Published online Apr 7, 2025. doi: 10.3748/wjg.v31.i13.102527
Revised: February 13, 2025
Accepted: March 6, 2025
Published online: April 7, 2025
Processing time: 163 Days and 17.7 Hours
Hepatocellular carcinoma (HCC) is a prevalent malignant tumor with a poor prognosis, which is often associated with chronic hepatitis B virus infection in China. Our previous study has shown that long non-coding RNA semaphorin 6A-antisense RNA 1 (SEMA6A-AS1) was significantly downregulated in hepatitis B virus-related HCC and associated with poor prognosis.
To explore the underlying mechanism of SEMA6A-AS1 in HCC progression.
The expression levels of SEMA6A-AS1 and SEMA6A were detected using quan
Downregulation of SEMA6A-AS1 in HCC was negatively correlated with SEMA6A protein expression. SEMA6A was upregulated in HCC and correlated with high alpha-fetoprotein level, high Edmondson-Steiner grade and poor prognosis. SEMA6A-AS1 significantly inhibited the proliferation, migration and invasion of HCC cells by combining with SEMA6A mRNA and promoting its degradation. SEMA6A protein promoted the proliferation, migration and invasion of HCC cells by regulating the actin cytoskeleton.
Our findings suggest that SEMA6A-AS1 can inhibit HCC progression through decreasing SEMA6A expression by promoting its mRNA degradation. SEMA6A-AS1 may be a prognostic biomarker and therapeutic target for HCC.
Core Tip: This study investigated the association of semaphorin 6A-antisense RNA 1 (SEMA6A-AS1) and SEMA6A with clinical features and prognosis of hepatocellular carcinoma (HCC), and their functional implications in HCC cell proliferation, migration and invasion. Further investigation has revealed that SEMA6A-AS1 can reverse the progression of HCC by decreasing SEMA6A expression through mRNA stabilization and actin cytoskeleton regulation. These findings expand the understanding of molecular mechanisms of SEMA6A-AS1 in HCC progression and provide novel therapeutic targets for HCC.
- Citation: Yu SM, Zhang M, Li SL, Pei SY, Wu L, Hu XW, Duan YK. Long noncoding RNA semaphorin 6A-antisense RNA 1 reduces hepatocellular carcinoma by promoting semaphorin 6A mRNA degradation. World J Gastroenterol 2025; 31(13): 102527
- URL: https://www.wjgnet.com/1007-9327/full/v31/i13/102527.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i13.102527
Liver cancer is the third leading cause of cancer death worldwide and China accounts for nearly half of the global burden of liver cancer[1]. Hepatocellular carcinoma (HCC) contributes to 80% of all liver cancer cases globally and chronic hepatitis B virus (HBV) infection predominates as a risk factor in China[2]. Although the outcome of patients with HCC has improved with the approval of new agents and the establishment of immune checkpoint inhibitor-based therapies as standard of care, the long-term prognosis remains unsatisfactory[3]. Therefore, predictive biomarkers and molecular targeted therapy are urgently needed to improve the diagnosis and treatment of HCC. Long noncoding RNA (lncRNA) is a class of non-coding RNAs characterized by containing more than 200 nucleotides and lacking protein-coding capacity. Recently, emerging evidence indicated that lncRNAs play important roles in the diverse pathogenesis and progression of HCC[4,5]. For example, lncRNA down syndrome critical region 8[6] and small nucleolar RNA host gene 5[7] have been found to upregulate Wnt and result in HCC growth, while lnc-epidermal growth factor receptor[8] and lncRNA nuclear paraspeckle assembly transcript 1[9] can promote HCC development by activating epidermal growth factor receptor and c-Met, respectively. In addition, lncRNA-6195 represses the growth of HCC by combining with enolase 1 and inhibiting its enzymatic activity[10]. LncRNA tumor protein P73 antisense RNA 1 (AS1)[11] and MIR4435-2 host gene[12] aggravate HCC cell malignant behaviors by activating the transforming growth factor beta signaling pathway. Although the functions of many lncRNAs involved in HCC have been investigated, most dysregulated lncRNAs lack functional characterization and may play critical roles in HCC development and progression.
Previously, we demonstrated that lncRNA SEMA6A-AS1 was downregulated in HBV-related HCC and low SEMA6A-AS1 expression level was significantly associated with poor prognosis and malignant phenotypes such as capsular invasion, high Edmondson-Steiner grade and tumor-node-metastasis (TNM) stage[13]. Semaphorin 6A (SEMA6A) is a multifunctional protein involved in the formation of malignant tumors by forming a complex with plexins to regulate the actin cytoskeleton, motility, and proliferation[14,15]. However, the function of SEMA6A in HCC is still unclear. The aim of this study was to investigate the function and underlying mechanism of SEMA6A-AS1 and SEMA6A in HCC progression.
The HCC and adjacent non-tumor tissue samples and clinicopathological characteristics used in this study were collected and analyzed in our previous studies[10,13]. All enrolled patients were followed up for 2 years after operation, and the overall survival time was the period from the operation to death or the end of follow-up.
According to the manufacturer’s instructions, total RNA was extracted from tissues and HCC cells utilizing Total RNA kit II (Omega, R6934) and subsequently reverse-transcribed into cDNA utilizing Prime Script RT reagent kit with gDNA Eraser (Takara, RR047A) The expression levels of SEMA6A-AS1, SEMA6A and glyceraldehyde-3-phosphate dehydrogenase (as the internal control) were measured by quantitative polymerase chain reaction (qPCR) with SYBR Premix Ex Taq II Kit (Takara, RR820A) on ABI 7500 Fast real-time PCR platform. All primer sequences were listed in Supplementary Table 1.
The LM3, Hep3B and 293T cell lines were obtained from Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in the dulbecco’s modified eagle medium containing 10% fetal bovine serum (HyClone, Utah, United States), 100 units/mL penicillin and 100 mg/mL streptomycin at 37 °C, 5% CO2 and 95% humidity.
The SEMA6A-AS1 expression plasmid vector (pc-SEMA6A-AS1) and the SEMA6A expression plasmid vector (pc-SEMA6A) were constructed by inserting a full-length SEMA6A-AS1 or SEMA6A mRNA into the BamHI/EcoRI site of pcDNA3.1. Short hairpin RNAs (shRNAs) against SEMA6A-AS1 were designed (RIBOBIO, China) and produced using the Lenti-X HTX packaging system (Clontech, United States) in 293T cells. The target sequences of shRNA are listed in Supplementary Table 2.
The SEMA6A-AS1 or SEMA6A overexpressing cells and control cells were obtained by transfecting pc-SEMA6A-AS1, pc-SEMA6A or pcDNA3.1 into cells with Lipo8000 (Beyotime, C0533) and selecting with G418 (Beyotime, ST081). The SEMA6A-AS1 knockdown cells and control cells were obtained by transfecting SEMA6A-AS1 shRNA or control shRNA into cells with polybrene (Genomeditech, China) and selecting with puromycin (Beyotime, ST551).
Growth curve and colony formation assay were performed to assess cell proliferation ability. Wound-healing and transwell assay were performed to assess cell migration ability. And transwell Matrigel invasion assay was performed to assess cell invasion ability.
Cell cycle and apoptosis assays were performed on flow cytometer platform (BD Biosciences, United States) using Cell Cycle and Apoptosis Analysis Kit (Beyotime, C1052) and data were analyzed using FlowJo X software.
Total protein lysates were prepared from cells using Cell lysis buffer for Western and immunoprecipitation (Beyotime, P0013J) with protease inhibitors (Beyotime, P1005). The antibodies to SEMA6A and glyceraldehyde-3-phosphate dehydrogenase were respectively obtained from Abcam (ab154938, United States) and ABclonal Technology (A19056, China). The signals were visualized using Clarity Max Western Enhanced Chemiluminescence Substrate (Bio-Rad, #1705062) and scanned by Image Lab software.
Immunohistochemistry for SEMA6A was performed on paraffin-embedded tissue sections with rabbit anti-human SEMA6A (Abcam, United States) as primary antibody and goat anti-rabbit immunoglobulin G horseradish peroxidase-conjugated secondary antibody (Abcam, United States). Proteins were visualized in situ with 3,3’-diaminobenzidine, and photographs were then captured with microscope (LEICA, Germany) and analyzed using Image-Pro Plus 6.0 software.
Tagged RNA affinity purification (TRAP) assay was performed using the TRAP kit (Bes5106, BersinBio, China) according to the manufacturer’s instructions. In TRAP-RNA assay, RNAs obtained by SEMA6A-AS1 pulldown were reversely transcribed and then subjected to qPCR for SEMA6A mRNA level. In TRAP-RNA binding protein assay, proteins obtained by SEMA6A-AS1 pulldown were assayed by Western blotting to confirm the presence of SEMA6A.
Potential SEMA6A-AS1 and SEMA6A mRNA binding site (BS) was predicted based on the base complementation principle, and BS or mutant-form BS (Mut-BS) of SEMA6A-AS1 were respectively inserted into the dual luciferase reporter vectors (pmirGLO). BS, Mut-BS, pc-SEMA6A and pcDNA3.1 plasmids were respectively transfected into Hep3B cells of the corresponding subgroups using Lipo 8000 (Beyotime, C0533). Dual luciferase reporter assays were performed utilizing Dual Luciferase Reporter Gene Assay Kit (Beyotime, RG027) to measure the luciferase activity of firefly and renilla 48 hours after transfection. The sequences of BS and Mut-BS are listed in Supplementary Table 3.
Cells were seeded on poly-L-lysine-coated coverslips, fixed in 4% paraformaldehyde, permeabilized with 0.25% Triton X-100, blocked with 1% bovine serum albumin and then incubated with rabbit anti-SEMA6A (Abcam, ab154938) overnight at 4 °C. Next day, coverslips were incubated with Alexa Fluor 594-conjugated anti-rabbit immunoglobulin G (Thermo, A32754) and phalloidin-Alexa Fluor 488 (Beyotime, C2201S) for 1 hour at room temperature in dark and then mounted onto microscope slides using Antifade Mounting Medium with DAPI (Beyotime, P0131). Photographs were captured with immunofluorescence microscope (Axio Imager M2, Zeiss, Germany).
SPSS 24.0 and GraphPad Prism 6 software were used for statistical analysis. Paired sample t-test was performed to analyze the expression levels of SEMA6A-AS1 and SEMA6A in HCC and adjacent liver tissues. Pearson’s correlation analysis was used to analyze the correlation of SEMA6A-AS1 and SEMA6A with HBV-related HCC. Kaplan-Meier method and a log-rank test were used to calculate the survival curves. Pearson’s χ2-square test and Fisher’s exact test were performed to analyze the association of SEMA6A expression level with clinicopathological characteristics. Student’s t-test was used for comparison between groups. P < 0.05 was considered statistically significant, and data are expressed as mean ± SD. All experiments in vitro were repeated at least three times.
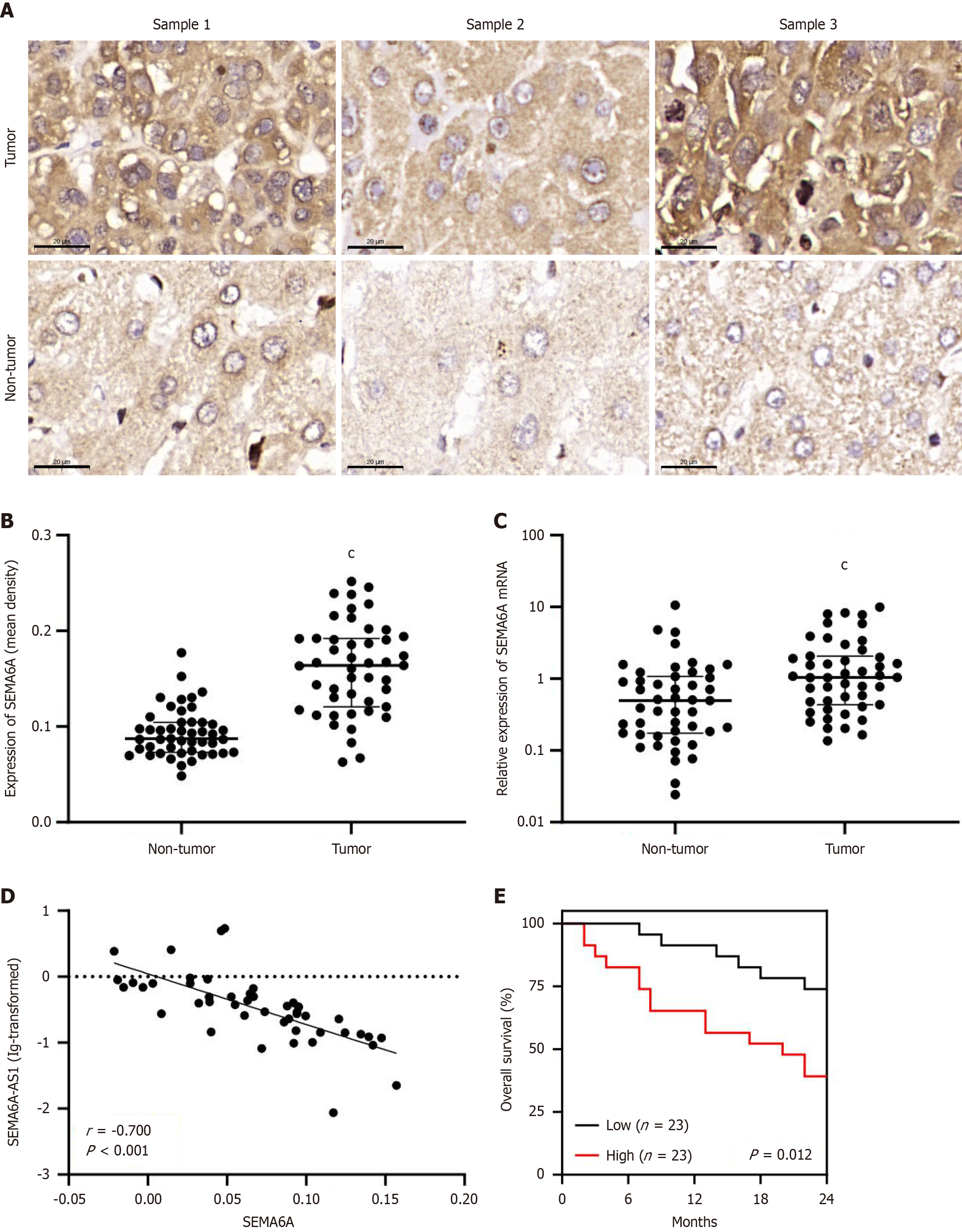
In a previous study, we demonstrated that SEMA6A-AS1 was downregulated in HBV-related HCC and associated with poor prognosis and malignant phenotypes such as capsular invasion, differentiation grade, and TNM stage[13]. We hypothesized that as an antisense lncRNA, SEMA6A-AS1 might participate in the occurrence and progression of HCC by regulating the expression of its antisense protein-coding counterpart, SEMA6A. Previous results from quantitative qPCR showed that in 47 pairs of HCC and adjacent non-tumor liver samples, SEMA6A-AS1 was significantly downregulated in the HCC samples[13]. To investigate the expression profile of SEMA6A, immunohistochemistry analysis was performed using the same 47 pairs of clinical samples collected in the above study. Both protein (Figure 1A and B) and mRNA (Figure 1C) levels of SEMA6A in HCC tissues were significantly higher than those in adjacent non-tumorous hepatic tissues. Specifically, SEMA6A expression levels in 89.36% (42 pairs) of the 47 pairs of clinical samples were higher in HCC tissues than in corresponding non-tumorous liver tissues (Supplementary Figure 1A). Pearson correlation analysis showed that in HCC tissues, SEMA6A-AS1 Levels were negatively correlated with SEMA6A protein (Figure 1D) and mRNA (Supplementary Figure 1B) levels. The expression levels of SEMA6A-AS1 were significantly lower in HCC cell lines (LM3, Hep3B, HepG2 and Huh7) compared to normal liver cell lines such as L02, while SEMA6A mRNA was expressed at a higher level in HCC cell lines than in L02 cells (Supplementary Figure 1C). Further analysis of clinical data showed that the serum alpha-fetoprotein levels and Edmondson-Steiner grades of HCC were significantly higher in the high SEMA6A group, suggesting that the high expression of SEMA6A in HCC is associated with a low level of differentiation (Table 1). Kaplan-Meier analysis of 46 enrolled patients (one patient died due to postoperative hemorrhage and was excluded) revealed a strong correlation between upregulated SEMA6A levels and reduced overall survival (Figure 1E). These findings prompted a deeper investigation of the relationship between SEMA6A-AS1, SEMA6A expression, and HCC progression.
| Variable | Low SEMA6A expression levels | High SEMA6A expression levels | P value | |
| All cases | 23 | 24 | ||
| Age, years | ≤ 55 | 16 | 19 | 0.450 |
| > 55 | 7 | 5 | ||
| Gender | Male | 22 | 23 | 0.975 |
| Female | 1 | 1 | ||
| HBV-DNA, copies/mL | ≤ 103 | 14 | 17 | 0.471 |
| > 103 | 9 | 7 | ||
| Tumor size, cm | ≤ 5 | 13 | 8 | 0.110 |
| > 5 | 10 | 16 | ||
| Tumors, n | Solitary | 19 | 18 | 0.524 |
| Multiple | 4 | 6 | ||
| Cirrhosis | Yes | 17 | 17 | 0.813 |
| No | 6 | 7 | ||
| AFP, ng/mL | ≤ 20 | 10 | 3 | 0.041 |
| > 20 | 13 | 21 | ||
| TNM classification | I/II/III | 7/13/3 | 4/11/9 | 0.064 |
| Edmondson grade1 | I/II/III/IV | 5/16/2/0 | 0/19/4/1 | 0.022 |
| ECOG status | 0 | 21 | 22 | 0.965 |
| 1 | 2 | 2 | ||
| BCLC stage | 0/A/B/C | 4/10/2/7 | 1/9/3/11 | 0.132 |
| Vascular invasion | Yes | 7 | 11 | 0.278 |
| No | 16 | 13 | ||
| Capsular invasion | Yes | 7 | 13 | 0.100 |
| No | 16 | 11 | ||
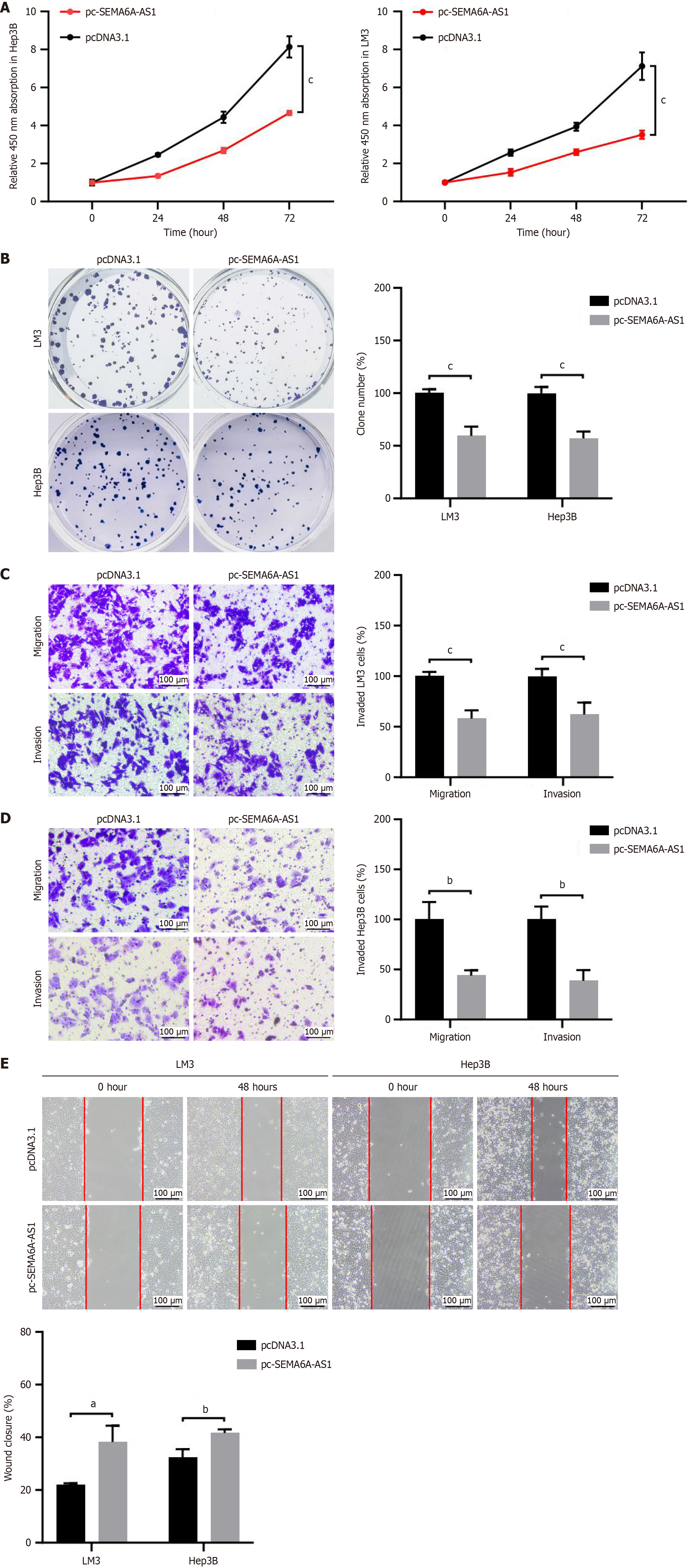
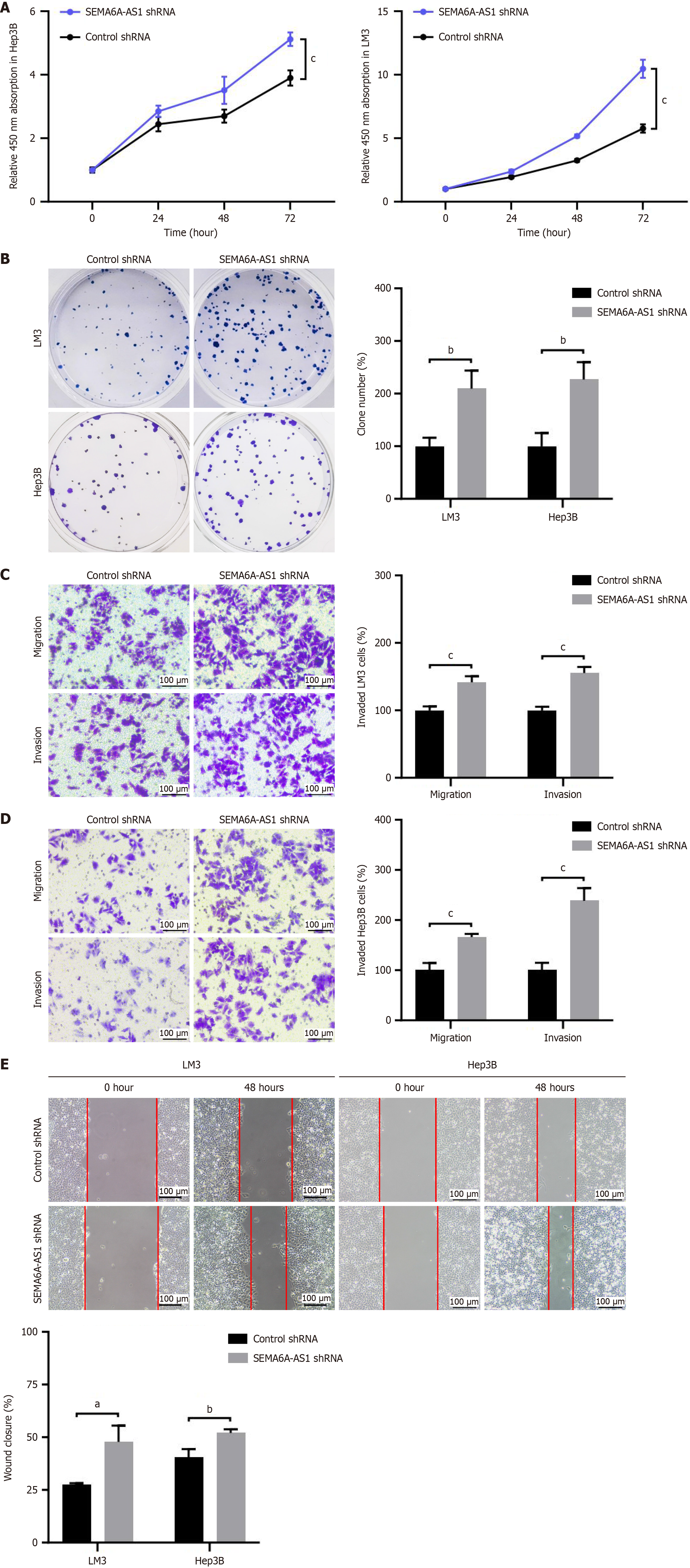
To verify the function of SEMA6A-AS1 in HCC, the effects of SEMA6A-AS1 on cell proliferation, apoptosis, migration, and invasion were assessed by gain- and loss-of-function studies in LM3 and Hep3B cell lines. SEMA6A-AS1 stably overexpressed or inhibited LM3 and Hep3B cell lines were constructed and then verified by qPCR (Supplementary Figure 1D). Functional assays revealed that overexpression of SEMA6A-AS1 prevented HCC cell proliferation (Figure 2A and B), migration and invasion (Figure 2C-E), compared with the negative control group. However, no significant difference was found in terms of cell apoptosis between the SEMA6A-AS1 overexpressed and control groups (Supplementary Figure 2A). To further confirm these findings, functional assays were also performed with SEMA6A-AS1 knockdown LM3 and Hep3B cell lines. As expected, SEMA6A-AS1 downregulation significantly promoted the cell proliferation (Figure 3A and B), migration and invasion (Figure 3C-E) abilities of HCC cells. The above results demonstrated that SEMA6A-AS1 can inhibit HCC by suppressing the proliferation, migration and invasion abilities of HCC cells.
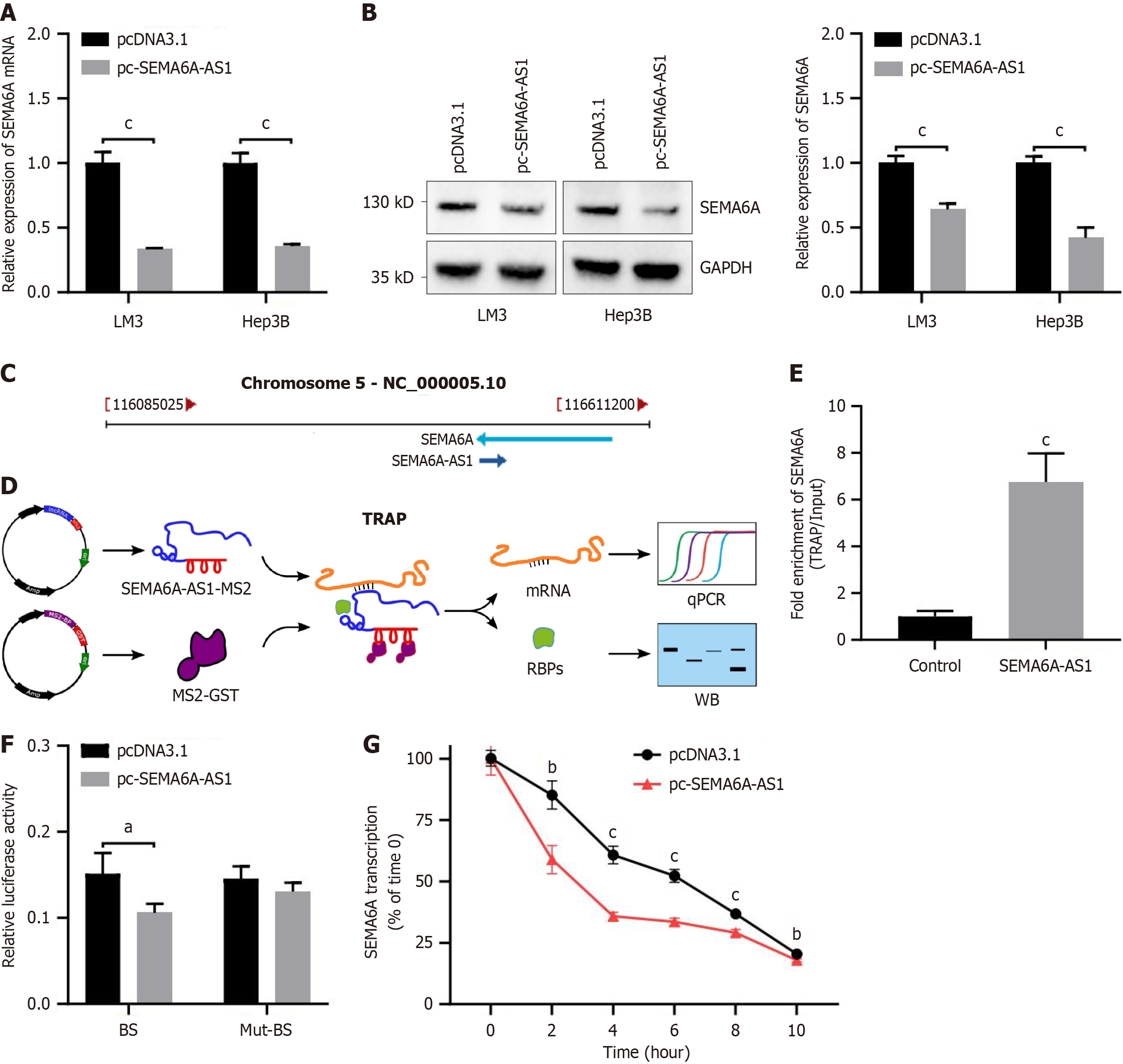
Based on previous findings, we hypothesized that SEMA6A-AS1 is involved in the development of HCC by regulating SEMA6A expression. Results from qPCR (Figure 4A) and Western blot (Figure 4B) showed that the expression levels of SEMA6A were downregulated by SEMA6A-AS1 overexpression in Hep3B and LM3 cells. The genes encoding SEMA6A-AS1 and SEMA6A are both located on chromosome 5 (5q23.1), transcribed in opposite directions and share a region of 295 bp (Figure 4C). To further explore the relationship between SEMA6A-AS1 and SEMA6A, TRAP assay was performed using a vector containing SEMA6A-AS1 (Figure 4D). The precipitated RNAs were analyzed by qPCR and revealed that SEMA6A-AS1 may directly combine with SEMA6A mRNA (Figure 4E). The Western blot results of RNA binding proteins showed that SEMA6A-AS1 did not bind to SEMA6A protein (Supplementary Figure 2B). To verify the association between SEMA6A-AS1 and SEMA6A mRNA, dual luciferase reporter analysis was performed with the luciferase reporter vector containing a potential BS or Mut-BS. Consistent with our speculation, we observed a significantly higher enrichment level of SEMA6A mRNA with the BS vector than with the Mut-BS vector (Figure 4F). These findings suggest that SEMA6A-AS1 could directly combine with SEMA6A mRNA and downregulate SEMA6A expression.
Recently, it was reported that, some antisense lncRNAs regulate the expression of corresponding proteins by binding and affecting the stability of mRNAs from homologous sense genes[16-18]. Thus, we hypothesized that SEMA6A-AS1 may regulate SEMA6A expression by binding and modulating the stability of SEMA6A mRNA. This hypothesis was verified by mRNA degradation assays with actinomycin D (mRNA synthesis inhibitor) and it was found that SEMA6A-AS1 indeed promoted SEMA6A mRNA degradation (Figure 4G). Based on these results, SEMA6A-AS1 regulated the degradation of SEMA6A mRNA, which subsequently affected SEMA6A protein expression.
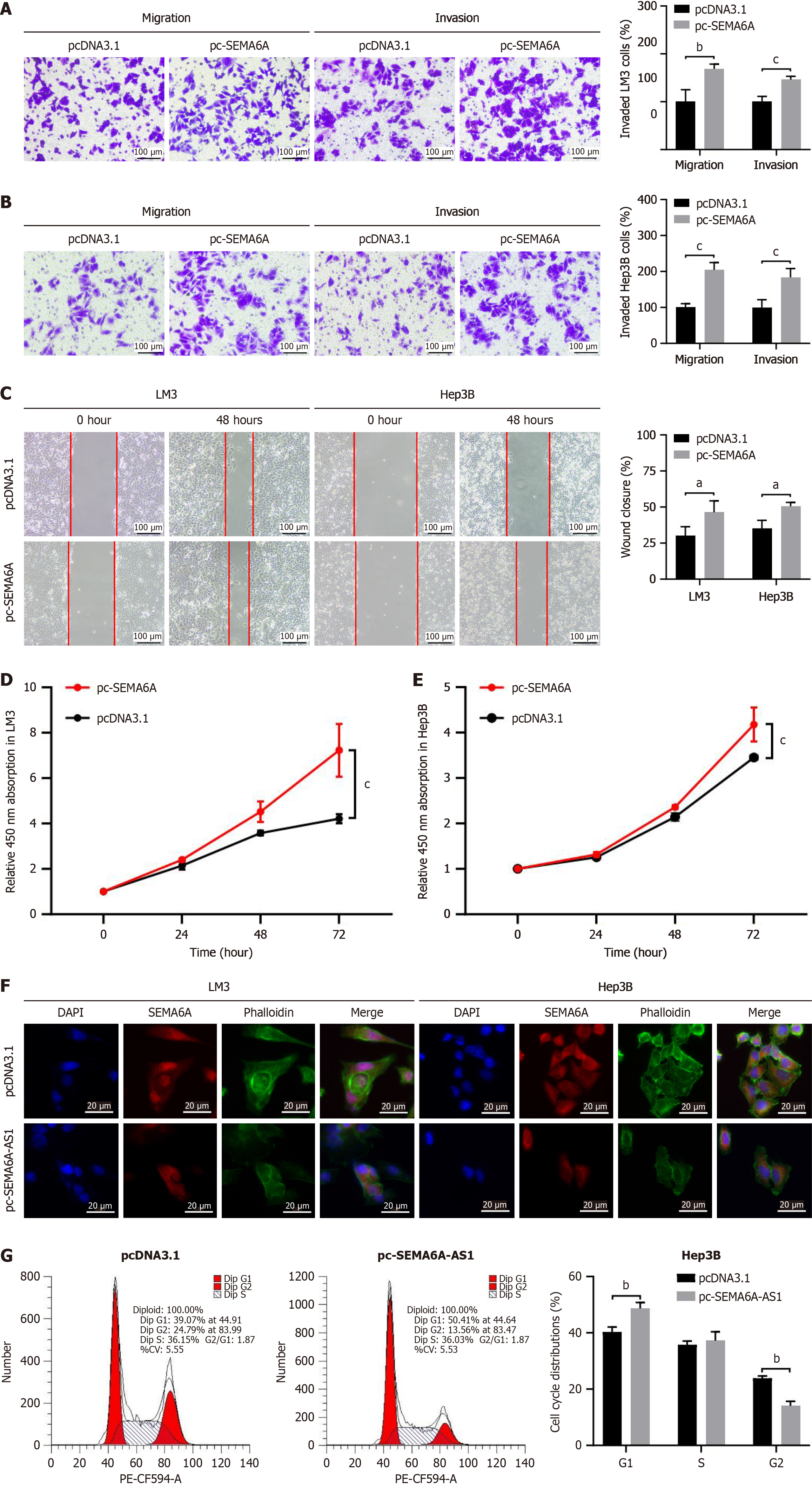
To determine the role of SEMA6A in HCC, the proliferation, migration, and invasion capabilities were estimated in a gain-of-function study. SEMA6A stably overexpressed in LM3 and Hep3B cell lines were constructed and then verified by qPCR and Western blot (Supplementary Figure 1E). Transwell and wound-healing assays revealed that SEMA6A overexpression enhanced the migration and invasion of LM3 and Hep3B cells (Figure 5A-C). Overexpression of SEMA6A promoted the proliferation of LM3 and Hep3B cells as assessed by the Cell Counting Kit-8 assay (Figure 5D and E). As reported previously, the knockdown of SEMA6A led to loss of actin stress fibers and actin cytoskeleton remodeling. Immunofluorescence results confirmed that overexpression of SEMA6A-AS1 induced a decrease in SEMA6A protein, and induced changes in the actin cytoskeleton as shown by loss of actin stress fibers (Figure 5F). Subsequently, a cell cycle assay was performed by flow cytometry and revealed that upregulation of SEMA6A-AS1 led to an increase in the proportion of cells in G1 phase and a decrease in the proportion of cells in G2 phase (Figure 5G and Supplementary Figure 2C). Taken together, our results support the notion that SEMA6A promotes cell proliferation, migration, and invasion through regulation of actin stress fibers and actin cytoskeleton remodeling.
The rapid growth and early metastasis of HCC make timely and effective treatment difficult and lead to poor prognosis[1,3]. A deeper exploration and understanding of the molecules and mechanisms involved in the regulation of growth, invasion, and metastasis of HCC will provide us with new perspectives and strategies for developing effective HCC treatments[19]. Our previous study demonstrated that SEMA6A-AS1 was downregulated in HBV-related HCC and associated with a poor prognosis and malignant phenotypes including capsular invasion, high Edmondson-Steiner grade and TNM stage[13]. Our present study revealed that SEMA6A-AS1 exhibits a negative correlation with SEMA6A expression and inhibited the proliferation, migration and invasion of HCC cells. In addition, SEMA6A was upregulated and positively correlated with shorter survival of HBV-related HCC patients. Mechanistically, SEMA6A-AS1 regulated SEMA6A expression by directly combining with SEMA6A mRNA and promoting its degradation, consequently leaded to loss of actin stress fibers and actin cytoskeleton remodeling.
Antisense lncRNAs are an important subset of lncRNAs that are expressed from the opposite strand of protein-coding and non-coding genes. Recent developments in the study of antisense lncRNAs suggest that whether antisense lncRNAs regulate expression, splicing, stability or translation of their sense transcript is influenced by the pattern and degrees of overlap between the sense-antisense pair[20]. For example, LncRNA glypican 3-AS1 (GPC3-AS1), which corresponds to the gene body region of GPC3, can promote HCC progression by recruiting P300/CBP-associated factor to the GPC3 gene body region, consequently inducing an increase in euchromatic histone marks and activating GPC3 transcription[21]. LncRNA GA-binding protein subunit beta-1-AS1 (GABPB1-AS1), which shares the same fragments in the first exon of GABPB1, can suppress GABPB1 translation by blocking GABPB1 mRNA recruitment to polysomes and binding with eukaryotic translation initiation factor 4A, leading to the downregulation of peroxiredoxin-5 peroxidase and promotion of ferroptosis in HCC cells[22]. LncRNA alkB homolog 3-AS1[23] and family with sequence similarity 83 member H-AS1[18,24] respectively enhance alkB homolog 3 and family with sequence similarity 83 member H mRNA stability to promote HCC cell proliferation and invasion. SEMA6A-AS1 is the antisense transcript of SEMA6A, and the two genes share the same exon region which is located on the 1-295 nt of SEMA6A-AS1 and the 1914-2208 nt of SEMA6A mRNA. In the pre
SEMA6A, a trans-membrane protein which was originally described as a ligand mediating axon guidance involved in central nervous system development[25,26], has recently been found to participate in tumor development by a growing number of studies. In clear cell renal cell carcinoma, SEMA6A is overexpressed and physically interacted with SEC62 and promoted clear cell renal cell carcinoma cell proliferation through SEC62-dependent β-catenin stabilization and activation[27]. In BRAF-mut melanoma cells, SEMA6A inhibits cell death and promotes cell chemotaxis and invasion via cytoskeletal remodeling by the RhoA-dependent activation of Yes-associated protein[15,28]. However, the role of SEMA6A in HCC has not been described previously. Our research found that SEMA6A was overexpressed in HCC and associated with poor prognosis, and played a pro-carcinogenic role by promoting HCC cell proliferation, invasion and migration. Current research suggests that stress fiber loss and cytoskeletal remodeling affect cell adhesion and anchoring-independent growth, thereby inhibiting cell migration and invasive capacity[28-30]. Additionally, cytoskeletal structures drive chromosomal separation and cell division in the cell cycle[30]. Thus, disruption of actin filaments can cause G1 arrest which leads to a slowdown in proliferation[31,32]. We found that SEMA6A-AS1 overexpression inhibits the level of SEMA6A and causes the loss of actin stress fibers and cytoskeletal remodeling in HCC cells. The proportion of cells in G1 phase also increased in SEMA6A-AS1 overexpressed HCC cells. Therefore, we speculate that regulation of the actin cytoskeleton by SEMA6A may be the molecular mechanism by which SEMA6A-AS1 inhibits the proliferation, invasion and migration abilities of HCC cells.
This study comprehensively investigated the clinical relevance of SEMA6A-AS1 and SEMA6A in the features and prognosis of HCC, and their functional implications in HCC cell proliferation, migration and invasion. We have revealed that SEMA6A-AS1 can reverse the progression of HCC by decreasing SEMA6A expression through mRNA stabilization and actin cytoskeleton regulation. These findings expand the understanding of the molecular mechanisms of HCC and provide potential novel prognostic biomarkers and therapeutic targets for HCC.
| 1. | Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, Jemal A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5690] [Cited by in RCA: 8198] [Article Influence: 8198.0] [Reference Citation Analysis (2)] |
| 2. | Rumgay H, Ferlay J, de Martel C, Georges D, Ibrahim AS, Zheng R, Wei W, Lemmens VEPP, Soerjomataram I. Global, regional and national burden of primary liver cancer by subtype. Eur J Cancer. 2022;161:108-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 258] [Article Influence: 86.0] [Reference Citation Analysis (0)] |
| 3. | Vogel A, Meyer T, Sapisochin G, Salem R, Saborowski A. Hepatocellular carcinoma. Lancet. 2022;400:1345-1362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1284] [Cited by in RCA: 1233] [Article Influence: 411.0] [Reference Citation Analysis (41)] |
| 4. | Huang Z, Zhou JK, Peng Y, He W, Huang C. The role of long noncoding RNAs in hepatocellular carcinoma. Mol Cancer. 2020;19:77. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 216] [Cited by in RCA: 361] [Article Influence: 72.2] [Reference Citation Analysis (0)] |
| 5. | Shi L, Peng F, Tao Y, Fan X, Li N. Roles of long noncoding RNAs in hepatocellular carcinoma. Virus Res. 2016;223:131-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 49] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 6. | Wang Y, Sun L, Wang L, Liu Z, Li Q, Yao B, Wang C, Chen T, Tu K, Liu Q. Long non-coding RNA DSCR8 acts as a molecular sponge for miR-485-5p to activate Wnt/β-catenin signal pathway in hepatocellular carcinoma. Cell Death Dis. 2018;9:851. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 112] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 7. | Li Y, Guo D, Zhao Y, Ren M, Lu G, Wang Y, Zhang J, Mi C, He S, Lu X. Long non-coding RNA SNHG5 promotes human hepatocellular carcinoma progression by regulating miR-26a-5p/GSK3β signal pathway. Cell Death Dis. 2018;9:888. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 109] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 8. | Jiang R, Tang J, Chen Y, Deng L, Ji J, Xie Y, Wang K, Jia W, Chu WM, Sun B. The long noncoding RNA lnc-EGFR stimulates T-regulatory cells differentiation thus promoting hepatocellular carcinoma immune evasion. Nat Commun. 2017;8:15129. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 176] [Cited by in RCA: 274] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 9. | Chen S, Xia X. Long noncoding RNA NEAT1 suppresses sorafenib sensitivity of hepatocellular carcinoma cells via regulating miR-335-c-Met. J Cell Physiol. 2019;234:14999-15009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 67] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 10. | Yu S, Li N, Huang Z, Chen R, Yi P, Kang R, Tang D, Hu X, Fan X. A novel lncRNA, TCONS_00006195, represses hepatocellular carcinoma progression by inhibiting enzymatic activity of ENO1. Cell Death Dis. 2018;9:1184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 46] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 11. | Chen J, Huang ZB, Liao CJ, Hu XW, Li SL, Qi M, Fan XG, Huang Y. LncRNA TP73-AS1/miR-539/MMP-8 axis modulates M2 macrophage polarization in hepatocellular carcinoma via TGF-β1 signaling. Cell Signal. 2020;75:109738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 12. | Li S, Hu X, Yu S, Yi P, Chen R, Huang Z, Huang Y, Huang Y, Zhou R, Fan X. Hepatic stellate cell-released CXCL1 aggravates HCC malignant behaviors through the MIR4435-2HG/miR-506-3p/TGFB1 axis. Cancer Sci. 2023;114:504-520. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 20] [Reference Citation Analysis (0)] |
| 13. | Yu S, Li N, Wang J, Fu Y, Huang Y, Yi P, Chen R, Tang D, Hu X, Fan X. Correlation of Long Noncoding RNA SEMA6A-AS1 Expression with Clinical Outcome in HBV-Related Hepatocellular Carcinoma. Clin Ther. 2020;42:439-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 14. | Rozbesky D, Verhagen MG, Karia D, Nagy GN, Alvarez L, Robinson RA, Harlos K, Padilla-Parra S, Pasterkamp RJ, Jones EY. Structural basis of semaphorin-plexin cis interaction. EMBO J. 2020;39:e102926. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 15. | Loria R, Laquintana V, Scalera S, Fraioli R, Caprara V, Falcone I, Bazzichetto C, Di Martile M, Rosanò L, Del Bufalo D, Bossi G, Sperduti I, Terrenato I, Visca P, Soddu S, Milella M, Ciliberto G, Falcioni R, Ferraresi V, Bon G. SEMA6A/RhoA/YAP axis mediates tumor-stroma interactions and prevents response to dual BRAF/MEK inhibition in BRAF-mutant melanoma. J Exp Clin Cancer Res. 2022;41:148. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 16. | Yuan S, Liu Q, Hu Z, Zhou Z, Wang G, Li C, Xie W, Meng G, Xiang Y, Wu N, Wu L, Yu Z, Bai L, Li Y. Long non-coding RNA MUC5B-AS1 promotes metastasis through mutually regulating MUC5B expression in lung adenocarcinoma. Cell Death Dis. 2018;9:450. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 17. | Zhao Y, Liu Y, Lin L, Huang Q, He W, Zhang S, Dong S, Wen Z, Rao J, Liao W, Shi M. The lncRNA MACC1-AS1 promotes gastric cancer cell metabolic plasticity via AMPK/Lin28 mediated mRNA stability of MACC1. Mol Cancer. 2018;17:69. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 135] [Cited by in RCA: 196] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 18. | Zhou M, Pan S, Qin T, Zhao C, Yin T, Gao Y, Liu Y, Zhang Z, Shi Y, Bai Y, Gong J, Guo X, Wang M, Qin R. LncRNA FAM83H-AS1 promotes the malignant progression of pancreatic ductal adenocarcinoma by stabilizing FAM83H mRNA to protect β-catenin from degradation. J Exp Clin Cancer Res. 2022;41:288. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 19. | Winkle M, El-Daly SM, Fabbri M, Calin GA. Noncoding RNA therapeutics - challenges and potential solutions. Nat Rev Drug Discov. 2021;20:629-651. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 884] [Cited by in RCA: 1026] [Article Influence: 256.5] [Reference Citation Analysis (0)] |
| 20. | Werner A, Kanhere A, Wahlestedt C, Mattick JS. Natural antisense transcripts as versatile regulators of gene expression. Nat Rev Genet. 2024;25:730-744. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 16] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 21. | Zhu XT, Yuan JH, Zhu TT, Li YY, Cheng XY. Long noncoding RNA glypican 3 (GPC3) antisense transcript 1 promotes hepatocellular carcinoma progression via epigenetically activating GPC3. FEBS J. 2016;283:3739-3754. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 119] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 22. | Qi W, Li Z, Xia L, Dai J, Zhang Q, Wu C, Xu S. LncRNA GABPB1-AS1 and GABPB1 regulate oxidative stress during erastin-induced ferroptosis in HepG2 hepatocellular carcinoma cells. Sci Rep. 2019;9:16185. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 148] [Cited by in RCA: 177] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 23. | Lu Q, Wang H, Lei X, Ma Q, Zhao J, Sun W, Guo C, Huang D, Xu Q. LncRNA ALKBH3-AS1 enhances ALKBH3 mRNA stability to promote hepatocellular carcinoma cell proliferation and invasion. J Cell Mol Med. 2022;26:5292-5302. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 24. | Ma YK, Shen TH, Yang XY. Upregulation of LncRNA FAM83H-AS1 in hepatocellular carcinoma promotes cell proliferation, migration and invasion by Wnt/β-catenin pathway. Eur Rev Med Pharmacol Sci. 2019;23:7855-7862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 25. | Mauti O, Domanitskaya E, Andermatt I, Sadhu R, Stoeckli ET. Semaphorin6A acts as a gate keeper between the central and the peripheral nervous system. Neural Dev. 2007;2:28. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 86] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 26. | Rünker AE, Little GE, Suto F, Fujisawa H, Mitchell KJ. Semaphorin-6A controls guidance of corticospinal tract axons at multiple choice points. Neural Dev. 2008;3:34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 81] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 27. | Ji J, Xu Y, Xie M, He X, Ren D, Qiu T, Liu W, Chen Z, Shi W, Zhang Z, Wang X, Wang W, Ma J, Qian Q, Jing A, Ma X, Qin J, Ding Y, Geng T, Yang J, Sun Z, Liu W, Liu S, Liu B. VHL-HIF-2α axis-induced SEMA6A upregulation stabilized β-catenin to drive clear cell renal cell carcinoma progression. Cell Death Dis. 2023;14:83. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 28. | Loria R, Bon G, Perotti V, Gallo E, Bersani I, Baldassari P, Porru M, Leonetti C, Di Carlo S, Visca P, Brizzi MF, Anichini A, Mortarini R, Falcioni R. Sema6A and Mical1 control cell growth and survival of BRAFV600E human melanoma cells. Oncotarget. 2015;6:2779-2793. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 59] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 29. | Liu B, Shen H, He J, Jin B, Tian Y, Li W, Hou L, Zhao W, Nan J, Zhao J, Shen J, Yu H, Wang Y, Shan G, Shi L, Cai X. Cytoskeleton remodeling mediated by circRNA-YBX1 phase separation suppresses the metastasis of liver cancer. Proc Natl Acad Sci U S A. 2023;120:e2220296120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 40] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 30. | Hall A. The cytoskeleton and cancer. Cancer Metastasis Rev. 2009;28:5-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 331] [Cited by in RCA: 387] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 31. | Heng YW, Koh CG. Actin cytoskeleton dynamics and the cell division cycle. Int J Biochem Cell Biol. 2010;42:1622-1633. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 207] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 32. | Chae HD, Kim SY, Park SE, Kim J, Shin DY. p53 and DNA-dependent protein kinase catalytic subunit independently function in regulating actin damage-induced tetraploid G1 arrest. Exp Mol Med. 2012;44:236-240. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |