Zhao YX, Zhao Z, Zhao ZJ, Zheng D, Zheng XL, Zheng X, Zhivotovsky B, Zhong Q, Zhou GZ, Zhou G, Zhou H, Zhou SF, Zhou XJ, Zhu H, Zhu H, Zhu WG, Zhu W, Zhu XF, Zhu Y, Zhuang SM, Zhuang X, Ziparo E, Zois CE, Zoladek T, Zong WX, Zorzano A, Zughaier SM. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12:1-222. [PubMed] [DOI] [Full Text] |
| 21. | Andrabi SA, Kim NS, Yu SW, Wang H, Koh DW, Sasaki M, Klaus JA, Otsuka T, Zhang Z, Koehler RC, Hurn PD, Poirier GG, Dawson VL, Dawson TM. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc Natl Acad Sci U S A. 2006;103:18308-18313. [PubMed] [DOI] [Full Text] |
| 22. | White E, DiPaola RS. The double-edged sword of autophagy modulation in cancer. Clin Cancer Res. 2009;15:5308-5316. [PubMed] [DOI] [Full Text] |
| 23. | Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132-141. [PubMed] [DOI] [Full Text] |
| 24. | Sprinkle TJ, Tippins RB, Kestler DP. Inhibition of bovine and human brain 2':3'-cyclic nucleotide 3'-phosphodiesterase by heparin and polyribonucleotides and evidence for an associated 5'-polynucleotide kinase activity. Biochem Biophys Res Commun. 1987;145:686-691. [PubMed] [DOI] [Full Text] |
| 25. | Christofferson DE, Li Y, Yuan J. Control of life-or-death decisions by RIP1 kinase. Annu Rev Physiol. 2014;76:129-150. [PubMed] [DOI] [Full Text] |
| 26. | Yan S, Li Q, Zhang D, Wang X, Xu Y, Zhang C, Guo D, Bao Y. Necroptosis pathway blockage attenuates PFKFB3 inhibitor-induced cell viability loss and genome instability in colorectal cancer cells. Am J Cancer Res. 2021;11:2062-2080. [PubMed] |
| 27. | Murphy JM, Czabotar PE, Hildebrand JM, Lucet IS, Zhang JG, Alvarez-Diaz S, Lewis R, Lalaoui N, Metcalf D, Webb AI, Young SN, Varghese LN, Tannahill GM, Hatchell EC, Majewski IJ, Okamoto T, Dobson RC, Hilton DJ, Babon JJ, Nicola NA, Strasser A, Silke J, Alexander WS. The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity. 2013;39:443-453. [PubMed] [DOI] [Full Text] |