Published online Mar 14, 2025. doi: 10.3748/wjg.v31.i10.103133
Revised: January 4, 2025
Accepted: February 5, 2025
Published online: March 14, 2025
Processing time: 107 Days and 22.3 Hours
Proteins play a central role in regulating biological functions, and various path
To elucidate the effects of SEL1L-mediated ERAD on Huh7 and explore the underlying mechanisms in vivo and in vitro.
Huh7 cells were treated with ERAD inhibitor to identify ERAD’s role. Cell counting kit-8, 5-ethynyl-2’-deoxy
The ERAD inhibitor suppressed cell proliferation and migration and promoted apoptosis. SEL1L-HRD1 signi
ERAD inhibition suppressed the proliferation and migration of Huh7 and promoted its apoptosis. EXT2 plays an important role and ERAD might be a potential treatment for Huh7 hepatocellular carcinoma.
Core Tip: The endoplasmic reticulum-associated degradation (ERAD) inhibitor suppressed cell proliferation and migration and promoted apoptosis. SEL1L-HRD1 significantly influenced Huh7 cell growth. SEL1L knockout suppressed tumor cell proliferation and migration, and enhanced apoptosis. Mass spectrometry revealed EXT2 is a primary substrate of ERAD. SEL1L knockout significantly increased the protein expression of EXT2, whereas EXT2 knockdown partially restored the effect of SEL1L knockout. In conclusion, this study showed the function and possible mechanisms of ERAD in hepatocellular carcinoma, providing new insights into more effective treatment and strategies for liver cancer.
- Citation: Chen JN, Wang L, He YX, Sun XW, Cheng LJ, Li YN, Yoshida S, Shen ZY. SEL1L-mediated endoplasmic reticulum associated degradation inhibition suppresses proliferation and migration in Huh7 hepatocellular carcinoma cells. World J Gastroenterol 2025; 31(10): 103133
- URL: https://www.wjgnet.com/1007-9327/full/v31/i10/103133.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i10.103133
There are approximately 400000 newly diagnosed cases of hepatocellular carcinoma (HCC) in China in 2022, making it the fourth most frequent tumor[1,2]. Due to late-stage diagnosis, many patients are unable to undergo surgical treatment, resulting in a mortality rate only slightly lower than that of lung cancer[2]. HCC can be caused by various risk factors, including hepatitis B or C virus infection and metabolic conditions, which can lead to chronic hepatic damage and progress to HCC[3,4]. Given the high incidence and mortality rates of HCC, exploring its molecular mechanisms and identifying new therapeutic targets is crucial.
Proteins are essential for life functions, with endoplasmic reticulum (ER) being crucial in regulating the protein production and processing. During the translation process, 10%-30% of synthesized proteins will be ubiquitinated and degraded[5]. Dealing with the increase of misfolded proteins is crucial for cell survival. The newly produced proteins enter the ER in an unfolded condition, where chaperone proteins and enzymes assist in their modification and folding into their natural conformation before transport to cellular organelles or secretion outside of the cell[6]. Factors, such as cancer, an altered immune response, metabolic regulation, and degenerative diseases, can cause an increase in the content of wrong proteins, leading to ER stress[7,8]. To combat ER stress, the unfolded protein response (UPR), ER-associated degradation (ERAD), and autophagy are employed[9,10]. While the UPR and autophagy have been widely studied regarding their roles in regulating proliferation, apoptosis, and metastasis in HCC, there is few research on the role of ERAD in liver cancer. Moreover, the inositol-requiring enzyme (IRE)-1α deficiency in the UPR pathway in podocytes has almost no effect on renal function in mice, only causing mild proteinuria symptoms after 1 year of age[11,12]. Yoshida et al[13] confirmed that the ERAD pathway could maintain ER homeostasis in podocytes, and inhibiting the ERAD pathway in mice can lead to severe congenital nephrotic syndrome, suggesting that it was possibly independent of the UPR pathway to regulate protein synthesis.
The SEL1L-HRD1 complex, a highly conserved form of ERAD in mammals, is crucial for maintaining ER homeostasis[14]. In mammals, SEL1L can maintain HRD1[15]. The systemic knockout of SEL1L leads to embryonic death in mice[16]. Furthermore, SEL1L is indispensable for pancreatic epithelial cell development[17]. Research has indicated that deleting SEL1L in adipocytes results in lipase retention in the ER, which contributes to delaying diet-induced fat[18]. However, the role of SEL1L in HCC remains unknown.
EXT2 and EXT1 form a glycosyltransferase that participates in heparan sulfate (HS) chain polymerization[19,20]. By alternately adding glucuronic acid and N-acetylglucosamine residues to the HS chain, it elongates to form HS proteoglycans (HSPGs)[19,20]. HSPGs, which are prevalent in the extracellular matrix, are crucial for tissue homeostasis and signaling[21]. A mutation in EXT2 is associated with hereditary multiple exostoses, a dominant skeletal disorder affecting endochondral bone growth. It characteristic is the development of multiple benign bone tumors covered by cartilage, frequently leading to bone deformities[22-24]. Although there have been studies proving that the EXT gene family are tumor suppressor genes[25], their role in HCC remains unclear.
This research is intended to clarify the effect of SEL1L on the HCC cell line Huh7. We identified EXT2 as a substrate of ERAD and proved that SEL1L-HRD1 ERAD could be a novel target for treating HCC. Nevertheless, the underlying mechanism requires further investigation.
The Huh7 was brought from Fenghui Biotechnology (Hunan Province, China). Cells were cultivated at 37 °C with 5% carbon dioxide in a humidified incubator using Gibco’s Dulbecco’s modified eagle medium supplemented with 10% fetal bovine serum and 1% penicillin streptomycin. They were passaged at a 1:3 split ratio upon reaching 85%-90% confluence.
A single guide RNA for SEL1L was chemically synthesized by Tsingke Biotech (Beijing, China). Briefly, after annealing, the polymerase chain reaction (PCR) product was connected to a linear carrier and added to a competent cell system. The E. coli was equally coated onto lysogeny broth agar plates and several single clones were chosen for sequencing. Pre-experimental sequencing confirmed that all oligonucleotides had the correct sequence. Monoclonal strains with the correct sequences were used for plasmid extraction using the TIANpure midi plasmid kit (Beijing, China). Transfection experiments were conducted using Lipofectamine 2000 reagent (Thermo Fisher, Shanghai, China). Transfected cells were selected with 2.5 μg/mL puromycin.
Following cell adhesion, 2000 cells were transfected into 96 well plates and evaluated by the cell counting kit-8 (CCK-8) assay (Dojindo, Japan). The wells were filled with CCK-8 working solutions (medium without fetal bovine serum and CCK-8 reagent at a 9:1 ratio), and they were left in the dark for two hours. The cell absorbance (optical density = 450 nm) was subsequently evaluated by a microplate reader (Agilent BioTek Cytation 5, Santa Clara, CA, United States).
The total RNA was obtained from Huh7 cell lines and xenograft tissues with AG RNAex Pro Reagent (AG21101, Hunan, China). Following RNA concentration assessment with the Nanodrop-2000, cDNA was produced from 1000 ng of RNA with the Evo M-MLV RT premix kit (AG11706, Accurate Biotechnology, Hunan Province, China). The final cDNA was subjected to quantitative real-time (qRT)-PCR on a LightCycler® 96 system (Roche, Basel, Switzerland). Glyceraldehyde-3-phosphate dehydrogenase served as the normalization reference for the mRNA level, and each mRNA level was assessed with the 2-△△CT method. Experiments were conducted in triplicate, and primer sequences were shown in Supplementary Table 1.
Briefly, cells were lysed in cold lysis buffer [4-(2-hydroxyethyl) piperazine-1-ethane-sulfon: 40 mmol/L, potential of hydrogen = 7.5, sodium chloride: 120 mmol/L, ethylene diamine tetraacetic acid: 1 mmol/L, pyrophosphate: 10 mmol/L, glycerophosphate: 10 mmol/L, sodium orthovanadate: 1.5 mmol/L, 3-3-(cholamidopropyl) dimethylamino propyl sulfonate: 0.3%, and a mixture of protease inhibitors] for 10 minutes. The lysates underwent a 15-minute, 13000 × g centrifugation at 4 °C. After boiling for five minutes, the supernatant was combined with 5 × sodium dodecyl sulfate buffer (Genstar, Beijing, China). Samples were detached by 8% tris-glycine gel electrophoresis and transferred onto a polyvinylidene fluoride membrane (Millipore, Billerica, MA, United States). For one hour at room temperature, 5% skim milk was used to block membranes. Membranes were incubated with specific antibodies for an entire night at 4 °C after being cleaned six times with Tris-buffered saline (TBST) for ten minutes each. The following day, they were treated with secondary antibodies for 1 hour at room temperature. After rinsing six times with TBST, protein bands were observed with the Minichemitm Chemiluminescent Imaging System (JUNYI, Beijing, China). The antibodies involved anti-heavy-chain binding protein (BiP) (66574-1-AP, Proteintech, Wuhan, Hubei Province, China), anti-SEL1L (A12073, Abclonal, Wuhan, Hubei Province, China), anti-IRE-1α (3294S, CST, Shanghai, China), anti-EXT2 (sc-514092, Santa Cruz, Shanghai, China), and anti-α-tubulin (AC012, Abclonal, Wuhan, Hubei Province, China).
After transfection, 6-well plates were seeded with 2 × 105 cells per well, and 10 μM 5-Ethynyl-2’-deoxyuridine (EdU) (C10310-3, RIBOBIO, Guangzhou, Guangdong Province, China) was added to the medium for one day. Cells were collected, fastened with 4% paraformaldehyde, and dealt with 2 mg/mL glycine and 0.5% Triton X-100, followed by incubation for 10 minutes in the dark with 1 × Apollo staining solution. Following three washes with 0.5% TritonX-100 and resuspension in 500 μL phosphate-buffered saline, fluorescence was assessed with a BD AccuriTM C6 Flow Cytometer (BD Biosciences, Palo Alto, CA, United States).
Apoptosis of Huh7 cells was detected using an annexin V-fluorescein isothiocyanate (FITC)/7-aminoactinomycin D (7-AAD) apoptosis kit (E-CK-A212, Elabscience, Wuhan, Hubei Province, China). The total of early (annexin V + 7-AAD-) and late apoptotic (annexin V + 7-AAD +) cells was used to characterize apoptotic cells.
Huh7 cells were incubated into 6-well plates in the 2 mL of culture medium (37 °C, 5% carbon dioxide), saturated humidity for half a month, fastened using 4% paraformaldehyde, and dyed using 0.1% crystal violet.
The upper chambers of 24-well transwell inserts (JetBio, Guangzhou, Guangdong Province, China) were seeded with 2 × 104 cells in the serum-free medium, while lower chambers contained the complete medium involving 10% fetal bovine serum. The top chambers were fastened with 4% paraformaldehyde for 25 minutes and dyed using 0.1% crystal violet for 20 minutes after a 24-hour incubation at 37 °C. Three random fields were chosen to count the migrated cells.
Proteins from scramble or SEL1L knockout cells were assessed with an Orbitrap Eclipse Tribrid mass spectrometer (Thermo Fisher, Waltham, MA, United States). The mass spectrometry data, with peaks marked by the data analysis software, were analyzed using Proteome Discoverer (Thermo Fisher, Waltham, MA, United States).
Huh7 cells, either scramble or SEL1L knockout, were exposed to 100 mg/mL cycloheximide (CHX) and harvested at 0, 3, and 6 hours. Protein expression levels were then assessed using western blotting.
Our research obtained ethical approval from the Ethics committee at Nankai University (approval No. 2025-SYDWLL-000002). Male BALB/c nude mice that were six weeks old were bought from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). They were divided into two groups at random and given subcutaneous injections of 1 × 106 scramble or SEL1L knockout cells into their posterior necks. Starting at 1-week post-implantation, the tumor xenograft dimensions were recorded bi-daily using vernier calipers. Volume = 0.5 × length × width2. Tumor xenografts were harvested from mice and weighed using a high-precision electronic balance.
GraphPad Prism 8 (San Diego, CA, United States) was used in the study. All data were shown as mean ± SEM. P values were calculated with unpaired Student’s t-tests for two groups or one-way analysis of variance for more than two groups.
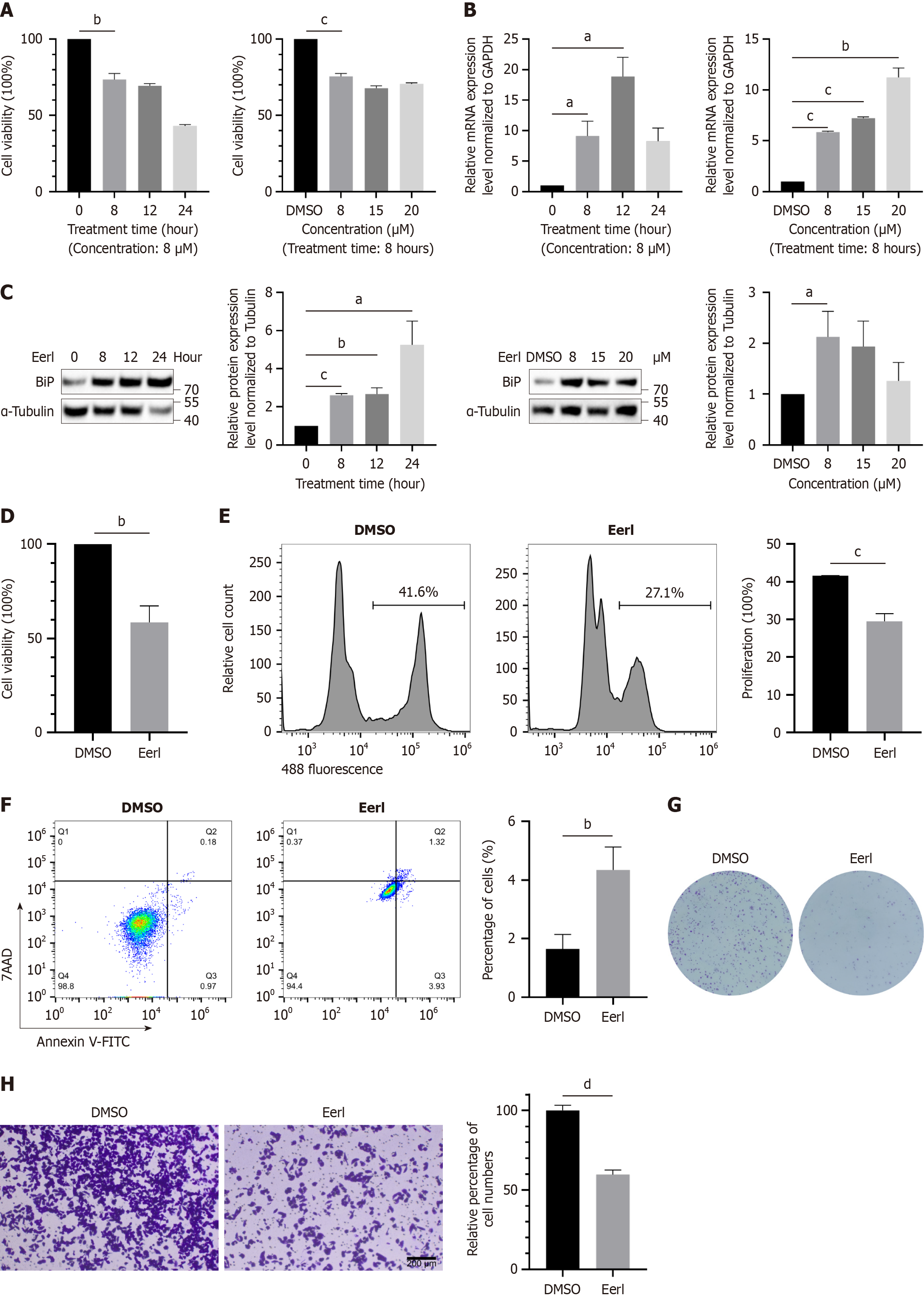
As a small molecule inhibitor of ERAD, Eeyarestatin I can not only suppress the deubiquitination process, but also exert its effect by inhibiting protein translocation[26,27]. As shown in Figure 1, the CCK-8 test showed that cell viability was affected by the concentration and duration of Eeyarestatin I treatment (Figure 1A). As expected, at both the RNA and protein levels, the marker of ER stress, BiP, showed a significant increase in expression after inhibitor treatment (Figure 1B and C). Subsequently, an inhibitor concentration of 8 μM and a treatment time of 8 hours were selected for further experiments. CCK-8 (Figure 1D) and EdU assays (Figure 1E) showed that the Eeyarestatin I treatment signi
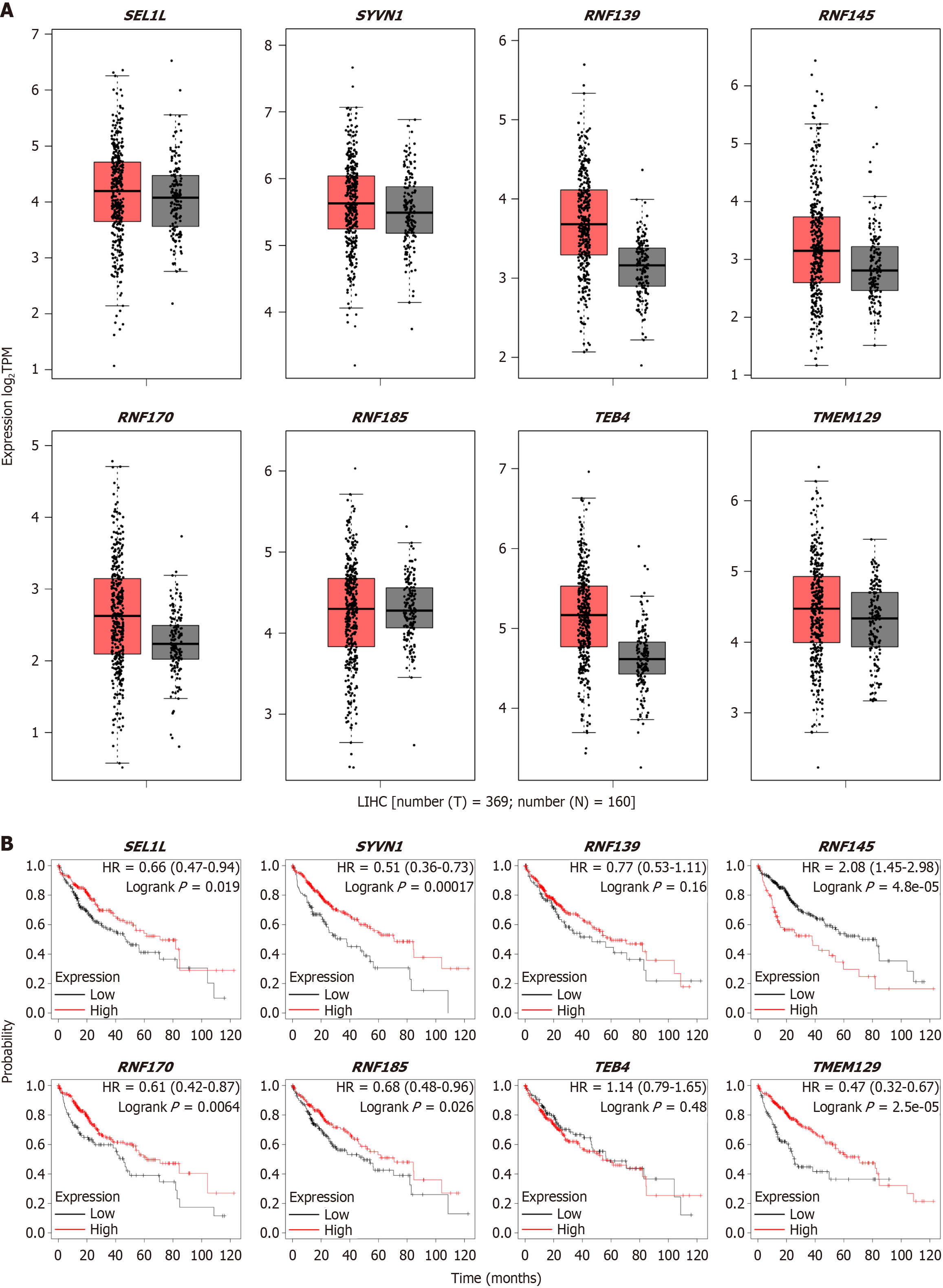
The gene expression profiling interactive analysis 2 online database was applied to obtain the level of various ERAD complexes in HCC cell lines and employed the Kaplan-Meier plot to assess their impact on survival. The SEL1L-HRD1 complex exhibited the highest expression level among all ERAD complexes, and there was no strong difference in its level between tumor and adjacent tissues (Figure 2A). The survival analysis indicated that high expression of ERAD complexes, including SEL1L, HRD1, RNF170, RNF185, and TMEM129, correlated with longer survival times, whereas elevated RNF145 expression was associated with a shorter survival time (Figure 2B). Furthermore, RNF139 and TEB4 showed no significant difference. SEL1L likely has a greater influence on HCC prognosis than other ERAD complexes.
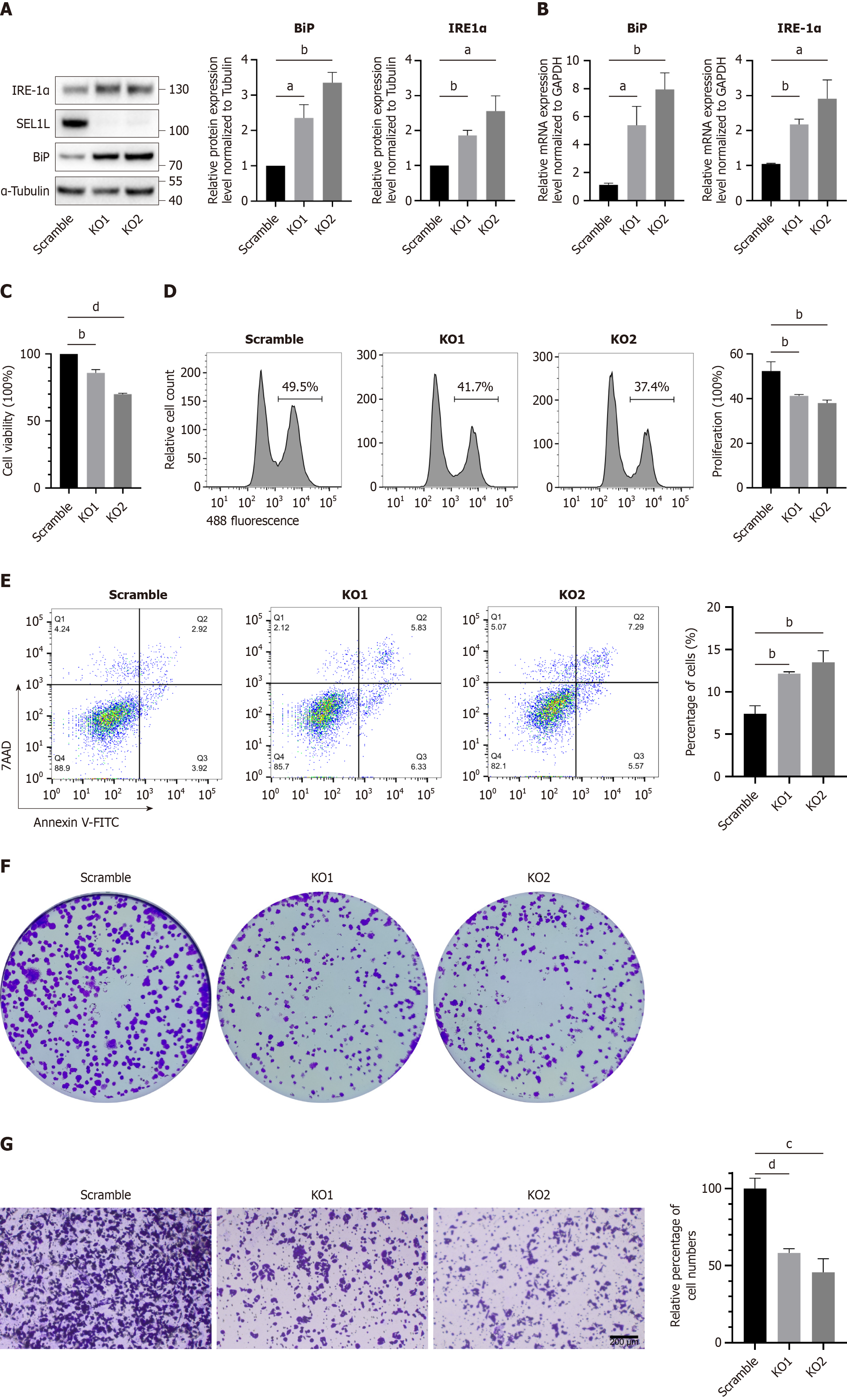
The SEL1L-HRD1 complex is crucial for degrading intracellular misfolded proteins, and we constructed a stable SEL1L knockout Huh7 cell line to verify its effect on Huh7 cells at the genetic level. The knockout efficiency was verified by western blotting. SEL1L knockout significantly increased BiP levels, similar to ERAD inhibitor treatment (Figure 3A and B). Furthermore, the CCK8 (Figure 3C) and EdU experiments (Figure 3D) verified that cell viability and proliferation ability decreased after SEL1L knockout and apoptosis levels increased (Figure 3E), similar to ERAD inhibitor treatment. The results of the colony formation (Figure 3F) and transwell experiments (Figure 3G) were also similar to those obtained with Eeyarestatin I treatment, with a decrease in the colony formation rate and migration abilities of Huh7 cells. However, compared with inhibitor treatment, the effect of SEL1L knockout was less pronounced.
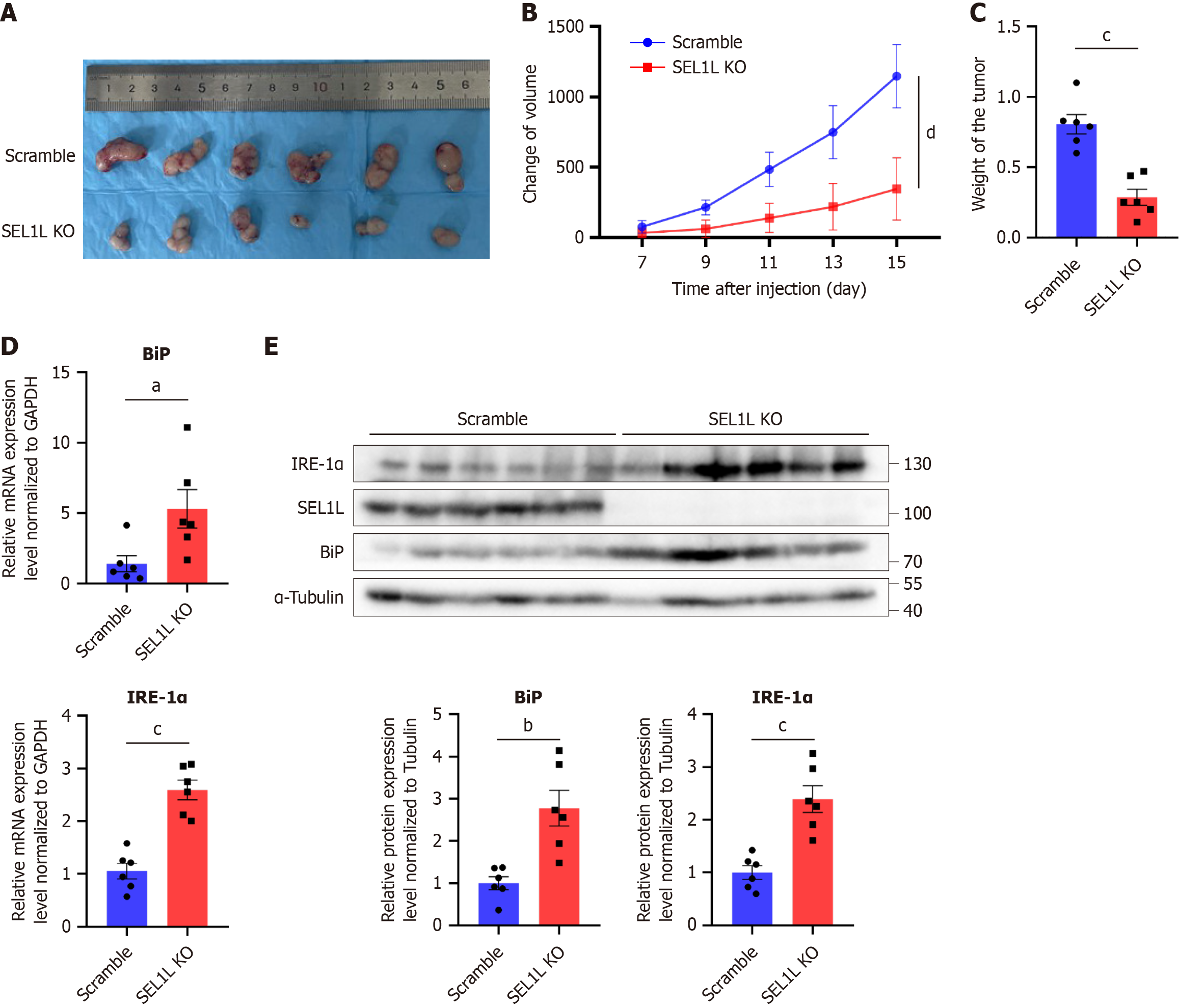
We used SEL1L knockout cells to establish the tumor model and found that SEL1L knockout significantly reduced the volume and weight of subcutaneous tumors compared to the control group (Figure 4A-C). The qPCR and western blotting results were also consistent with in vitro experiments, showing that knockout of SEL1L increases the expression of the ER stress marker, BiP and one downstream of UPR pathway, IRE-1α (Figure 4D and E).
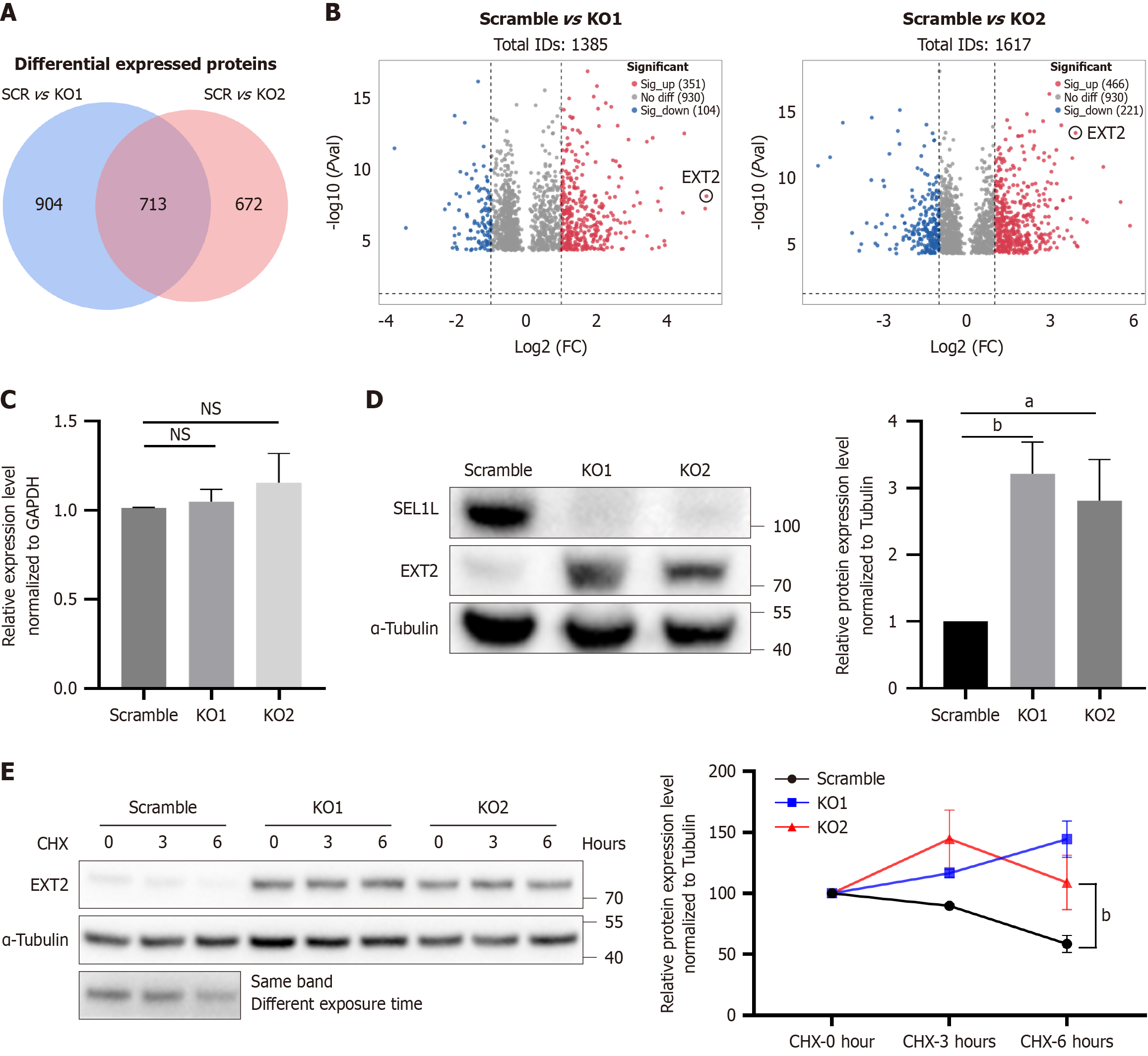
To elucidate specific downstream proteins affected by SEL1L knockout, we performed mass spectrometry analysis. There were more than 700 differentially expressed proteins in the two groups, among which EXT2 had the most significant increase in expression following SEL1L knockout (Figure 5A and B). To rule out the possibility that increased expression was due to changes at the mRNA level, we measured EXT2 mRNA expression, finding no substantial alterations (Figure 5C). Western blotting analysis revealed an approximately three-fold increase in EXT2 protein expression following SEL1L knockout (Figure 5D). When protein synthesis was suppressed using CHX, it was observed that the degradation rate of EXT2 protein was significantly reduced after SEL1L knockout (Figure 5E). Thus, it can be concluded that EXT2 is degraded through the ERAD pathway and is one of the substrates of ERAD.
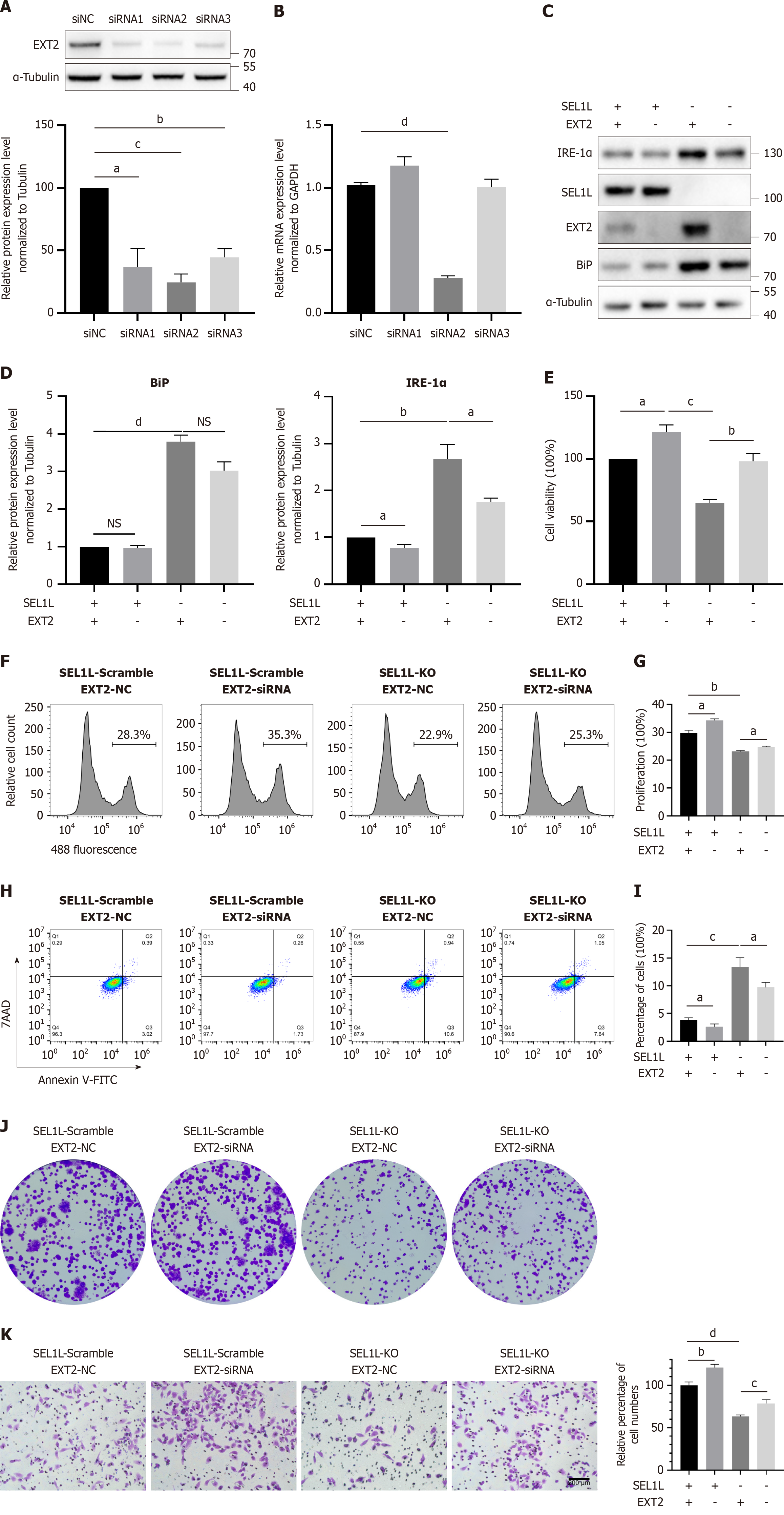
To demonstrate that the effect of SEL1L knockout is caused by the increased expression of EXT2, we performed rescue experiments using siRNA knockdown of EXT2. First, we verified the knockdown efficiency of the siRNA and ultimately selected the most efficient siRNA (si2) for subsequent experiments (Figure 6A). PCR detection confirmed that the siRNA reduced the expression of EXT2 at the transcriptional level (Figure 6B). Moreover, the increased contents of BiP and IRE-1α in SEL1L knockout cells after the knockdown of EXT2 decreased (Figure 6C and D). CCK8 (Figure 6E) and EdU detection (Figure 6F and G) showed a significant recovery in the cell viability and proliferation ability between SEL1L knockout cells and cells with knockdown of EXT2. Similarly, there were significant differences in apoptosis (Figure 6H and I), colony formation (Figure 6J), and transwell results (Figure 6K). Therefore, we believe that EXT2 is the most differentially expressed protein after SEL1L knockout, as one of its substrates, and its knockdown can partly reverse the effects of SEL1L knockout.
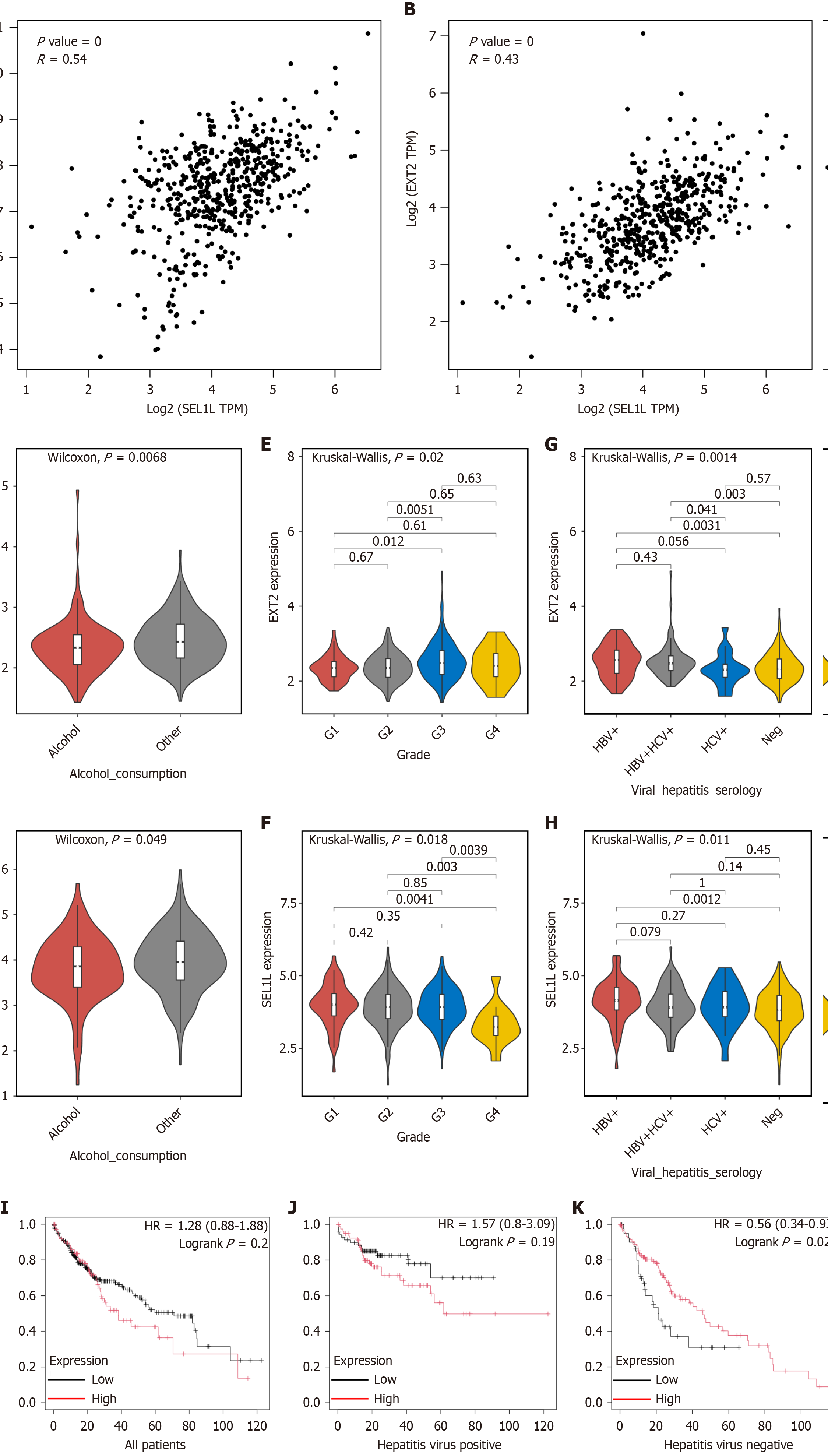
The Cancer Genome Atlas database revealed a correlation between EXT2 and SEL1L expressions, similar to that between SEL1L and BiP expressions (Figure 7A and B). Differences in SEL1L and EXT2 gene expression were compared among different subgroups based on clinical factors such as alcohol use, neoplasm histologic grade, and viral hepatitis serology. We found that the expression of EXT2 and SEL1L genes is significantly reduced in patients with alcohol consumption (Figure 7C and D). The expression level of EXT2 is significantly higher in G3 patients than in G1 and G2 patients (Figure 7E). Conversely, the expression level of SEL1L is notably lower in G4 patients than in G1, G2, and G3 patients (Figure 7F). The expression levels of EXT2 and SEL1L were significantly increased in hepatitis B virus positive patients (Figure 7G and H). Survival analysis revealed that the expression level of EXT2 was not significantly correlated with prognosis in all patients, while high SEL1L expression related with a better prognosis (Figure 2B and Figure 7I). However, among hepatitis virus-negative patients, those with high EXT2 expression exhibited longer survival times (Figure 7J and K). Moreover, this interesting phenomenon warrants further exploration.
HCC is a highly malignant cancer characterized by abnormalities in multiple genes and signaling pathways. Our study demonstrated that SEL1L knockout inhibited HCC cell activity while inducing apoptosis. This may be caused by the increased content of misfolded proteins in tumor cells, potentially originating from mutations, abnormal expression, or post-translational modifications. ERAD activity decreased after SEL1L knockout, and these misfolded proteins could not be degraded normally in the ER, resulting in biological effects such as ER stress and apoptosis.
We identified EXT2 as one of the substrates of ERAD in the Huh7 cell line. EXT2 has been identified as a substrate of ERAD in both human HEK293T cells and mouse brown adipose tissue[28], showing the feature of EXT2 as a substrate for ERAD is conserved in different cells and tissues. As anticipated, knocking down EXT2, which exhibited the highest expression increase following SEL1L knockout, only partially reversed the growth inhibition induced by SEL1L knockout. One possible reason for this phenomenon is that there are many ERAD substrates upregulated after SEL1L knockout, and knocking down EXT2 alone cannot eliminate the accumulation of misfolded proteins caused by decreased ERAD activity. The second point is that it may be due to the additional EXT2 being folded incorrectly and not functioning properly. Further investigations are needed to clarify the underlying mechanism.
The analysis of human liver tissue specimens indicates that SEL1L levels are slightly raised in cancer tissues than in adjacent tissues. Data from The Cancer Genome Atlas indicated that patients with high SEL1L expression exhibited a better prognosis. We speculate that this may be related to the enhanced protein synthesis and metabolic ability in the tumor tissue. While EXT2 expression levels and overall prognosis showed no significant correlation across all patients, higher EXT2 expression was associated with better prognosis specifically in patients without hepatitis virus infection. This may provide more treatment strategies for patients with liver cancer caused by non-hepatitis virus infection.
The ER can intricately regulate the protein quality control system. Liu et al[29] found that knocking out SEL1L reduced the survival rate of HepG2 cells, which they suggested is related to impaired mitochondrial function. However, Bhattacharya et al[30] suggested that the SEL1L-HRD1 complex inhibits liver cell proliferation and tumorigenesis in mice. The tumor suppressor factor WNT5A, as a substrate of ERAD, exists in the ER and forms high molecular weight aggregates following SEL1L knockout, thereby losing its inhibitory effect on liver cell proliferation[30]. Previous studies have indicated that SEL1L is essential for HRD1 stability[18]. Furthermore, research indicates that HRD1 promotes HCC cell proliferation[31]. HRD1 facilitates angiogenesis and augments the HCC cell activity[31]. We speculate that the regulatory effect of ERAD on HCC is similar to a dynamic equilibrium state, which can boost HCC activity under mild stress; however, the opposite effect occurs after exceeding a certain degree. As one of the pathways for protein synthesis and degradation, crosstalk exists between UPR and ERAD. UPR regulates the expression of numerous ERAD genes. As a branch of the UPR, ATF6 can regulate the expression of ERAD genes, including SEL1L[32]. Meanwhile, IRE-1α and ATF6 were substrates of SEL1L-HRD1 ERAD complex both in vitro and in vivo[33,34]. Furthermore, SEL1L deficiency in oligodendrocytes reduces myelin sheath thickness in the adult central nervous system by activating protein kinase RNA-like endoplasmic reticulum kinase (PERK), which subsequently inhibits myelin protein translation[35]. These indicate that UPR and ERAD have close relation. In cancer, the PERK branch in the UPR can induce cancer activity by regulating redox homeostasis, thereby promoting tumor cell survival in adverse environments[36]. Inhibiting IRE-1α in the body can slow down the progression of diet-induced, obesity-induced liver cancer[37]. Another study showed that inhibiting the endonuclease activity of IRE-1α with exogenous drugs reduced liver cancer activity in the animal model[38]. Aran et al[39] showed that a cluster of differentiation 5 L binds to BiP to activate UPR and autophagy, promoting liver cancer cell proliferation and inhibiting cancer cell apoptosis. Our results also found an increase in IRE-1α expression after knocking out SEL1L, indicating that UPR will have a certain degree of compensatory enhancement after ERAD activity is inhibited. Therefore, in this case, a major focus of future research will be developing technical methods to eliminate the impact of UPR changes on ERAD.
However, our research has some limitations. First, we used only the Huh7 cell line, and no EXT2 rescue experiments were conducted in vivo. Additionally, the knock-out of SEL1L to inhibit ERAD in primary liver cells should be explored to investigate whether the effects of ERAD are cancer-cell-specific. Considering that the commonly used LO2 cell line is considered Hela cells, we did not validate it in our study. Future studies could consider using organoid models to investigate the role of SEL1L experiments in the liver. Further investigations are required to clarify the exact impact of SEL1L in HCC.
In summary, our research demonstrated that both pharmacological and genetic inhibition of ERAD would inhibit proliferation and migration, induce apoptosis, and finally inhibit cancer cell growth in Huh7 cells. EXT2 was identified as a substrate of ERAD and plays a role in SEL1L knockout cells.
Our study revealed that ERAD inhibition suppressed the proliferation and migration of Huh7 and promoted its apoptosis. EXT2 plays an important role and ERAD might be a potential treatment of Huh7 HCC. Both the pharmacological and genetic inhibition of ERAD suppressed the proliferation and migration of Huh7 and promoted its apoptosis. As one of the downstream substrates of ERAD, EXT2 plays an important role in the growth regulation of Huh7. Therefore, ERAD might be a potential treatment for HCC, providing new insights into more effective strategies for liver cancer.
We want to express our sincere gratitude to PhD Kong DJ and PhD Zhang WQ at Nankai University for their invaluable assistance. Without their generous support, this research would not have been possible.
| 1. | Zheng R, Zhang S, Zeng H, Wang S, Sun K, Chen R, Li L, Wei W, He J. Cancer incidence and mortality in China, 2016. J Natl Cancer Cent. 2022;2:1-9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 852] [Cited by in RCA: 944] [Article Influence: 314.7] [Reference Citation Analysis (1)] |
| 2. | Xia C, Dong X, Li H, Cao M, Sun D, He S, Yang F, Yan X, Zhang S, Li N, Chen W. Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chin Med J (Engl). 2022;135:584-590. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1790] [Cited by in RCA: 2189] [Article Influence: 729.7] [Reference Citation Analysis (1)] |
| 3. | El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118-1127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2881] [Cited by in RCA: 3088] [Article Influence: 220.6] [Reference Citation Analysis (0)] |
| 4. | Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S, Lencioni R, Koike K, Zucman-Rossi J, Finn RS. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7:6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4432] [Cited by in RCA: 3847] [Article Influence: 961.8] [Reference Citation Analysis (3)] |
| 5. | Braakman I, Hebert DN. Protein folding in the endoplasmic reticulum. Cold Spring Harb Perspect Biol. 2013;5:a013201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 302] [Cited by in RCA: 389] [Article Influence: 32.4] [Reference Citation Analysis (0)] |
| 6. | Guerriero CJ, Brodsky JL. The delicate balance between secreted protein folding and endoplasmic reticulum-associated degradation in human physiology. Physiol Rev. 2012;92:537-576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 277] [Cited by in RCA: 336] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 7. | Kim H, Bhattacharya A, Qi L. Endoplasmic reticulum quality control in cancer: Friend or foe. Semin Cancer Biol. 2015;33:25-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 63] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 8. | Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519-529. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4586] [Cited by in RCA: 5055] [Article Influence: 280.8] [Reference Citation Analysis (0)] |
| 9. | Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13:89-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2364] [Cited by in RCA: 3047] [Article Influence: 234.4] [Reference Citation Analysis (0)] |
| 10. | Sha H, He Y, Yang L, Qi L. Stressed out about obesity: IRE1α-XBP1 in metabolic disorders. Trends Endocrinol Metab. 2011;22:374-381. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 75] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 11. | Kaufman DR, Papillon J, Larose L, Iwawaki T, Cybulsky AV. Deletion of inositol-requiring enzyme-1α in podocytes disrupts glomerular capillary integrity and autophagy. Mol Biol Cell. 2017;28:1636-1651. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 12. | Hassan H, Tian X, Inoue K, Chai N, Liu C, Soda K, Moeckel G, Tufro A, Lee AH, Somlo S, Fedeles S, Ishibe S. Essential Role of X-Box Binding Protein-1 during Endoplasmic Reticulum Stress in Podocytes. J Am Soc Nephrol. 2016;27:1055-1065. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 40] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 13. | Yoshida S, Wei X, Zhang G, O'Connor CL, Torres M, Zhou Z, Lin L, Menon R, Xu X, Zheng W, Xiong Y, Otto E, Tang CA, Hua R, Verma R, Mori H, Zhang Y, Hu CA, Liu M, Garg P, Hodgin JB, Sun S, Bitzer M, Qi L. Endoplasmic reticulum-associated degradation is required for nephrin maturation and kidney glomerular filtration function. J Clin Invest. 2021;131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 44] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 14. | Christianson JC, Olzmann JA, Shaler TA, Sowa ME, Bennett EJ, Richter CM, Tyler RE, Greenblatt EJ, Harper JW, Kopito RR. Defining human ERAD networks through an integrative mapping strategy. Nat Cell Biol. 2011;14:93-105. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 359] [Cited by in RCA: 416] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 15. | Sun S, Shi G, Han X, Francisco AB, Ji Y, Mendonça N, Liu X, Locasale JW, Simpson KW, Duhamel GE, Kersten S, Yates JR 3rd, Long Q, Qi L. Sel1L is indispensable for mammalian endoplasmic reticulum-associated degradation, endoplasmic reticulum homeostasis, and survival. Proc Natl Acad Sci U S A. 2014;111:E582-E591. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 164] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 16. | Francisco AB, Singh R, Li S, Vani AK, Yang L, Munroe RJ, Diaferia G, Cardano M, Biunno I, Qi L, Schimenti JC, Long Q. Deficiency of suppressor enhancer Lin12 1 like (SEL1L) in mice leads to systemic endoplasmic reticulum stress and embryonic lethality. J Biol Chem. 2010;285:13694-13703. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 83] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 17. | Li S, Francisco AB, Munroe RJ, Schimenti JC, Long Q. SEL1L deficiency impairs growth and differentiation of pancreatic epithelial cells. BMC Dev Biol. 2010;10:19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 18. | Sha H, Sun S, Francisco AB, Ehrhardt N, Xue Z, Liu L, Lawrence P, Mattijssen F, Guber RD, Panhwar MS, Brenna JT, Shi H, Xue B, Kersten S, Bensadoun A, Péterfy M, Long Q, Qi L. The ER-associated degradation adaptor protein Sel1L regulates LPL secretion and lipid metabolism. Cell Metab. 2014;20:458-470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 100] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 19. | Leisico F, Omeiri J, Le Narvor C, Beaudouin J, Hons M, Fenel D, Schoehn G, Couté Y, Bonnaffé D, Sadir R, Lortat-Jacob H, Wild R. Structure of the human heparan sulfate polymerase complex EXT1-EXT2. Nat Commun. 2022;13:7110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 20] [Reference Citation Analysis (0)] |
| 20. | Li H, Chapla D, Amos RA, Ramiah A, Moremen KW, Li H. Structural basis for heparan sulfate co-polymerase action by the EXT1-2 complex. Nat Chem Biol. 2023;19:565-574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 21. | Heinritz W, Hüffmeier U, Strenge S, Miterski B, Zweier C, Leinung S, Bohring A, Mitulla B, Peters U, Froster UG. New mutations of EXT1 and EXT2 genes in German patients with Multiple Osteochondromas. Ann Hum Genet. 2009;73:283-291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 22. | Philippe C, Porter DE, Emerton ME, Wells DE, Simpson AH, Monaco AP. Mutation screening of the EXT1 and EXT2 genes in patients with hereditary multiple exostoses. Am J Hum Genet. 1997;61:520-528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 85] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 23. | Francannet C, Cohen-Tanugi A, Le Merrer M, Munnich A, Bonaventure J, Legeai-Mallet L. Genotype-phenotype correlation in hereditary multiple exostoses. J Med Genet. 2001;38:430-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 133] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 24. | Xu L, Xia J, Jiang H, Zhou J, Li H, Wang D, Pan Q, Long Z, Fan C, Deng HX. Mutation analysis of hereditary multiple exostoses in the Chinese. Hum Genet. 1999;105:45-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 37] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 25. | Bernard MA, Hall CE, Hogue DA, Cole WG, Scott A, Snuggs MB, Clines GA, Lüdecke HJ, Lovett M, Van Winkle WB, Hecht JT. Diminished levels of the putative tumor suppressor proteins EXT1 and EXT2 in exostosis chondrocytes. Cell Motil Cytoskeleton. 2001;48:149-162. [PubMed] [DOI] [Full Text] |
| 26. | McKibbin C, Mares A, Piacenti M, Williams H, Roboti P, Puumalainen M, Callan AC, Lesiak-Mieczkowska K, Linder S, Harant H, High S, Flitsch SL, Whitehead RC, Swanton E. Inhibition of protein translocation at the endoplasmic reticulum promotes activation of the unfolded protein response. Biochem J. 2012;442:639-648. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 34] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 27. | Cross BC, McKibbin C, Callan AC, Roboti P, Piacenti M, Rabu C, Wilson CM, Whitehead R, Flitsch SL, Pool MR, High S, Swanton E. Eeyarestatin I inhibits Sec61-mediated protein translocation at the endoplasmic reticulum. J Cell Sci. 2009;122:4393-4400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 89] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 28. | Wei X, Lu Y, Lin LL, Zhang C, Chen X, Wang S, Wu SA, Li ZJ, Quan Y, Sun S, Qi L. Proteomic screens of SEL1L-HRD1 ER-associated degradation substrates reveal its role in glycosylphosphatidylinositol-anchored protein biogenesis. Nat Commun. 2024;15:659. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 11] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 29. | Liu Q, Yang X, Long G, Hu Y, Gu Z, Boisclair YR, Long Q. ERAD deficiency promotes mitochondrial dysfunction and transcriptional rewiring in human hepatic cells. J Biol Chem. 2020;295:16743-16753. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 30. | Bhattacharya A, Wei J, Song W, Gao B, Tian C, Wu SA, Wang J, Chen L, Fang D, Qi L. SEL1L-HRD1 ER-associated degradation suppresses hepatocyte hyperproliferation and liver cancer. iScience. 2022;25:105183. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 31. | Ji F, Zhou M, Sun Z, Jiang Z, Zhu H, Xie Z, Ouyang X, Zhang L, Li L. Integrative proteomics reveals the role of E3 ubiquitin ligase SYVN1 in hepatocellular carcinoma metastasis. Cancer Commun (Lond). 2021;41:1007-1023. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 32. | Adachi Y, Yamamoto K, Okada T, Yoshida H, Harada A, Mori K. ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct Funct. 2008;33:75-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 299] [Cited by in RCA: 362] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 33. | Sun S, Shi G, Sha H, Ji Y, Han X, Shu X, Ma H, Inoue T, Gao B, Kim H, Bu P, Guber RD, Shen X, Lee AH, Iwawaki T, Paton AW, Paton JC, Fang D, Tsai B, Yates JR 3rd, Wu H, Kersten S, Long Q, Duhamel GE, Simpson KW, Qi L. IRE1α is an endogenous substrate of endoplasmic-reticulum-associated degradation. Nat Cell Biol. 2015;17:1546-1555. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 197] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 34. | Horimoto S, Ninagawa S, Okada T, Koba H, Sugimoto T, Kamiya Y, Kato K, Takeda S, Mori K. The unfolded protein response transducer ATF6 represents a novel transmembrane-type endoplasmic reticulum-associated degradation substrate requiring both mannose trimming and SEL1L protein. J Biol Chem. 2013;288:31517-31527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 70] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 35. | Wu S, Stone S, Nave KA, Lin W. The Integrated UPR and ERAD in Oligodendrocytes Maintain Myelin Thickness in Adults by Regulating Myelin Protein Translation. J Neurosci. 2020;40:8214-8232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 36. | Bobrovnikova-Marjon E, Grigoriadou C, Pytel D, Zhang F, Ye J, Koumenis C, Cavener D, Diehl JA. PERK promotes cancer cell proliferation and tumor growth by limiting oxidative DNA damage. Oncogene. 2010;29:3881-3895. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 246] [Cited by in RCA: 237] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 37. | Wu J, Qiao S, Xiang Y, Cui M, Yao X, Lin R, Zhang X. Endoplasmic reticulum stress: Multiple regulatory roles in hepatocellular carcinoma. Biomed Pharmacother. 2021;142:112005. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 35] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 38. | Pavlović N, Calitz C, Thanapirom K, Mazza G, Rombouts K, Gerwins P, Heindryckx F. Inhibiting IRE1α-endonuclease activity decreases tumor burden in a mouse model for hepatocellular carcinoma. Elife. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 39. | Aran G, Sanjurjo L, Bárcena C, Simon-Coma M, Téllez É, Vázquez-Vitali M, Garrido M, Guerra L, Díaz E, Ojanguren I, Elortza F, Planas R, Sala M, Armengol C, Sarrias MR. CD5L is upregulated in hepatocellular carcinoma and promotes liver cancer cell proliferation and antiapoptotic responses by binding to HSPA5 (GRP78). FASEB J. 2018;32:3878-3891. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |