Published online Apr 14, 2025. doi: 10.3748/wjg.v31.i14.99897
Revised: February 19, 2025
Accepted: March 17, 2025
Published online: April 14, 2025
Processing time: 251 Days and 19.7 Hours
Extramural venous invasion (EMVI) is a critical prognostic factor in gastric cancer (GC); however, its detection and underlying molecular mechanisms remain underexplored.
To investigate the relationship between EMVI and expression of the circular RNA hsa_circ_0097977 in orthotopic GC mouse models.
A retrospective analysis was conducted in addition to a preclinical animal study, involving 13 GC patients and 24 orthotopic GC mouse models, respectively. EMVI was assessed using axial T2-weighted fat suppression sequences on a 9.4T magnetic resonance imaging (MRI) with histopathological confirmation as the gold standard for EMVI. The impact of hsa_circ_0097977 on EMVI and GC cell function was evaluated. Statistical analyses comprised consistency, area under the curve analysis, correlation, χ2/Fisher exact, and Mann-Whitney U/t-tests, with significance set at P < 0.05.
EMVI was accurately detected using 9.4T MRI in orthotopic mouse models with an area under the curve of 0.843 (sensitivity 78.6%, specificity 90.0%). MRI detected EMVI was the only imaging factor associated with distant metastasis (P = 0.04). Furthermore, knockdown of hsa_circ_0097977 was the only factor associated with EMVI (P = 0.043, 0.038) and led to reduced invasion and increased apoptosis in GC cells.
EMVI, a risk factor for distant metastasis in GC, is detectable by 9.4T MRI and regulated by hsa_circ_0097977, making it a potential therapeutic target.
Core Tip: This study demonstrates that extramural venous invasion (EMVI), a risk factor for distant metastasis in gastric cancer (GC), can be accurately detected using 9.4T magnetic resonance imaging in orthotopic GC mouse models, with histopathological confirmation. The expression of hsa_circ_0097977, a circular RNA, was found to regulate EMVI, and its knockdown reduced tumor cell invasion and promoted apoptosis, suggesting its potential as a therapeutic target in GC. These findings highlight the clinical relevance of EMVI detection and the role of hsa_circ_0097977 in GC progression.
- Citation: Feng CZ, Gou XY, Liu YQ, Xin YW, Zhang YL, Zhao HM, Wei SC, Hong N, Wang Y, Cheng J. Extramural venous invasion in gastric cancer: 9.4T magnetic resonance imaging assessment and circular RNA functional analysis. World J Gastroenterol 2025; 31(14): 99897
- URL: https://www.wjgnet.com/1007-9327/full/v31/i14/99897.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i14.99897
Gastric cancer (GC) is a significant global health issue, particularly in Eastern Asia; Unfortunately, nearly 80% of patients are diagnosed at an advanced stage with a poor prognosis[1,2]. Despite advances in systemic therapy and surgery, the high heterogeneity of GC underscores the urgent need for novel diagnostic biomarkers and therapeutic targets.
Extramural venous invasion (EMVI) refers to the invasion of cancer cells beyond the muscularis propria and into the venous lumen[3]. EMVI in patients with GC detected in routine medical imaging examinations has been demonstrated to be a significant predictor of distant metastasis and recurrence[4-6]. However, the diagnostic performance of EMVI remains uncertain due to challenges in obtaining sufficient pathological samples. This knowledge gap underscores the urgent need to focus research on the genetic basis of EMVI, which could be a foundation for improved diagnostic, prognostic, and therapeutic interventions for GC patients.
Recent studies have highlighted the potential of radiogenomics in linking imaging phenotypes to gene expression[7]. Our previous studies have demonstrated that EMVI-related mRNA signature has a significant impact on overall survival[8], and response to anti-programmed cell death 1 drugs[9] in GC patients. Compared with mRNA, circular RNA (circRNA) is a unique class of single-stranded RNAs in eukaryotic cells known for their stability and tissue-specific expression patterns[10]. circRNAs are pivotal in the development and progression of GC influencing critical processes such as cell proliferation, invasion, and metastasis[11]. For instance, hsa_circ_0136666 promotes GC progression and immune escape by modulating the miR-375/PRKDC axis and programmed death ligand 1 phosphorylation[12]. Given their multifaceted roles, circRNAs hold substantial potential as biomarkers for diagnosing, monitoring, and predicting the prognosis of GC, while also presenting a promising avenue for targeted therapy research.
Our dual objectives were: (1) To confirm the accuracy of 9.4T magnetic resonance imaging (MRI) in detecting EMVI using histopathology as the gold standard; and (2) To investigate whether hsa_circ_0097977 regulates EMVI through cell functional assays. This combined approach aims to uncover both imaging and molecular mechanisms underlying EMVI, offering new strategies for precision diagnosis and therapy in GC.
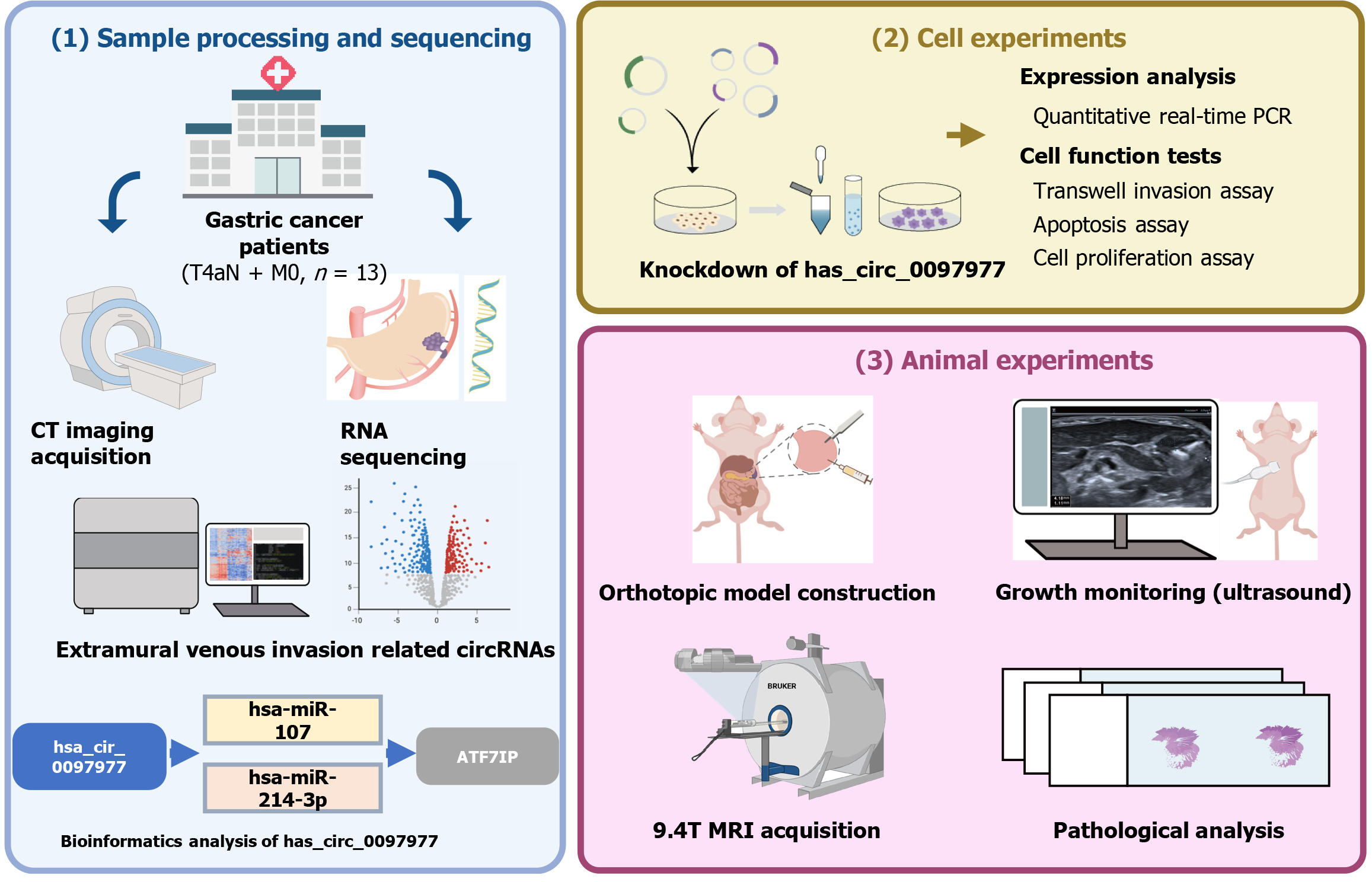
The overall workflow is depicted in Figure 1.
The institutional review board of Peking University People's Hospital approved this study (Approval No. 2019PHB171-01), and the requirement for informed consent was waived. GC samples were retrospectively collected between January 2018 and January 2019 from 13 patients with locally advanced GC. All patients had a pathological stage of T4aN + M0[1]. All patients underwent preoperative chest and abdominal contrast enhanced computed tomography (CT). None of the patients had received chemotherapy or radiotherapy before surgery, and no evidence of other malignancies was found. Tumor samples were collected based on EMVI status, immediately frozen in liquid nitrogen, and stored in the institute biobank until use. Total RNA was isolated from the GC samples. After quality control tests were performed, the whole circRNA expression of these samples was sequenced using the Illumina NextSeq system. Differentially expressed genes, defined as |log2fold change| > 1 and an adjusted P value of < 0.05, were screened using the R software with the limma package. circRNAs with high read count scores and included in the Circbase database (http://www.circbase.org/) were selected. Then the CircBank database (http://www.circbank.cn, circbank_miRNA_all_v1) was used to predict the miRNAs sponged by selected circRNAs, and the multiMiR R package (version 1.20.0) was used to predict miRNA target genes and analyze miRNAs-target interactions with their disease and drug associations.
Human GC cell lines MKN-74, BGC-823, NCI-N87, SGC-7901, MKN-45, HGC-27, and gastric epithelial immortalized GES-1 cells were obtained from Gekai Biotechnology Co., Ltd. (China) and the Central Laboratory of our hospital. The expressions of target genes were determined by quantitative real-time polymerase chain reaction (qRT-PCR). To construct stable cell lines with low-expression of target circRNA [knockdown (KD) group], we transfected the lentivirus (GV493) into GC cells following the supplier’s instructions. In the negative control (NC) group, GC cells were transfected with the control siRNA (CON313). The transfected cells were selected with puromycin (Sigma, United States), and PCR was used to measure transfection efficiency.
This study received official approval from the Institutional Animal Care and Use Committee of Peking University People's Hospital (IACUC protocol No. 2019PHE058). The study was conducted in compliance with the Animal Research: Reporting of in Vivo Experiments guidelines.
We selected two cell lines with the highest expression levels of the target genes. The animal sample size was based on previous reports[13]. A total of 24 female BALB/c nude mice, aged 4 weeks and weighing 18 ± 2 g, were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. The mice were acclimated for one week and then randomly assigned to four experimental groups (BGC-823 NC, BGC-823 KD, NCI-N87 NC, and NCI-N87 KD), with six mice per group. Throughout the experimental period, the mice were provided with standard laboratory diet and sterilized water ad libitum. Each group of six mice was housed in individual, standard, and sanitized cages under an environmentally controlled system, maintaining a temperature of 22 ± 2 °C, humidity of approximately 60%, and a 12:12-hour light-dark cycle.
The mice were subjected to orthotopic transplantation after randomization based on their size. Anesthesia was induced by exposing the mice to 4% isoflurane (RWD Life Science Co., Ltd., China) which was maintained at 2% isoflurane in oxygen[14]. After skin disinfection around the surgical area, a 5-10 mm incision was made overlying the abdomen. Forceps were used to exteriorize the stomach. Cells were injected (1 × 107 GC cells/30 μL phosphate-buffered saline, 29G insulin syringe) into the serous coat (subserosa) of the stomach walls of BALB/c nude mice[15]. The stomach was returned to the peritoneum, and the abdominal wall was closed with a wound suture in one layer.
Ultrasound examinations were performed using HRU with a 24-MHz center frequency and a spatial resolution of 75 µm (Aplio i800, Canon Medical Systems, Toshiba, Japan). The mice were placed on a heated table and maintained under anesthesia with 2% isoflurane in oxygen.
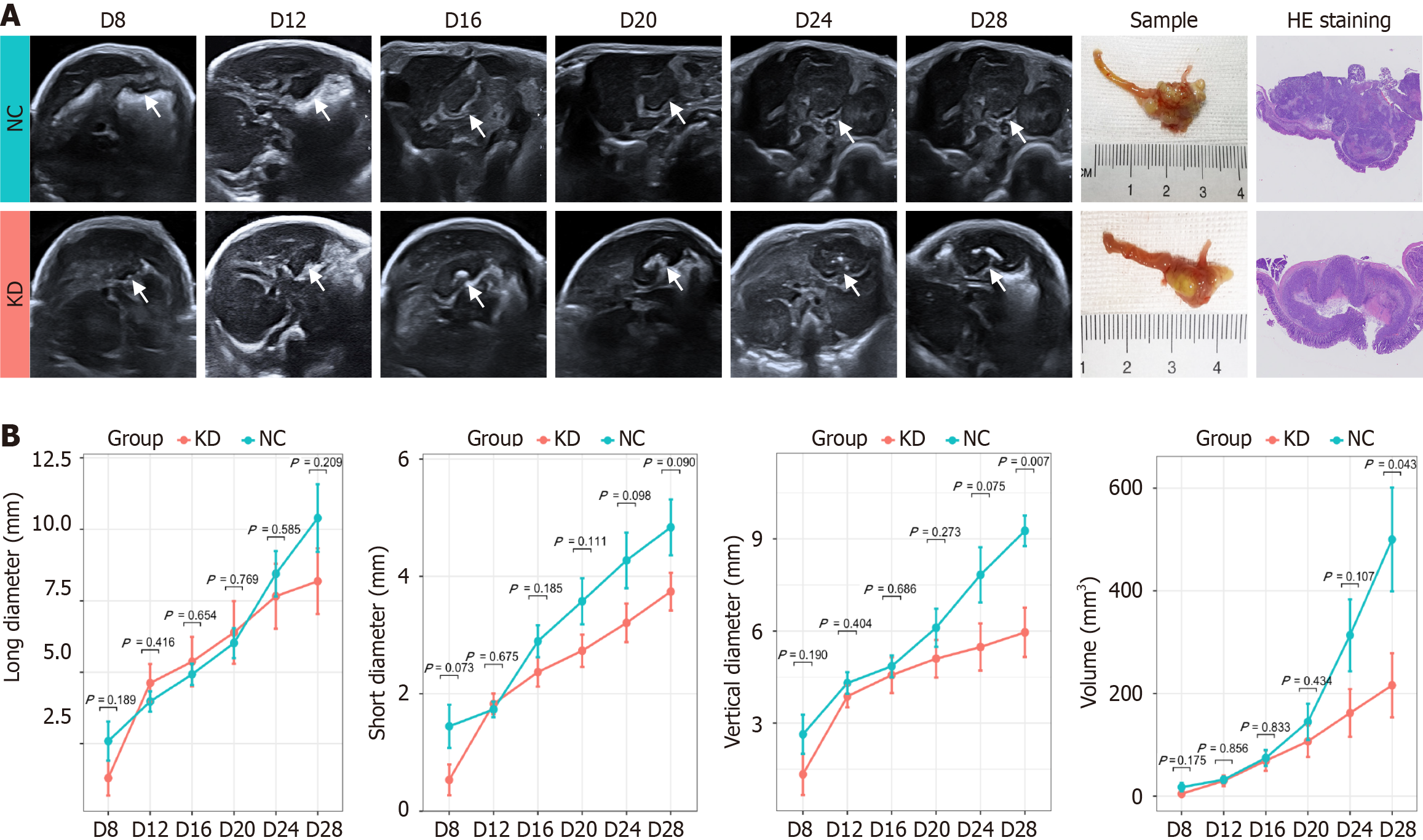
Examinations were performed by two independent physicians (Liu YQ and Xin YW), each with more than five years of experience in ultrasonography of mouse models. Repeated measurements were randomly performed in 6 mice by both physicians on two separate occasions to evaluate interobserver reliability. The remainder of the mice were examined by only one physician. The animals in each group were monitored by ultrasound every four days starting from the eighth day after injection (Day 8, 12, 16, 24, 28). The presence or absence of tumors was recorded, along with their size (long, short, and vertical diameter). Mice were sacrificed when the maximum diameter of the tumor reached 20 mm[13]; the remaining mice were sacrificed on day 28.
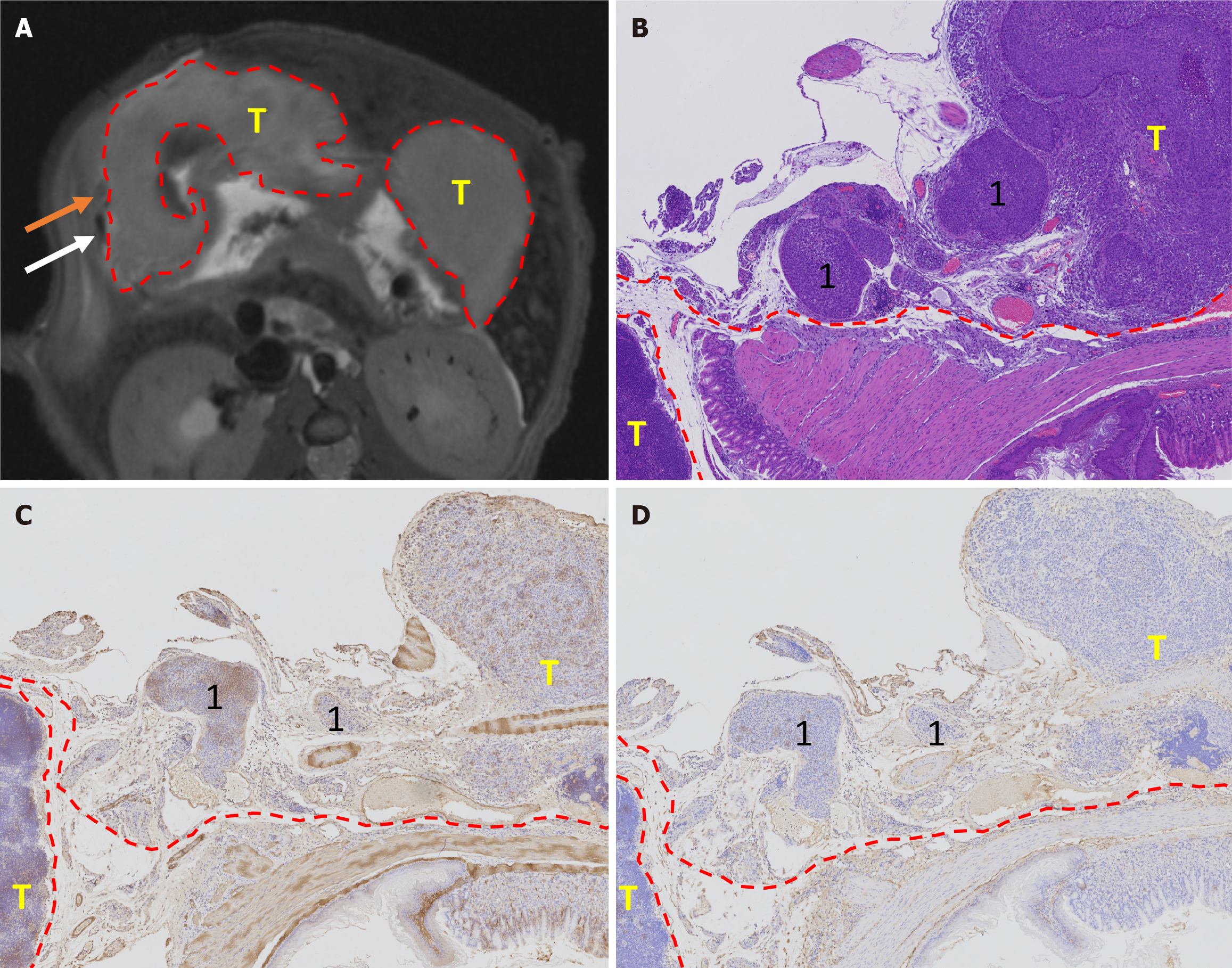
Twenty-four mice from each established orthotopic model underwent preclinical 9.4T MRI (Bruker BioSpin MRI GmbH, BioSpec 94/20, Ettlingen, Germany). Mice were anesthetized with isoflurane and 2% oxygen[14]. The MRI was performed using a single channel transceive coil (40 mm × 40 mm) in the prone position. Body temperature was maintained at 37 ± 0.5 °C with a thermostatically controlled water bed and monitored along with the respiratory rate by an MRI-compatible remote-monitoring system (Model 1030 Monitoring Gating System, SA Instruments, United States). An axial T2-weighted fat suppression sequence was performed with the following parameters: Time of repetition = 1500 millisecond, time of echo = 24 millisecond, average = 1, slice thickness = 0.7 mm, slice gap = 0.3 mm, field of view = 30 mm × 30 mm, matrix = 320 × 320, excitation angle = 90°, and duration 10-15 minutes. All data were analyzed using ParaVision® 6.0.1 software.
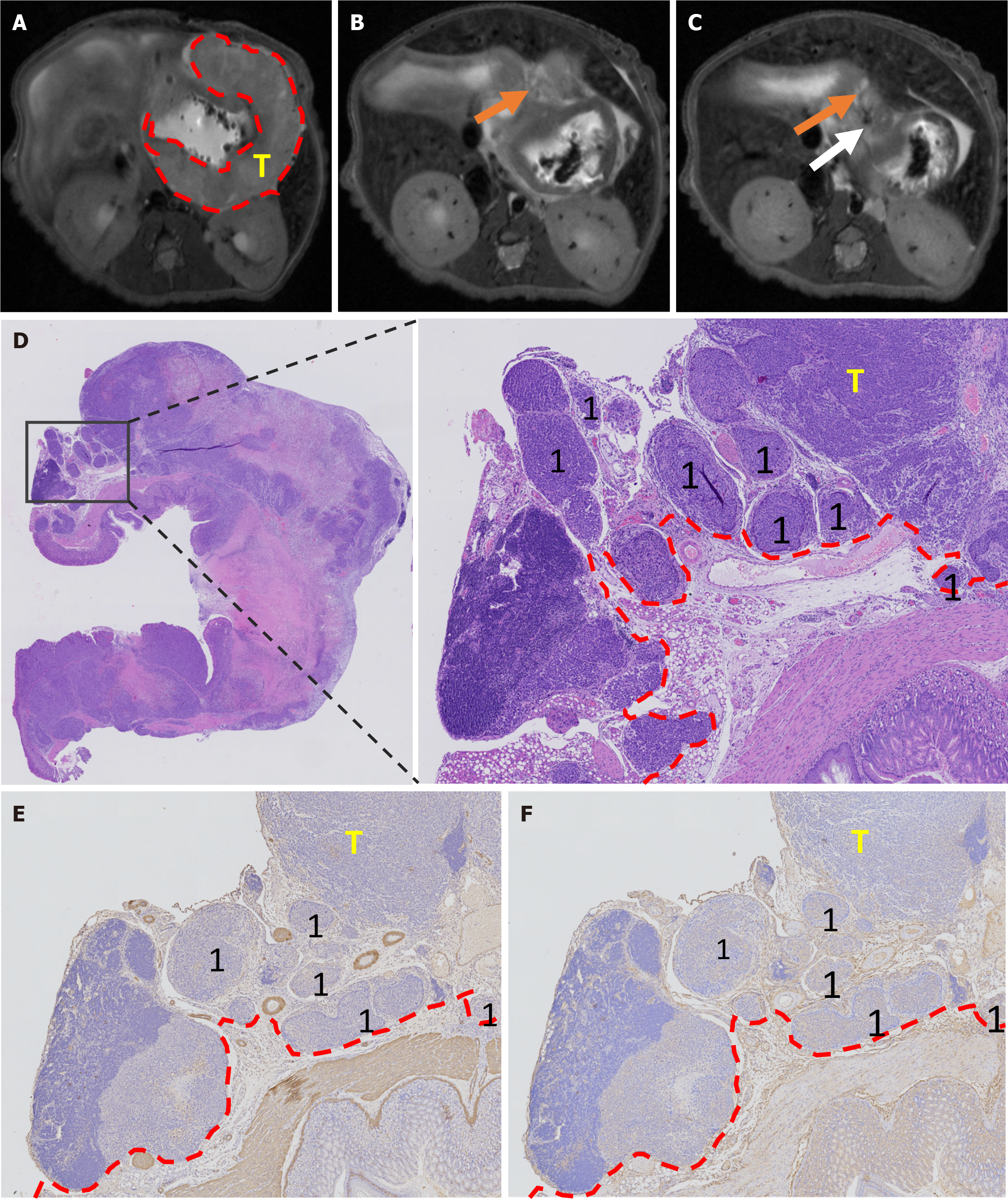
Two board radiologists (Cheng J and Feng CZ with 18 and 7 years of experience, respectively) reviewed various tumor-related parameters, including size (long diameter, mm), signal intensity (heterogeneous/homogeneous), ulcer formation (yes/no), MRI-detected EMVI (mrEMVI) (yes/no), lymph node involvement (yes/no), and distant metastasis (yes/no). mrEMVI was defined as vessels with tubular or serpiginous structures corresponding to the tumor and an intermediate signal[16,17]. In addition, the vessels may show irregular or nodular borders. Although mrEMVI may or may not occur continuously with the primary tumor, mrEMVI should be considered whenever a tumor is near a vessel[18].
After the MRI examination, the mice were euthanized under anesthesia. The stomach was dissected out and fixed in 10% formaldehyde for 24 hours. Tumor size was measured (long diameter, mm), and the tumor tissue was embedded in paraffin for immunohistochemical staining. Four μm-sections were used for staining with hematoxylin and eosin (H&E), CD34 (Abcam, United Kingdom), alpha-smooth muscle actin (SMA) (Abcam, United Kingdom). After a joint reading of H&E, CD34, and SMA staining, the existence of pathology-EMVI (pEMVI) was confirmed. pEMVI was identified as positive when the tumor spread within the lumen of veins beyond the muscular propria layer[19,20]. Lymph node and distant metastases were confirmed on H&E-stained slides from suspected tissues.
Cells in the logarithmic phase were trypsinized and seeded into a 6-well plate. After 48 hours, 0.25% trypsin (without ethylenediaminetetraacetic acid) was used to detach and collect the cells. The cells were washed twice with phosphate-buffered saline by centrifugation at 1300 rpm for 5 minutes, and 5 × 105 cells were collected. The cells were then resuspended in 500 µL of binding buffer, and 10 µL of Annexin V-APC was added and mixed well. The reaction was performed at room temperature for 5-15 minutes in the dark, and a flow cytometer (FACSCalibur; Becton-Dickinson, United States) was used to detect apoptosis.
A single-cell suspension of cells in the logarithmic phase was prepared, and 1000 cells per well were seeded in a 96-well plate with six replicate wells in each group. After 24 hours, 10 μL of cell counting kit-8 (CCK-8) solution (Beyotime Biotechnology, China) was added to the samples, and a blank control well containing only medium and CCK-8 solution was also included. Following incubation for 1 hour, the optical density (OD) absorbance value of each well was determined and recorded using a microplate reader at a wavelength of 450 nm. The plate was then measured at 24 hours for three consecutive days.
The agreement between two radiologists for mrEMVI and other imaging features was analyzed by kappa analysis and intraclass correlation coefficient. A correlation coefficient above 0.75 indicated good agreement. The same method was used to assess tumor growth monitoring consistency between the two ultrasound physicians. A receiver operating characteristic curve calculated the area under the curve (AUC) of mrEMVI according to the gold standard of pEMVI. Associations between imaging and histopathological factors were analyzed using correlation, χ2, or Fisher exact tests. Based on the results of the Shapiro-Wilk normality test and the homogeneity of variance test, either the Mann-Whitney U test or the t-test was used to analyze the differences between groups. All statistical analyses were performed using MedCalc computer software, Version 11.2 (Mariakierke, Belgium). All reported P values were two-sided, with a signi
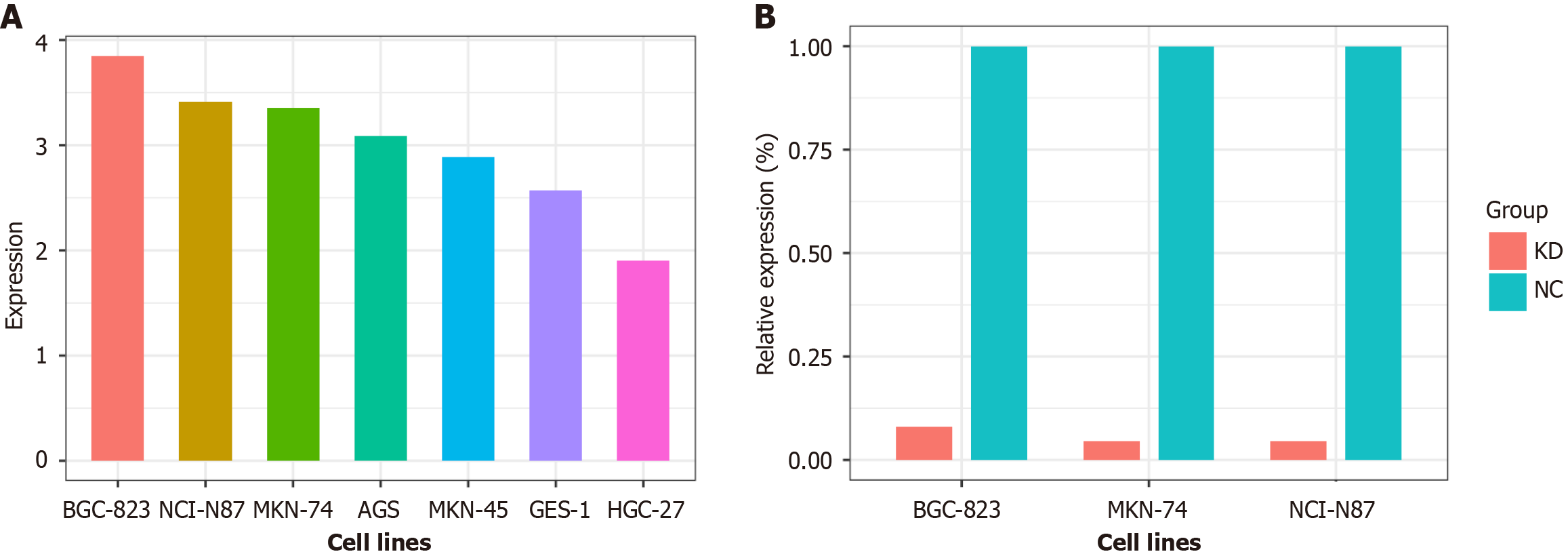
There were 11 differentially expressed circRNAs between EMVI positive and negative samples (Table 1). Among them, only hsa_circ_0097977 showed significantly high expression among multiple GC cells (P < 0.05) by the qRT-PCR test (Figure 2A). Therefore, we selected hsa_circ_0097977 for further experiments. hsa_circ_0097977 contained 1565 nucleotides, and was located in chr12: 14576842-14578407, and its host gene symbol was ATF7IP, according to the Circbase database. In the CircBank database, 137 miRNAs were found that may be sponged by hsa_circ_0097977, and the information on 12 miRNAs has been verified in all miRecords, miRTarBase, and TarBase databases. Using the multiMiR package, we discovered that half of the 12 target miRNAs were linked to GC in the PhenomiR database, whereas two miRNAs were associated with GC in the miR2Disease database. Phenomir and miR2Disease database analyses revealed that “hsa-miR-107” and “hsa-miR-214-3p” are among the miRNAs most strongly associated with GC. The target genes of hsa-miR-107 and hsa-miR-214-3p were predicted using the miRecords, miRTarBase, and TarBase databases. A total of 557 overlapping target genes were obtained using a Venn diagram, including multiple ATF7IP family members.
| circRNAs | Gene name | log2FC | FC | P value | q value | CPM for EMVI positive group | CPM for EMVI negative group |
| hsa_circ_0058218 | ARPC2 | 3.951 | 15.469 | 0.019 | 0.526 | 7.976 | 4.025 |
| hsa_circ_0057109 | PDK1 | 3.140 | 8.818 | 0.033 | 0.571 | 8.282 | 5.141 |
| hsa_circ_0082335 | KLHDC10 | 3.103 | 8.595 | 0.035 | 0.571 | 8.228 | 5.124 |
| hsa_circ_0097977 | ATF7IP | 3.066 | 8.373 | 0.033 | 0.571 | 8.261 | 5.195 |
| hsa_circ_0000283 | CSTF3 | 3.027 | 8.150 | 0.012 | 0.526 | 8.767 | 5.740 |
| hsa_circ_0007813 | DHTKD1 | 2.719 | 6.584 | 0.007 | 0.463 | 9.244 | 6.525 |
| hsa_circ_0006760 | PRUNE | -4.573 | 0.042 | 0.003 | 0.345 | 4.025 | 8.598 |
| hsa_circ_0027857 | ACTR6 | -3.120 | 0.115 | 0.005 | 0.389 | 5.737 | 8.857 |
| hsa_circ_0002387 | TNIK | -2.977 | 0.127 | 0.000 | 0.104 | 7.248 | 10.224 |
| hsa_circ_0071185 | LRBA | -2.370 | 0.193 | 0.031 | 0.571 | 6.639 | 9.009 |
| hsa_circ_0005882 | STK39 | -2.303 | 0.203 | 0.005 | 0.389 | 7.380 | 9.683 |
Following the KD of hsa_circ_0097977 in BGC-823, NCI-N87 and MKN74 cells, the relative expression levels of hsa_circ_0097977 were significantly reduced to 8.0%, 4.5% and 4.6%, respectively, compared to the NC group (all P < 0.05; Figure 2B).
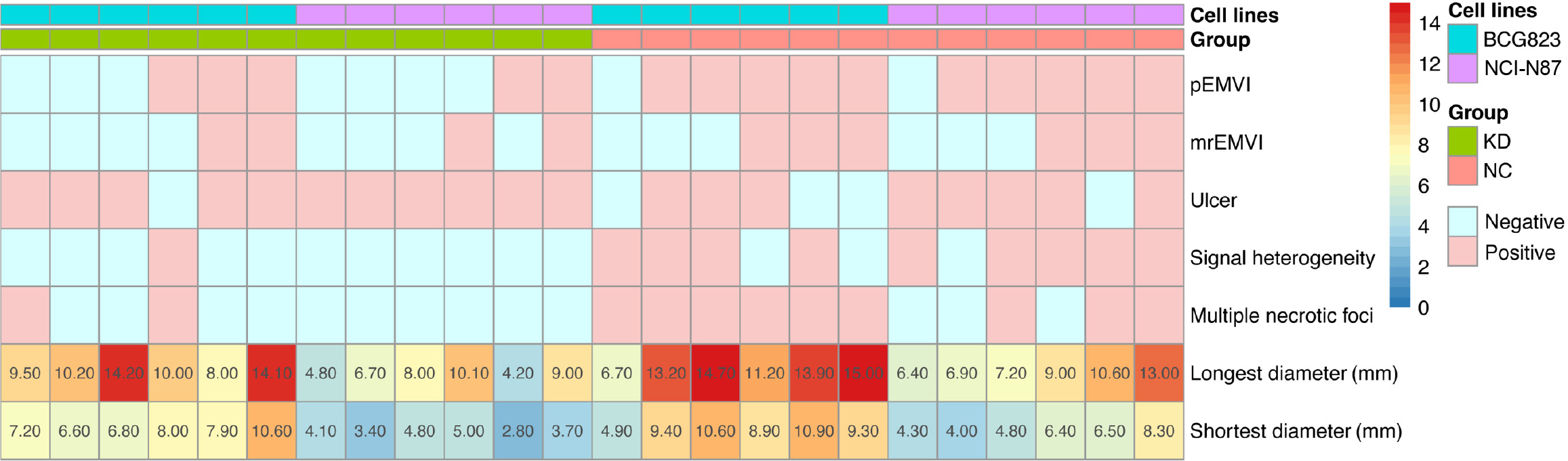
The tumor formation rates of both BGC-823 and NCI-N87 cell lines were 100% (24/24). The agreement between the two ultrasound physicians was greater than 0.75. By monitoring the growth of the NCI-N87 cell line in the orthotopic tumor model dynamically, we found that the growth rate of orthotopic tumors in the NC group was generally faster than that in the KD group (Figure 3). However, there were no significant differences in tumor size between the NC and KD groups. The two radiologists agreed well in terms of mrEMVI (0.83) and other imaging characteristics analyses (0.77-0.91). The median long diameter [interquartile range (IQR)] of the tumors measured on MRI was 9.8 mm (7.0-13.1 mm). The prevalence rate of mrEMVI was 50.0% (12/24) (Figures 4 and 5). The median long diameter (IQR) of the tumors in pathological specimens was 12 mm (8.5-13.0 mm). The incidence of pEMVI was 58.3% (14/24). Ulcer formation was found in 75.0% (18/24) of cases, and heterogeneity of signal intensity was found in 41.7% (10/24). mrEMVI was associated with tumor size and KD of hsa_circ_0097977. pEMVI was only associated with the KD of hsa_circ_0097977 (Figure 6). The AUC of mrEMVI was 0.843 (sensitivity 78.6%, specificity 90.0%) using pEMVI as the reference. All cases were detected with lymph node metastases and proved by histopathological examination. Additionally, 20.8% (5/24) of cases were diagnosed with pathologically proven distant metastasis (peritoneal and liver metastasis). Among multiple MRI features, mrEMVI was the only factor associated with distant metastasis (P = 0.04). No adverse events were observed in any of the experimental groups.
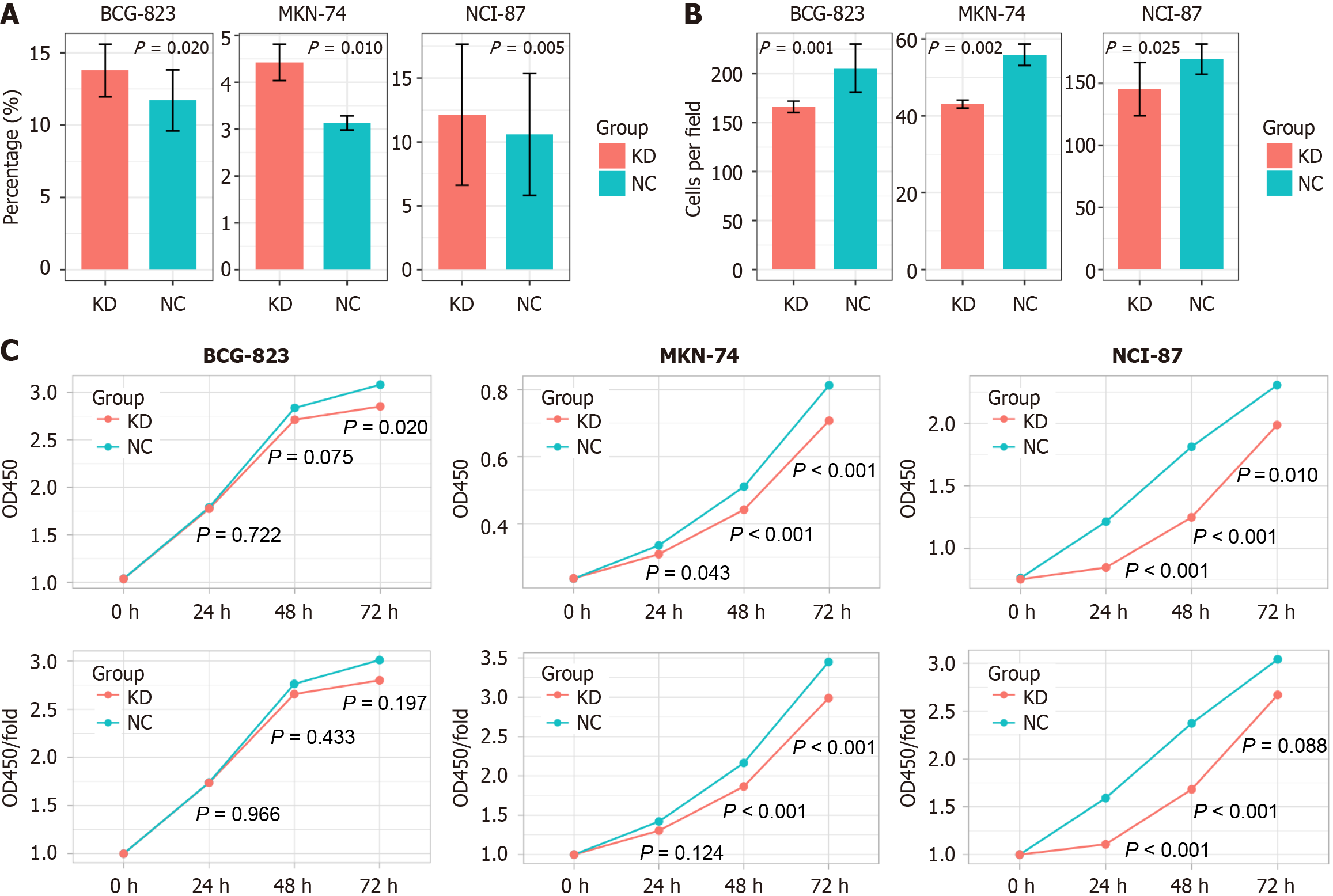
The percentages of apoptotic cells in the KD groups of BGC-823, NCI-N87, and MKN-74 cells were all significantly higher than that in the NC groups (all P < 0.05; Figure 7, Table 2). The number and percentage of invasive cells in the BGC-823-KD, NCI-N87-KD, and MKN-74-KD groups were all significantly lower than those in the NC groups. BGC-823-KD showed lower OD 450 only at 72 hours (P = 0.020); NCI-87-KD had lower OD450 at all time points (P < 0.05) and lower OD450/fold at 24 and 48 hours (P < 0.001); MKN-74-KD had lower OD450 at all time points (P < 0.05) and lower OD450/fold at 48 and 72 hours (P < 0.001).
| BGC-823 | MKN-74 | NCI-N87 | |
| Apoptosis assay | |||
| Apoptotic cells (%) | P = 0.020 | P = 0.010 | P = 0.005 |
| Transwell invasion assay | |||
| Cells per field | P = 0.001 | P = 0.002 | P = 0.025 |
| Cell proliferation assay | |||
| OD450-24 hours | P = 0.722 | P = 0.043 | P < 0.001 |
| OD450-48 hours | P = 0.075 | P < 0.001 | P < 0.001 |
| OD450-72 hours | P = 0.020 | P < 0.001 | P = 0.010 |
| OD450/fold-24 hours | P = 0.966 | P = 0.124 | P < 0.001 |
| OD450/fold-48 hours | P = 0.433 | P < 0.001 | P < 0.001 |
| OD450/fold-72 hours | P = 0.197 | P < 0.001 | P = 0.088 |
The main findings in this study are as follows: (1) EMVI can be detected using 9.4T MRI in orthotopic GC mouse models and subsequently confirmed by histopathology; (2) EMVI is the only imaging feature associated with distant metastasis in orthotopic GC models; and (3) The expression of EMVI and cell function have been observed to be modulated through the KD of hsa_circ_0097977.
To our knowledge, this is the first preclinical imaging histopathological correlation study to explore the diagnostic value of MRI for EMVI in orthotopic GC models. In this study, EMVI was clearly depicted on MRI, and its promising diagnostic accuracy was confirmed through imaging-histopathological correlation, with results similar to those of mrEMVI in rectal cancer[21]. Previous studies on the incidence of EMVI have shown considerable variability, with consistent evidence of underreporting of GC[22]. Pathologically, the underassessment of EMVI stems from the focus on obtaining sufficient lymph nodes from perigastric tissue post-resection for GC[23]. In addition, imaging limitations arise because conventional CT/MRI cannot fully detect EMVI. In this study, the relatively high detection accuracy may be attributed to the application of complete sampling, serial sectioning, and blood vessel staining[24]. Also, 9.4T MRI was chosen for its superior spatial resolution and signal-to-noise ratio, which enables the precise detection of EMVI features that may be missed by clinical MRI (typically 1.5T or 3T) for GC. Although the ultra-high resolution of 9.4T MRI is not directly translatable to current clinical imaging technologies, our study suggests that optimizing MRI protocols, such as enhancing spatial resolution and incorporating specific imaging sequences, could improve the detection of EMVI in GC. This could lead to better risk stratification and personalized treatment strategies for GC patients.
In this study, 20.8% (5/24) of the models were confirmed to have synchronous distant metastasis with pathological examination. mrEMVI was the only relevant imaging factor, whereas tumor size, ulcer formation, and signal uniformity were not significantly correlated with metastasis. This finding is consistent with multiple clinical studies of EMVI in gastrointestinal tumors[25], which confirms that EMVI is a relatively independent risk factor. Furthermore, according to Lord et al’s report[26], only the combined tumor deposits and EMVI status detected on MRI retained prognostic significance on multivariable analysis for overall recurrence and distant recurrence for rectal cancer, rather than TN stage and circumferential resection margin.
This study innovatively revealed the correlation between hsa_circ_0097977 and EMVI, as well as its functional role in GC cells. The association analysis showed that the only common associated factor of mrEMVI and pEMVI was the KD of hsa_circ_0097977. Simultaneously, the results of tumor growth monitoring provided further insight, confirming that KD of hsa_circ_0097977 decreased tumor growth, albeit without significant differences, compared with the NC group. In further cell function tests, hsa_circ_0097977 accelerated the proliferation and invasion of GC cells.
The genetic analysis results provide a possible explanation for these findings. Hsa_circ_0097977 originates from the ATF7IP locus. ATF7IP shows elevated expression in human malignancies, including GC, while exhibiting minimal expression in non-cancerous tissues[27,28]. The association between ATF7IP and Sp1 constitutes a transcription factor complex that regulates genes crucial for cellular proliferation, cell cycle progression, and angiogenesis[29]. Based on bioinformatics analysis of database information, we identified two miRNAs, "hsa-miR-107" and "hsa-miR-214-3p", which have been verified to be sponged by hsa_circ_0097977 and are closely associated with GC. Hsa-miR-214-3p plays a sig
This study innovatively identified a novel therapeutic target and biomarker for GC and further validated previously proposed potential targets. The correlation between hsa_circ_0097977 and EMVI, as well as its impact on GC cell function, suggests that hsa_circ_0097977 itself may serve as a promising therapeutic target and biomarker for GC particularly in patients with EMVI. Targeting hsa_circ_0097977 may inhibit tumor growth and metastasis, potentially reducing recurrence risk and improving survival rates when combined with existing therapies such as chemotherapy or immunotherapy. Detection of hsa_circ_0097977 Levels could serve as a biomarker for early diagnosis and prognosis, existing diagnostic or prognostic models, and help identify high-risk patients for more aggressive treatment strategies. Additionally, ATF7IP has been associated with microsatellite instability in colon cancer and antitumor immune responses[33,34], highlighting its potential significance in GC immunotherapy. Inhibition of miR-214-3p can increase the sensitivity of vascular endothelial cells to Apatinib, thereby promoting the antiangiogenic effect of Apatinib, suggesting a potential combination therapy for advanced GC[32].
This study has several limitations. First, this was a preclinical, small sample size, and single-center study, further verification by an imaging-histopathological study based on GC patients is necessary but remains challenging and may not be fully reproducible at the patient level. Second, animal MRI does not use contrast agents due to the high soft tissue resolution of MRI. Third, hsa_circ_0097977 is a new circRNA that requires further exploration and verification of downstream pathways, such as larger animal studies, clinical trials, or bioinformatics analyses.
EMVI of GC, a risk factor for distant metastasis, can be detected by imaging examination and confirmed by histopathological analysis. Simultaneously, its expression can be regulated by circRNA.
| 1. | Amin MB, Edge S, Greene F, Byrd DR, Brookland RK, Washington MK, Gershenwald JE, Compton CC. AJCC Cancer Staging Manual. 8th ed. Chicago, IL: American College of Surgeons, 2017. |
| 2. | American Cancer Society. Cancer Facts & Figures 2021. Atlanta: American Cancer Society, 2021. |
| 3. | Betge J, Pollheimer MJ, Lindtner RA, Kornprat P, Schlemmer A, Rehak P, Vieth M, Hoefler G, Langner C. Intramural and extramural vascular invasion in colorectal cancer: prognostic significance and quality of pathology reporting. Cancer. 2012;118:628-638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 196] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 4. | Feng C, Cheng J, Zeng X, Zhang Y, Hong N, Ye Y, Wang Y. Development and evaluation of a ceMDCT-based preoperative risk stratification model to predict disease-free survival after radical surgery in patients with gastric cancer. Abdom Radiol (NY). 2021;46:4079-4089. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 5. | Zhu Y, Zhou Y, Zhang W, Xue L, Li Y, Jiang J, Zhong Y, Wang S, Jiang L. Value of quantitative dynamic contrast-enhanced and diffusion-weighted magnetic resonance imaging in predicting extramural venous invasion in locally advanced gastric cancer and prognostic significance. Quant Imaging Med Surg. 2021;11:328-340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 6. | Yang YT, Dong SY, Zhao J, Wang WT, Zeng MS, Rao SX. CT-detected extramural venous invasion is corelated with presence of lymph node metastasis and progression-free survival in gastric cancer. Br J Radiol. 2020;93:20200673. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 7. | Zhou M, Leung A, Echegaray S, Gentles A, Shrager JB, Jensen KC, Berry GJ, Plevritis SK, Rubin DL, Napel S, Gevaert O. Non-Small Cell Lung Cancer Radiogenomics Map Identifies Relationships between Molecular and Imaging Phenotypes with Prognostic Implications. Radiology. 2018;286:307-315. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 144] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 8. | Gao B, Feng C, Chai F, Wei S, Hong N, Ye Y, Wang Y, Cheng J. CT-detected extramural venous invasion-related gene signature for the overall survival prediction in patients with gastric cancer. Cancer Med. 2021;10:7816-7830. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 9. | Yang H, Gou X, Feng C, Zhang Y, Chai F, Hong N, Ye Y, Wang Y, Gao B, Cheng J. Computed tomography-detected extramural venous invasion-related gene signature: a potential negative biomarker of immune checkpoint inhibitor treatment in patients with gastric cancer. J Transl Med. 2023;21:4. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 10. | Li J, Sun D, Pu W, Wang J, Peng Y. Circular RNAs in Cancer: Biogenesis, Function, and Clinical Significance. Trends Cancer. 2020;6:319-336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 524] [Cited by in RCA: 473] [Article Influence: 94.6] [Reference Citation Analysis (0)] |
| 11. | Shan C, Zhang Y, Hao X, Gao J, Chen X, Wang K. Biogenesis, functions and clinical significance of circRNAs in gastric cancer. Mol Cancer. 2019;18:136. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 172] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 12. | Miao Z, Li J, Wang Y, Shi M, Gu X, Zhang X, Wei F, Tang X, Zheng L, Xing Y. Hsa_circ_0136666 stimulates gastric cancer progression and tumor immune escape by regulating the miR-375/PRKDC Axis and PD-L1 phosphorylation. Mol Cancer. 2023;22:205. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 13. | Yoon SJ, Park J, Shin Y, Choi Y, Park SW, Kang SG, Son HY, Huh YM. Deconvolution of diffuse gastric cancer and the suppression of CD34 on the BALB/c nude mice model. BMC Cancer. 2020;20:314. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 104] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 14. | Hildebrandt IJ, Su H, Weber WA. Anesthesia and other considerations for in vivo imaging of small animals. ILAR J. 2008;49:17-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 172] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 15. | Li Y, Li B, Xiang CP, Zhang Y, Li YY, Wu XL. Characterization of gastric cancer models from different cell lines orthotopically constructed using improved implantation techniques. World J Gastroenterol. 2012;18:136-143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 9] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 16. | Liu LH, Lv H, Wang ZC, Rao SX, Zeng MS. Performance comparison between MRI and CT for local staging of sigmoid and descending colon cancer. Eur J Radiol. 2019;121:108741. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 17. | Lee ES, Kim MJ, Park SC, Hur BY, Hyun JH, Chang HJ, Baek JY, Kim SY, Kim DY, Oh JH. Magnetic Resonance Imaging-Detected Extramural Venous Invasion in Rectal Cancer before and after Preoperative Chemoradiotherapy: Diagnostic Performance and Prognostic Significance. Eur Radiol. 2018;28:496-505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 58] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 18. | Fernandes MC, Gollub MJ, Brown G. The importance of MRI for rectal cancer evaluation. Surg Oncol. 2022;43:101739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 79] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 19. | Li X, Jonnagaddala J, Yang S, Zhang H, Xu XS. A retrospective analysis using deep-learning models for prediction of survival outcome and benefit of adjuvant chemotherapy in stage II/III colorectal cancer. J Cancer Res Clin Oncol. 2022;148:1955-1963. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 20. | Ale Ali H, Kirsch R, Razaz S, Jhaveri A, Thipphavong S, Kennedy ED, Jhaveri KS. Extramural venous invasion in rectal cancer: overview of imaging, histopathology, and clinical implications. Abdom Radiol (NY). 2019;44:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 21. | Kim TH, Woo S, Han S, Suh CH, Vargas HA. The Diagnostic Performance of MRI for Detection of Extramural Venous Invasion in Colorectal Cancer: A Systematic Review and Meta-Analysis of the Literature. AJR Am J Roentgenol. 2019;213:575-585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 22. | Chand M, Palmer T, Blomqvist L, Nagtegaal I, West N, Brown G. Evidence for radiological and histopathological prognostic importance of detecting extramural venous invasion in rectal cancer: recommendations for radiology and histopathology reporting. Colorectal Dis. 2015;17:468-473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 23. | Burgart LJ, Chopp WV, Jain D. Protocol for the Examination of Specimens From Patients With Carcinoma of the Stomach, Version: 4.4.0.0. 2023. [cited 13 March 2025]. Available from: https://documents.cap.org/documents/Stomach_4.4.0.0.REL_CAPCP.docx. |
| 24. | Kirsch R, Messenger DE, Riddell RH, Pollett A, Cook M, Al-Haddad S, Streutker CJ, Divaris DX, Pandit R, Newell KJ, Liu J, Price RG, Smith S, Parfitt JR, Driman DK. Venous invasion in colorectal cancer: impact of an elastin stain on detection and interobserver agreement among gastrointestinal and nongastrointestinal pathologists. Am J Surg Pathol. 2013;37:200-210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 91] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 25. | Tripathi P, Guo W, Rao S, Zeng M, Hu D. Additional value of MRI-detected EMVI scoring system in rectal cancer: applicability in predicting synchronous metastasis. Tumori. 2020;106:286-294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 26. | Lord AC, D'Souza N, Shaw A, Rokan Z, Moran B, Abulafi M, Rasheed S, Chandramohan A, Corr A, Chau I, Brown G. MRI-Diagnosed Tumor Deposits and EMVI Status Have Superior Prognostic Accuracy to Current Clinical TNM Staging in Rectal Cancer. Ann Surg. 2022;276:334-344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 123] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 27. | Yasuda Y, Tokunaga K, Koga T, Sakamoto C, Goldberg IG, Saitoh N, Nakao M. Computational analysis of morphological and molecular features in gastric cancer tissues. Cancer Med. 2020;9:2223-2234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 28. | Moon SW, Mo HY, Choi EJ, Yoo NJ, Lee SH. Mutational Alterations of DNA Methylation-related Genes CTCF, ZFP57, and ATF7IP Genes in Colon Cancers. Appl Immunohistochem Mol Morphol. 2022;30:e16-e20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 29. | Liu L, Ishihara K, Ichimura T, Fujita N, Hino S, Tomita S, Watanabe S, Saitoh N, Ito T, Nakao M. MCAF1/AM is involved in Sp1-mediated maintenance of cancer-associated telomerase activity. J Biol Chem. 2009;284:5165-5174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 48] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 30. | Wang M, Zhao C, Shi H, Zhang B, Zhang L, Zhang X, Wang S, Wu X, Yang T, Huang F, Cai J, Zhu Q, Zhu W, Qian H, Xu W. Deregulated microRNAs in gastric cancer tissue-derived mesenchymal stem cells: novel biomarkers and a mechanism for gastric cancer. Br J Cancer. 2014;110:1199-1210. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 190] [Article Influence: 17.3] [Reference Citation Analysis (1)] |
| 31. | Yang Y, Li Z, Yuan H, Ji W, Wang K, Lu T, Yu Y, Zeng Q, Li F, Xia W, Lu S. Reciprocal regulatory mechanism between miR-214-3p and FGFR1 in FGFR1-amplified lung cancer. Oncogenesis. 2019;8:50. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 32. | Wang W, Wang T, Zhang Y, Deng T, Zhang H, Ba YI. Gastric cancer secreted miR-214-3p inhibits the anti-angiogenesis effect of apatinib by suppressing ferroptosis in vascular endothelial cells. Oncol Res. 2024;32:489-502. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 33. | Hu H, Khodadadi-Jamayran A, Dolgalev I, Cho H, Badri S, Chiriboga LA, Zeck B, Lopez De Rodas Gregorio M, Dowling CM, Labbe K, Deng J, Chen T, Zhang H, Zappile P, Chen Z, Ueberheide B, Karatza A, Han H, Ranieri M, Tang S, Jour G, Osman I, Sucker A, Schadendorf D, Tsirigos A, Schalper KA, Velcheti V, Huang HY, Jin Y, Ji H, Poirier JT, Li F, Wong KK. Targeting the Atf7ip-Setdb1 Complex Augments Antitumor Immunity by Boosting Tumor Immunogenicity. Cancer Immunol Res. 2021;9:1298-1315. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 34. | Sin JH, Kashyap S, Acenas D, Cortez JT, Lee J, Marson A, Matloubian M, Waterfield MR. ATF7ip Targets Transposable Elements for H3K9me3 Deposition to Modify CD8(+) T Cell Effector and Memory Responses. J Immunol. 2022;208:1155-1169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 1.0] [Reference Citation Analysis (0)] |