Published online Mar 14, 2025. doi: 10.3748/wjg.v31.i10.101014
Revised: December 19, 2024
Accepted: February 5, 2025
Published online: March 14, 2025
Processing time: 176 Days and 1.3 Hours
Although an association between gut microbiota and cholestatic liver disease (CLD) has been reported, the precise functional roles of these microbes in CLD pathogenesis remain largely unknown.
To explore the function of gut microbes in CLD pathogenesis and the effects of gut microbiota on intestinal barrier and bile acid (BA) metabolism in CLD.
Male C57BL/6J mice were fed a 0.05% 3,5-diethoxycarbonyl-1,4-dihydrocollidine diet for 2 weeks to induce CLD. The sterile liver tissues of mice were then meticulously harvested, and bacteria in homogenates were identified through culture methods. Furthermore, 16S ribosomal DNA sequencing was employed to analyze sterile liver samples collected from eight patients with primary biliary cholangitis (PBC) and three control individuals with hepatic cysts. The functional roles of the identified bacteria in CLD pathogenesis were assessed through microbiota transfer experiments, involving the evaluation of changes in intestinal per
Ligilactobacillus murinus (L. murinus) and Lactococcus garvieae (L. garvieae) were isolated from the bacterial culture of livers from CLD mice. L. murinus was prevalently detected in PBC patients and controls, whereas L. garvieae was detected only in patients with PBC but not in controls. Mice inoculated with L. garvieae exhibited increased susceptibility to experimental CLD, with both in vitro and in vivo indicating that L. garvieae disrupted the intestinal barrier function by down-regulating the expression of occludin and zonula occludens-1. Moreover, L. garvieae administration significantly upregulated the expression of the apical sodium-dependent BA transporter in the terminal ileum and increased serum BA levels.
L. garvieae contributes to excessive BA-induced hepatobiliary injury and liver fibrosis by increasing intestinal permeability and enhancing BA reabsorption.
Core Tip: Mice gavaged with Lactococcus garvieae (L. garvieae) exhibited marked aggravation of cholestasis, portal edema, portal infiltrates, liver fibrosis during cholestatic liver disease (CLD) compared with control mice. Mechanically, L. garvieae treatment decreased the expression of intestinal tight junction proteins and enhanced bile acid reabsorption in CLD mice.
- Citation: Liu M, Ji YL, Hu YJ, Su YX, Yang J, Wang XY, Chu HY, Zhang X, Dong SJ, Yang H, Liu YH, Zhou SM, Guo LP, Ran Y, Li YN, Zhao JW, Zhang ZG, Piao MY, Zhou L. Lactococcus garvieae aggravates cholestatic liver disease by increasing intestinal permeability and enhancing bile acid reabsorption. World J Gastroenterol 2025; 31(10): 101014
- URL: https://www.wjgnet.com/1007-9327/full/v31/i10/101014.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i10.101014
Cholestatic liver disease (CLD) is characterized by hepatic accumulation of bile acids (BAs)[1]. Excess cytotoxic BAs in the liver can cause liver fibrosis, cirrhosis and even liver carcinogenesis[2]. In particular, primary biliary cholangitis (PBC), formerly known as primary biliary cirrhosis, is a common immune-mediated CLD, occurring in approximately 0.1% of women aged > 40 years[3,4]. The precise pathogenesis of PBC still remains unclear, but genetic predisposition and various environmental factors may contribute to the development and perpetuation of bile duct injury[5-7]. Recently, the gut microbiota has been recognized as a pivotal environmental risk factor for PBC, with the associated mechanisms including molecular mimicry, autoantigen modification, intestinal barrier disruption and intestinal bacterial translocation[4,8-11].
Exploration of the liver and ductal bile microbiomes represents a novel direction in PBC microbiota research. Conventionally, the liver and bile were considered sterile, but more recent reports have revealed that a unique microbiome in individuals with and without hepatobiliary disorders[11,12]. Specifically, bile samples from PBC patients have revealed the presence of specific bacteria, mainly including Enterococcus faecium, Staphylococcus aureus, and Streptococcus pneumoniae[13]. It is very likely that the imbalances in intestinal microbiota and loss of intestinal barrier integrity during cholestasis may promote microbial translocation to the liver[14-16]. However, the study of bile communities is challenging because of the invasive procedure of bile sampling, usually requiring endoscopic retrograde cholangiopancreatography. Addi
The gut barrier prevents the translocation of enteric bacteria and microbial antigens into the portal circulation[17]. The integrity of the intestinal epithelial layer is stabilized by tight junctions comprising occludin, zonula occludens-1 (ZO-1), cingulin, and junctional adhesion molecule-A[18]. Following the disruption of the intestinal barrier, gut bacteria and their products, including lipopolysaccharides or microbial RNAs, may enter the liver via the portal circulation, thus inducing liver inflammation and fibrosis[19]. Furthermore, bacteria that are beneficial under normal physiological conditions can also cause inflammation and elicit organ injury following intestinal barrier disruption[11,20]. Disrupted intestinal epithelial barriers and increased permeability have been reported in several liver disorders, including PBC and autoimmune hepatitis[21-23]. Moreover, a leaky gut and increased intestinal permeability contributed to the initiation and progression of hepatobiliary disease[22,24,25]. However, the precise mechanisms underlying leaky intestine remains incompletely understood.
BAs are signaling molecules primarily synthesized from cholesterol in the liver and secreted into the intestine via the biliary system[26]. Approximately 95% of BAs are reabsorbed in the ileum and transported back through the bloodstream back to the liver and recycled. Intrahepatic BA accumulation can be reduced by decreasing BA synthesis in the liver or decreasing BA reuptake from the ileum[27]. BA synthesis in the liver is regulated by two main metabolic pathways. Notably, cytochrome P450 family 7 subfamily A member 1 (CYP7A1) regulates the classic pathway and limits the de novo BA synthesis, whereas cytochrome P450 family 27 subfamily A member 1 (CYP27A1) is an important enzyme in the alternative pathway[28,29]. BA reuptake from the intestine involves the ileal bile acid transporter or apical sodium-dependent bile acid transporter (ASBT). Therefore, BA reabsorption can be blocked using their inhibitors and several agents exhibiting this property have been identified[26,30]. Gut microbiome plays a critical role in BA metabolism, whereas BAs simultaneously alter the gut microbiome, both representing the important elements of the gut-liver axis[31]. Therefore, microbial modulation may represent a novel therapeutic approach to cholestasis.
The present study aimed to investigate the functional link between hepatobiliary bacteria and CLD. Our findings revealed a specific enrichment of the gut bacterium Lactococcus garvieae (L. garvieae) in the livers of patients with PBC and CLD mice. Importantly, inoculation with L. garvieae aggravated experimental CLD by exacerbating intestinal barrier disruption and promoting BA reabsorption. Furthermore, L. garvieae induced barrier injury in Caco-2 monolayers, and incited inflammatory responses in RAW264.7 cells and intrahepatic bile duct epithelial cells in vitro. Our study provides valuable insights into the environmental risk factors in CLD pathogenesis.
Eleven participants, including eight patients with PBC and three controls with hepatic cysts were recruited from the Gastroenterology Department at Tianjin Medical University General Hospital. PBC diagnosed based on the presence of at least two of the following three criteria: (1) Cholestatic liver function tests; (2) Compatible or diagnostic liver histology; and (3) Positivity for AMA or AMA-M2. The inclusion criteria for controls were as follows: (1) Cyst diameter < 4 cm with clear cystic fluid; (2) Normal liver function test results; (3) Normal histopathology of the liver tissue next to the cyst; and (4) Absence of the hepatitis B/C virus antigen. Sterile liver biopsy specimens were obtained from all the participants. This study was approved by the Ethics Committee of the General Hospital of Tianjin Medical University (No. IRB2021-KY-318).
Eight-week-old male C57BL/6J mice were purchased from the Beijing Animal Study Center and housed in specific pathogen-free facilities at the Animal Center of the Tianjin Medical University, with a controlled 12-hour dark/light cycle at 24 ± 2 ºC and 50% ± 5% relative humidity. All animals had free access to sterile water and diet. And all the mice were acclimatized for 1 week and then randomized into four groups (n = 5 per group). The mice were fed a 0.05% 3,5-diethoxycarbonyl-1,4-dihydrocollidine (DDC) diet for 2 weeks to induce CLD and were gavaged L. garvieae or phosphate-buffered saline (PBS) once every other day. This study complied with the 1986 Animals (Scientific Procedures) Act of the United Kingdom, ensuring the ethical treatment of laboratory animals. The experimental protocol for animals was reviewed and approved by the Animal Ethical and Welfare Committee of Tianjin Medical University (approval No. IRB2021-DWFL-001).
CLD mice were sterilely dissected, and their liver tissues were harvested and cultured on Columbia blood agar plates. Single colonies were harvested and grown in the MRS medium. DNA was extracted from bacterial pellets and bacterial 16S ribosomal DNA (16S rDNA) was amplified using specific primers 27F_ (16S-F) and 1492R_ (16S-R). Subsequently, sequences were identified using Basic Local Alignment Search Tool. The isolates stored in 50% glycerol at -80 °C.
All mice were administered an antibiotic cocktail (250 mg vancomycin, 250 mg metronidazole, 500 mg neomycin and 500 mg ampicillin) every other day for 1 week to deplete the native gut microbiota before L. garvieae and Ligilactobacillus murinus (L. murinus) colonization. The bacteria were grown overnight in MRS medium, harvested via centrifugation (10 minutes, 4000 rpm, 4 °C), and resuspended in 200 μL sterile PBS. Mice in the experimental group were orally gavaged with L. garvieae (BSN307) and L. murinus (NBRC 14221), respectively, at a density of 2 × 109 CFU/mL every other day for 3 weeks, whereas mice in the PBS and DDC + PBS groups were orally administered 0.2 mL of sterile PBS. Successful colonization was confirmed based on mouse fecal samples. Briefly, total fecal bacterial DNA was extracted from fecal samples using a Stool Genomic DNA Extraction Kit (Solarbio, China) and subjected to 1.5% agarose gel electrophoresis. After the first week of inoculation, the DDC diet was administered for 2 weeks. On day 28, all the animals were eutha
Liver samples were collected from patients with PBC and controls using a sterile technique. DNA was extracted from the liver samples was extracted using the TGuide S96 Magnetic Bead Method Genomic DNA Extraction Kit. And 16S rDNA sequencing was performed on a NovaSeq 6000 platform in paired-end 250 bases (PE250) mode, which generated 50000-100000 reads per sample. After the identification and removal of chimeric sequences, the original subreads were corrected to generate circular consensus sequencing using SMRT Link (version 8.0). The sequences were further clustered into operational taxonomic units (97% sequence similarity) using the USEARCH (version 10.0). Subsequently, a table was generated for each taxonomic level.
HIBEpiC-immortalized (ZQY009) cells were cultured in epithelial cell medium containing 2.0% fetal bovine serum (FBS), 1.0% epithelial cell growth supplement (EpiCGS) and 1.0% antibiotic solution (P/S) (SclenCell, United States). Human Caco-2 cells (BNCC 338148) were cultured in Minimum Essential Medium (MEM) (Gibco, United States) supplemented with 20% FBS and 1.0% nonessential amino acids. The macrophage cell line RAW264.7 (ATCC SC-6003) was cultured in Dulbecco's Modified Eagle Medium-high glucose (Gibco, United States) supplemented with 10% FBS and 1.0% nonessential amino acids. All cells were grown in a humidified incubator with 95% air and 5% CO2 at 37 °C. Cells were seeded into 12-well plates at a density of 1 × 105 cells /well and treated with L. garvieae supernatant at different concentrations (1:100, 1:50, and 1:10) for 12 hours for subsequent analysis.
Alanine aminotransferase (ALT), serum alkaline phosphatase (ALP) and gamma-glutamyl transferase (GGT) activities were measured by standard procedures at the Institute of Clinical Chemistry of the Tianjin Medical University General Hospital.
Total RNA was extracted from liver and ileum using SteadyPure Quick RNA Extraction Kit (Accurate Biology, China) and 1 μg of RNA was reverse-transcribed into cDNA using HiScript III RT SuperMix for qPCR (Vazyme, China) according to the manufacturer’s directions. The obtained cDNA was analyzed by real-time-qPCR with the SYBR Green qPCR Master Mix (GLPBIO, United States) and specific primers (Tsingke Biotechnology Co, Ltd, China). The primers sequences for target genes were listed in Table 1. The relative mRNA levels were determined using the 2-ΔΔCt method, normalized to glyceraldehyde 3-phosphate dehydrogenase.
| Gene | Forward sequence | Reverse sequence |
| Mouse ZO-1 | GGGCCATCTCAACTCCTGTA | AGAAGGGCTGACGGGTAAAT |
| Mouse occludin | ACTATGCGGAAAGAGTTGACAG | GTCATCCACACTCAAGGTCAG |
| Mouse TNFα | GAAGTTCCCAAATGGCCTCC | GTGAGGGTCTGGGCCATAGA |
| Mouse IL-1β | CTTTGAAGTTGACGGACCC | TGAGTGATACTGCCTGCCTG |
| Mouse IL-6 | CTAGGTTTGCCGAGTAGATCTC | GACAAAGCCAGAGTCCTTCAGAGA |
| Mouse α-SMA | AGCCATCTTTCATTGGGATGG | CCCCTGACAGGACGTTGTTA |
| Mouse COL1A1 | CCCTGGTCCCTCTGGAAATG | GGACCTTTGCCCCCTTCTTT |
| Mouse COL1A2 | TGCGTACCTGGATGAGGAGA | GAGCAGCCATCGACTAGGAC |
| Mouse COL3A1 | CCCACAGCCTTCTACACCTG | CCAGGGTCACCATTTCTCCC |
| Mouse COL4A2 | CGGCGTAATCTCAAAAGGCG | GGCCTCTGCTTCCTTTCTGT |
| Mouse TGFβ | GCTGAACCAAGGAGACGGAAT | GCTGATCCCGTTGATTTCCA |
| Mouse CCL2 | AGCTGTAGTTTTTGTCACCAAGC | GTGCTGAAGACCTTAGGGCA |
| Mouse CCL5 | TGCTGCTTTGCCTACCTCTC | TCCTTCGAGTGACAAACACGA |
| Mouse CCL20 | CGACTGTTGCCTCTCGTACA | GCTTCATCGGCCATCTGTCT |
| Mouse P16 | GCTCAACTACGGTGCAGATTC | GCACGATGTCTTGATGTCCC |
| Mouse ASBT | GCGACATGGACCTCAGTGTTA | GTTCCCGAGTCAACCCACAT |
| Mouse OATP | CCTGGAGCAGCAATATGGAAA | CCAAGGCATACTGGAGGCAA |
| Mouse NTCP | TACCTCCTCCCTGATGCCTTTC | TGCGTCTGCAGCTTGGATTTA |
| Mouse BSEP | CAATGTTCAGTTCCTCCGTTCA | TTTGGTGTTGTCCCCGTGCTTG |
| Mouse MDR1 | CCCCCGAGATTGACAGCTAC | ACTCCACTAAATTGCACATTTCCTTC |
| Mouse CYP7A1 | TACTAGATAGCATCATCAAGGAGGCTC | CCATCCTCAAGGTGCAGAGTG |
| Mouse CYP8B1 | GGTACGCTTCCTCTATCGCC | GAGGGATGGCGTCTTATGGG |
| Mouse CYP27A1 | GAAGCCATCACCTATATC | ATAGACTGAGTTCTGGAA |
| Mouse GAPDH | TTGATGGCAACAATCTCCAC | CGTCCCGTAGACAAAATGGT |
| Human ZO-1 | TGCTGAGTCCTTTGGTGATG | AATTTGGATCTCCGGGAAGAC |
| Human occludin | GTTGCGGCGAGCGGATTG | TGGACTTTCAAGAGGCCTGG |
| Human TNFα | GCTGCACTTTGGAGTGATCG | GCTTGAGGGTTTGCTACAACA |
| Human CCL2 | TCAAACTGAAGCTCGCACTC | TGGGGCATTGATTGCATCTGG |
| Human CCL5 | CACCCTGCTGCTTTGCCTA | CATCCTTGACCTGTGGACGACT |
| Human GAPDH | GCTCCTCCTGTTCGACAGTCA | ACCTTCCCCATGGTGTCTGA |
Caco-2 cells were lysed using RIPA buffer containing protein phosphatase and protease inhibitors (Solarbio, China). The obtained proteins were separated on 10% SDS-PAGE gels and transferred onto polyvinylidene difluoride membranes by electro-blotting. The membranes were incubated overnight at 4 °C with primary antibodies against anti-ZO-1 and anti-occludin (Abmart, China), diluted 1:1000 in TBS-T. Subsequently, secondary anti rabbit-HRP antibodies were applied at room temperature for one hour. Protein bands were detected using an ECL plus detection system (Solarbio, China).
Paraffin-embedded liver and ileum tissues were cut into 5 μm sections and mounted onto glass slides. Sections were stained with haematoxylin and eosin for histopathology. Moreover, liver tissues were stained with Sirius red for immunohistochemistry analysis. All sections were analyzed with a light microscopy.
Paraffin-embedded ileum sections were cut, deparaffinized and dehydrated, followed by blocked with goat serum. The slides were incubated overnight at 4 °C with anti-ZO-1 and anti-occludin antibodies (Abcam, United States), respectively. After washing three times in PBS for five minutes, the slides were incubated with a secondary antibody for 60 minutes at room temperature. Nuclei were stained by DAPI. Fluorescence images were observed and photographed using the fluorescence microscope DM5000 B (Leika, Germany).
Statistical analyses were performed SPSS 22.0 and GraphPad Prism software version 9.0.0 (GraphPad software Inc). The measurement data were performed as the mean ± SD. Values were compared using a two-tailed Student’s t-test (parametric data) for two-group comparisons, and one-way ANOVA for differences among more than two groups. P value of < 0.05 was considered statistically significant.
Chronic liver disorders, such as CLD and autoimmune hepatitis, can contribute to imbalances in the intestinal microbiota and loss of intestinal barrier integrity, resulting in the translocation of gut commensals to the periphery[4,14,22,24,32]. To assess commensal imbalances in CLD progression, liver samples obtained from mice with DDC-induced cholestatic cholangitis and from patients with PBC were used for bacteriological culture and 16S rDNA analysis, respectively. L. murinus and L. garvieae were identified as the predominant bacterial species in the livers of CLD mice using the standard bacterial culture technique, whereas no bacteria were detected in the controls (Figure 1A and B). Next, we determined whether these bacteria were prevalent in the livers of patients with PBC. Interestingly, full-length 16S rDNA gene sequencing revealed bacteria in the livers of both patients with PBC and controls. The microbiota of all the samples was dominated by three major phyla: Proteobacteria, Firmicutes and Bacteroidota (Figure 1C). Measurement of the Chao1, Ace, Shannon, and Simpson indices revealed significantly increased hepatic microbiota richness in patients with PBC than that in the controls (Supplementary Figure 1). Notably, at the genus level, seven out of the eight patients with PBC harbored Lactococcus, a rate higher than that of the controls (one of the three), whereas Ligilactobacillus was present in all samples (Figure 1D). At the species level, L. murinus was commonly detected in patients with PBC and controls, while L. garvieae was detected only in patients with PBC but not in controls (Figure 1E). Collectively, these findings indicated microbiota enrichment in the livers during CLD, with a potential association between L. garvieae prevalence and this conditions.
We gavaged CLD mice with L. garvieae and L. murinus to further explore whether L. garvieae and L. murinus isolated from CLD mice livers influenced the hepatobiliary pathological features (Figure 2A). Compared with control mice, CLD mice exhibited severe changes in liver morphology, including bile duct hyperplasia and cholestasis, portal edema, and portal infiltrates (Figure 2B). These changes in liver morphology were markedly aggravated following L. garvieae administration. Furthermore, body weight of CLD mice significantly decreased at 3-4 weeks of the modeling process, with a further decreasing trend observed in L. garvieae-treated CLD mice, although the differences were less pronounced (Figure 2C). Notably, serum ALP, GGT and ALT levels were significantly higher in CLD mice than in control mice. L. garvieae treatment significantly increased serum levels of ALP and GGT levels, whereas ALT levels did not significantly change in CLD mice (Figure 2D). Furthermore, mice inoculated with L. murinus did not exhibit increased susceptibility to experimental CLD (Supplementary Figure 2). Therefore, L. garvieae colonization in the early stages of CLD contributes to inflammatory cell infiltration, cholestasis, and bile duct injury.
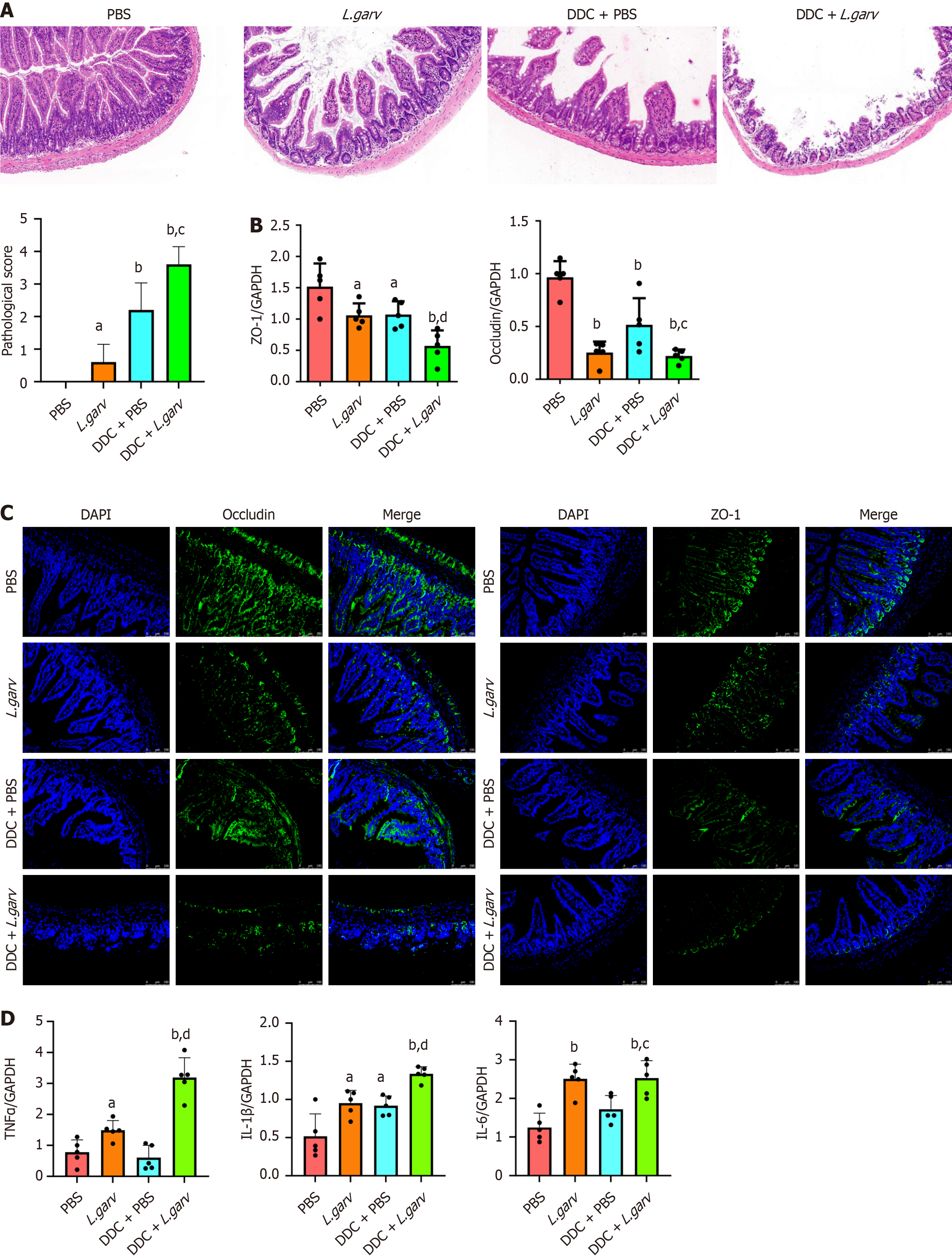
We explored whether L. garvieae aggravates cholestatic cholangitis by targeting the impaired intestinal barrier function. Hematoxylin and eosin staining of the small intestines from the four groups revealed significantly destroyed intestinal structure was in the L. garvieae and DDC + PBS groups compared with that in the control group (Figure 3A). Intestinal tract lesions were more severe in the DDC + L. garvieae group than in the DDC + PBS group.
Tight junctions in the intestinal epithelial and endothelial cells are formed by tight junctional proteins such as ZO-1 and occludin. Loss of expression of these junctional proteins was observed in the L. garvieae and DDC + PBS groups compared with the control group. L. garvieae treatment further decreased ZO-1 and occludin expression in the small intestine of DDC-fed mice (Figure 3B). Consistently, immunostaining for tight junction proteins revealed lower ZO-1 and occludin expression in the L. garvieae-treated mice than in the corresponding control mice (Figure 3C). Additionally, the expression of cytokines that participate in barrier disruption, including tumor necrosis factor alpha (TNFα), interleukin (IL)-6, and IL-1β, significantly increased in the gut mucosa of the L. garvieae-treated mice compared with that in the corresponding control group (Figure 3D). Overall, L. garvieae administration aggravated intestinal barrier disruption in mice with CLD.
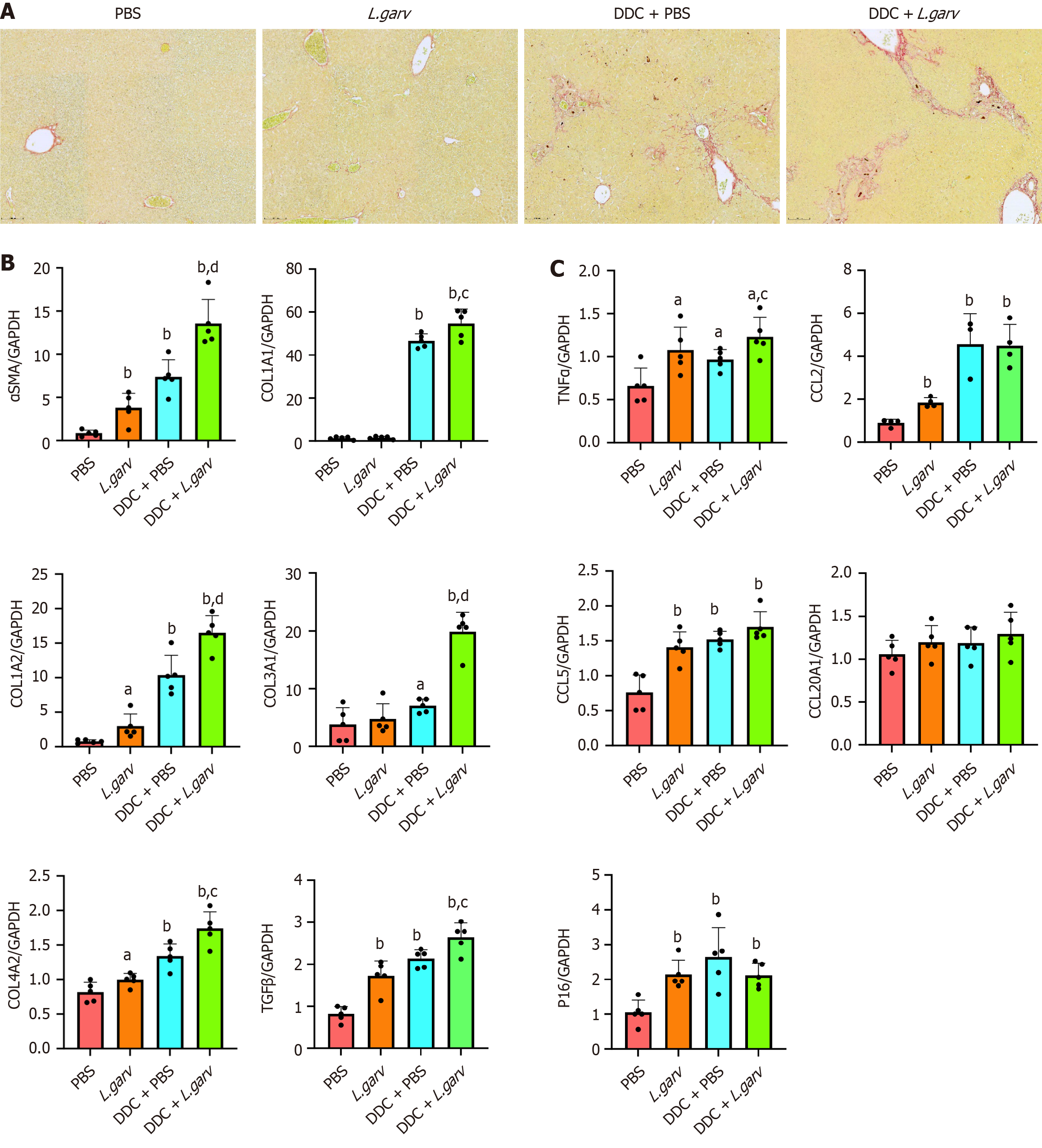
We evaluated the effect of L. garvieae on liver fibrosis and the inflammatory response in CLD mice. CLD mice displayed marked liver fibrosis, which was further aggravated by early monocolonization of L. garvieae (Figure 4A). Subsequently, we examined the expression of genes associated with liver fibrosis to further determine the effect of L. garvieae on hepatic fibrosis. Notably, mRNA expression of liver fibrosis markers, such as α-SMA, COL1A1, COL1A2, COL3A1, COL4A2, and TGF-β significantly increased following L. garvieae treatment in CLD mice. Furthermore, the mRNA expression of α-SMA, COL1A2, COL4A2, and TGF-β in the livers of the L. garvieae group was also significantly increased compared with the PBS group (Figure 4B).
We also evaluated the inflammatory responses in the liver. DDC induced-cholestatic cholangitis was accompanied by significant upregulation of TNFα, CCL2, CCL5 and P16 mRNAs, L. garvieae monocolonization further increased the expression of TNFα. Moreover, the mRNA expressions of TNFα, CCL2, CCL5 and P16 were significantly increased following L. garvieae treatment in mice fed a standard chow diet (Figure 4C).
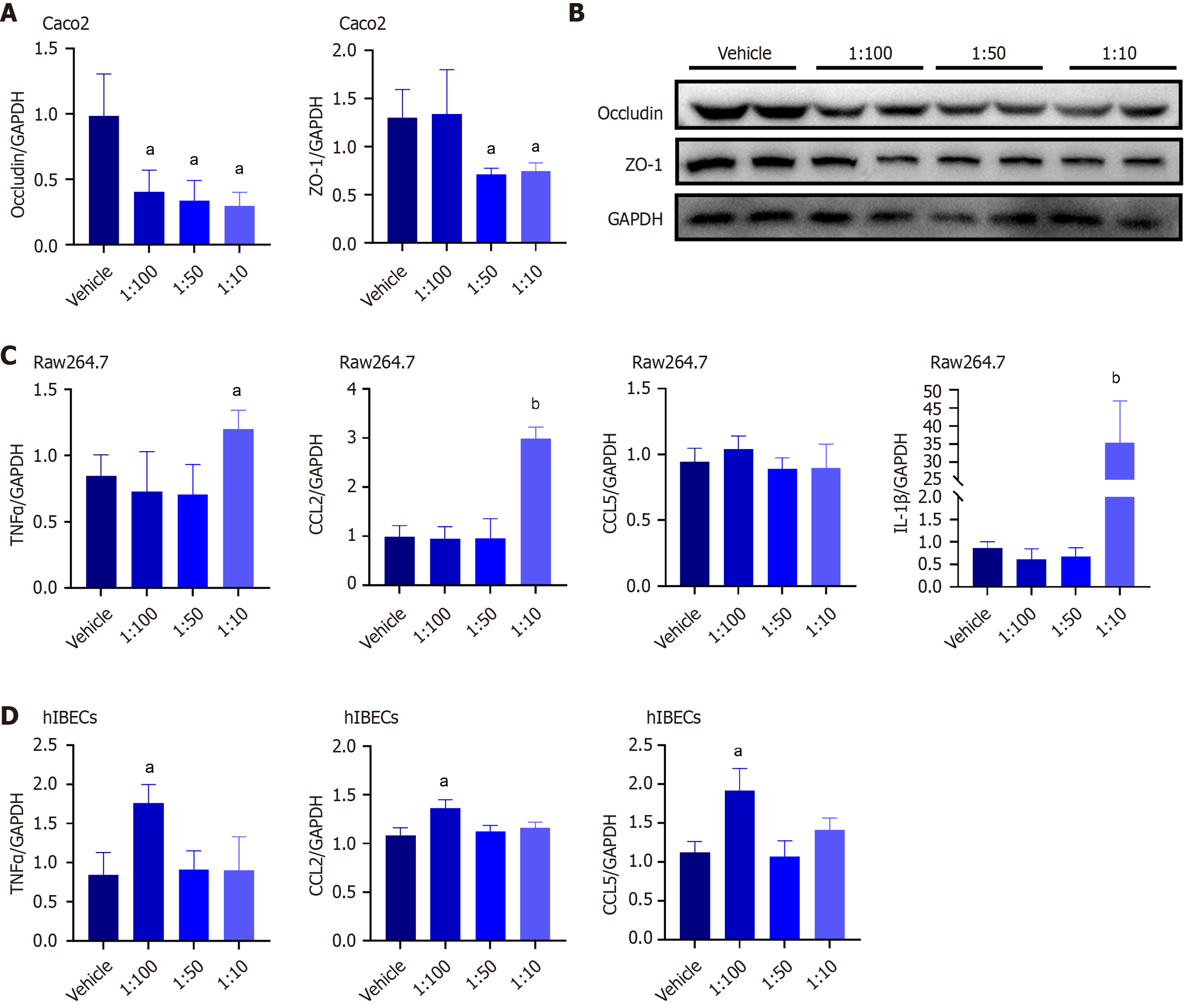
L. garvieae exacerbated intrahepatic cholestasis and liver fibrosis by increasing intestinal permeability and inducing liver inflammation in CLD mice. Consequently, we treated human intestinal epithelial Caco-2 cells with L. garvieae culture supernatants to further determine its effects on intestinal barrier function. Compared with the control group, the occludin mRNA expressions were significantly decreased in the groups exposed to L. garvieae culture supernatants. The mRNA expressions of ZO-1 were significantly decrease in the groups exposed to 1:50 or 1:10 L. garvieae culture supernatants compared with the control group (Figure 5A). Furthermore, L. garvieae culture supernatants decreased ZO-1 and occludin protein levels in Caco-2 cells (Figure 5B). Therefore, L. garvieae induced disruption of the intestinal epithelial barrier.
Next, we determined the effects of L. garvieae culture supernatants on the mRNA expressions of proinflammatory cytokines in RAW264.7 cells and human intrahepatic bile duct epithelial cells (hIBECs) using quantitative RT-PCR. TNFα, CCL2, and IL-1β expression were significantly increased in RAW264.7 cells exposed to 1:10 L. garvieae culture supernatants compared with the control group (Figure 5C). Furthermore, TNFα, CCL2, and CCL5 expression in hIBECs exposed to 1:100 L. garvieae culture supernatants were significantly increased compared with the control group (Figure 5D). Therefore, L. garvieae promoted inflammatory activation of RAW264.7 cells and hIBECs.
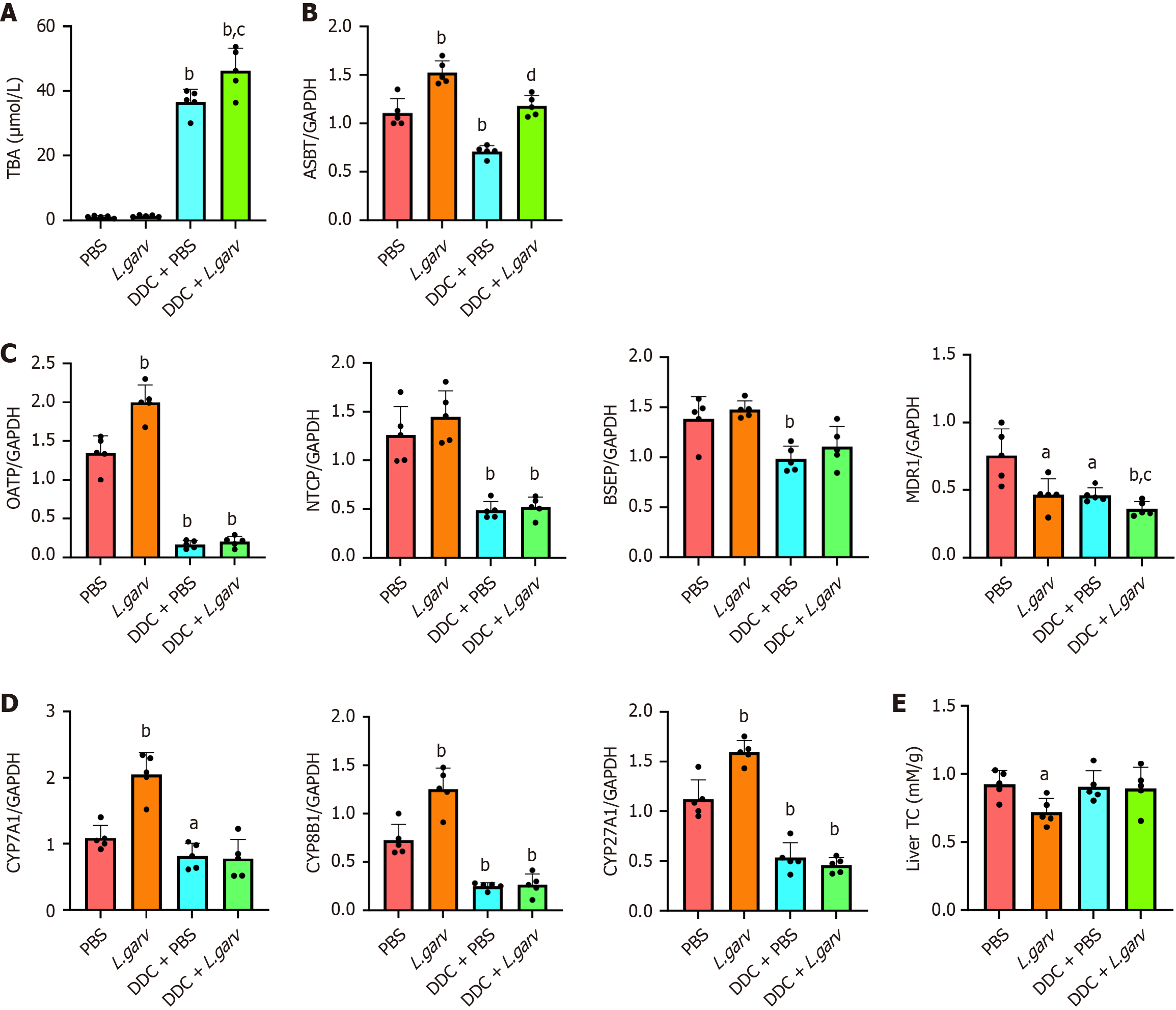
Various intestinal and hepatic membrane transporters play vital roles in the maintenance of BA homeostasis. Elevated serum total BA levels were observed in DDC mice, which were further upregulated by L. garvieae (Figure 6A). Correspondingly, ASBT expression markedly increased in L. garvieae-treated mice (Figure 6B). We also examined the expressions of genes involved in hepatic BAs metabolism. The expression of hepatic OATP was substantially increased in mice monocolonized with L. garvieae under standard chow-fed conditions. In contrast, the mRNA levels of other transporters (NTCP and BSEP) did not change significantly. Furthermore, L. garvieae decreased hepatic MDR1 mRNA levels in mice fed a standard chow or DDC diet (Figure 6C). The expression of hepatic CYP7A1, CYP8B1 and CYP27A1 was substantially increased in mice monocolonized with L. garvieae under standard chow-fed conditions (Figure 6D). Additionally, we assessed the effect of L. garvieae on liver cholesterol levels, as BAs are primarily synthesized from cholesterol. As expected, total cholesterol levels were significantly decreased following L. garvieae treatment in mice fed a standard chow diet (Figure 6E).
Collectively, these data suggested that L. garvieae disrupts BA homeostasis by enhancing their reabsorption in the intestines and promoting their synthesis in the liver under normal conditions. Furthermore, L. garvieae exacerbates BA disorders under cholestatic conditions.
Previous research has revealed an accumulating body of evidence reveals an association between intestinal microbiota dysbiosis and CLD[4,33,34]. Depletion of potentially beneficial microbiota and enrichment of opportunistic microbiota have been observed in patients with early-stage PBC[35]. Notably, rectal administration of microbial derivatives in rats contributed to a mixed inflammatory hepatobiliary infiltrate, particularly small duct cholangitis, resembling the histology of PBC[36]. However, the mechanism through which the gut microbiota contributes to the CLD pathogenesis remains elusive. In this study, we isolate L. murinus and L. garvieae from the livers of CLD mice through bacterial culturing. L. murinus was detected in both patients with PBC and controls, whereas L. garvieae was exclusively identified in patients with PBC. Inoculation with L. garvieae aggravated hepatobiliary inflammation by increasing intestinal barrier disruption and promoting BA reabsorption. Therefore, targeting the gut microbiota and restoring the intestinal barrier integrity could be a novel therapeutic approach for managing PBC. Accordingly, future studies should explore gut microbiota as a therapeutic target or biomarker of PBC progression.
Alterations in the liver microenvironment, BA composition biliary ducts, cholangiocytes, and intestinal barrier integrity during CLD can promote microbial translocation to biliary portals and liver tissues[14,16]. The liver and biliary microbiome may play a pathogenic role in PBC. Furthermore, the presence of bacteria in the majority of liver and bile duct cultures of patients with PBC and PSC supports the involvement of gut microbiota translocation in pathogenesis[13,37,38]. An increased T cell response is linked to the enrichment of gut Lactobacillus gasseri in the livers of CLD mice[14]. Moreover, Klebsiella pneumonia identified in the microbiota of patients with PSC disrupts the epithelial barrier, initiating bacterial translocation and liver inflammatory responses[25]. The present study revealed microbial enrichment in the liver during CLD. Particularly, the members of the Streptococcaceae family, specifically L. garvieae, were selectively observed in the liver microbiota of patients with PBC and CLD mice, whereas the members of the Lactobacillaceae family, L. murinus were detected in both patients with PBC and controls. The abundance of L. garvieae in CLD mice was associated with more severe cholestasis and liver fibrosis. Collectively, these results collectively emphasize the link between bacterial translocation and CLD, deepening our understanding of microbial involvement in CLD pathogenesis. Although our findings primarily focused on PBC, we believe that the mechanisms involving the gut-liver axis may have broader implications for other forms of chronic liver disease. Modulation of the gut microbiome through dietary interventions, probiotics, and fecal microbiota transplantation could offer novel therapeutic avenues to mitigate liver disease progre
The intestinal barrier plays a critical role in regulating interactions between the microbiome and liver, segregating the gut microbiota from host immune cells, thereby limiting the systemic spread of microbes and toxins. Disruption of the intestinal barriers is associated with CLD pathology[24,25]. The novel finding of our study is that L. garvieae modulates intestinal permeability by disrupting the tight junctions of intestinal epithelial cells, specifically altering ZO-1 and occludin expression. Furthermore, the gut permeability is increased by the elevated levels of inflammatory cytokines, such as TNFα, IL-6 and IL-1β in the mucosa layer. This observation is consistent with the findings of Len Verbeke et al[39], who reported that increased intestinal permeability and inflammation facilitate bacterial translocation in a rat model of cholestatic liver injury. Our findings indicate that disruption of the intestinal barrier by L. garvieae may escalate bacterial translocation, potentially contributing to the development of CLD. Collectively, the present study highlights a link between the intestinal barrier disruption and L. garvieae translocation in CLD.
The differential effects of various concentrations of bacterial supernatants on RAW264.7 cells and hIBECs underscore the nuanced interplay between microbial stimuli and host immune responses. Higher concentrations of bacterial supernatants can induce macrophages to release pro-inflammatory cytokines, including TNF-α, CCL2 and IL-1β. In contrast, hIBECs exhibited distinct responses to lower concentrations of bacterial supernatants. This phenomenon may be linked to apoptosis or functional exhaustion of bile duct cells induced by high concentrations of the supernatant.
Crosstalk between the intestinal microbiota and BAs directly impacts CLD. Furthermore, BAs influence the gut microbiota composition inhibiting the growth of specific bile-sensitive bacteria[40]. The absence of bile in the intestine facilitates transplantation of microbiota. L. garvieae aggravated BA reuptake from the intestine through ASBT and transported more BAs to hepatocytes through OATPs in our study. Additionally, L. garvieae suppressed MDR1 expression in the liver. MDR1 is the most prominent xenobiotic transporter responsible for eliminating metabolites, including BAs and potentially toxic products from liver cells into the biliary canaliculi[41]. Collectively, these regulatory actions mediated by L. garvieae collectively further worsen hepatocyte impairment and cholestasis. L. garvieae promoted CYP7A1, CYP8B1 and CYP27A1 expression in the chow diet mice group in our study; however, this phenomenon was not observed in the CLD mouse model. Therefore, excess BA accumulation and potentially toxic bile products may trigger a negative feedback regulation of BAs synthesis, thereby repressing the ability of L. garvieae to promote BAs synthesis. Overall, these findings support an important role in BAs transport, and suggest a direct association between L. garvieae and CLD mechanisms.
In conclusion, our study identifies the role of gut-derived L. garvieae in CLD. Understanding the intricate balance between the intestinal microbiota and the hepatobiliary microbiota is crucial for the clinical management of this disease. L. garvieae primarily disrupts the intestinal epithelial barrier and interferes with the enterohepatic circulation of BAs, subsequently leading to inflammation, cholestasis, and fibrosis. Our findings provide valuable insights into the translocation of gut microbiota and its role in CLD.
We thank all study participants who made this research possible.
| 1. | Jansen PL, Ghallab A, Vartak N, Reif R, Schaap FG, Hampe J, Hengstler JG. The ascending pathophysiology of cholestatic liver disease. Hepatology. 2017;65:722-738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 229] [Article Influence: 28.6] [Reference Citation Analysis (0)] |
| 2. | Schaap FG, Trauner M, Jansen PL. Bile acid receptors as targets for drug development. Nat Rev Gastroenterol Hepatol. 2014;11:55-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 451] [Cited by in RCA: 518] [Article Influence: 47.1] [Reference Citation Analysis (0)] |
| 3. | Griffiths L, Dyson JK, Jones DE. The new epidemiology of primary biliary cirrhosis. Semin Liver Dis. 2014;34:318-328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 48] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 4. | Tang R, Wei Y, Li Y, Chen W, Chen H, Wang Q, Yang F, Miao Q, Xiao X, Zhang H, Lian M, Jiang X, Zhang J, Cao Q, Fan Z, Wu M, Qiu D, Fang JY, Ansari A, Gershwin ME, Ma X. Gut microbial profile is altered in primary biliary cholangitis and partially restored after UDCA therapy. Gut. 2018;67:534-541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 296] [Article Influence: 42.3] [Reference Citation Analysis (0)] |
| 5. | Chen LP, Zhao H, Lyu B, Cheng JL. [Environmental factors and primary biliary cirrhosis]. Zhonghua Gan Zang Bing Za Zhi. 2016;24:541-544. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 6. | Webb GJ, Hirschfield GM. Using GWAS to identify genetic predisposition in hepatic autoimmunity. J Autoimmun. 2016;66:25-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 83] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 7. | Qiu F, Tang R, Zuo X, Shi X, Wei Y, Zheng X, Dai Y, Gong Y, Wang L, Xu P, Zhu X, Wu J, Han C, Gao Y, Zhang K, Jiang Y, Zhou J, Shao Y, Hu Z, Tian Y, Zhang H, Dai N, Liu L, Wu X, Zhao W, Zhang X, Zang Z, Nie J, Sun W, Zhao Y, Mao Y, Jiang P, Ji H, Dong Q, Li J, Li Z, Bai X, Li L, Lin M, Dong M, Li J, Zhu P, Wang C, Zhang Y, Jiang P, Wang Y, Jawed R, Xu J, Zhang Y, Wang Q, Yang Y, Yang F, Lian M, Jiang X, Xiao X, Li Y, Fang J, Qiu D, Zhu Z, Qiu H, Zhang J, Tian W, Chen S, Jiang L, Ji B, Li P, Chen G, Wu T, Sun Y, Yu J, Tang H, He M, Xia M, Pei H, Huang L, Qing Z, Wu J, Huang Q, Han J, Xie W, Sun Z, Guo J, He G, Eric Gershwin M, Lian Z, Liu X, Seldin MF, Liu X, Chen W, Ma X. A genome-wide association study identifies six novel risk loci for primary biliary cholangitis. Nat Commun. 2017;8:14828. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 103] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 8. | Carey EJ, Ali AH, Lindor KD. Primary biliary cirrhosis. Lancet. 2015;386:1565-1575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 357] [Cited by in RCA: 395] [Article Influence: 39.5] [Reference Citation Analysis (0)] |
| 9. | Bogdanos DP, Vergani D. Bacteria and primary biliary cirrhosis. Clin Rev Allergy Immunol. 2009;36:30-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 64] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 10. | Selmi C, Balkwill DL, Invernizzi P, Ansari AA, Coppel RL, Podda M, Leung PS, Kenny TP, Van De Water J, Nantz MH, Kurth MJ, Gershwin ME. Patients with primary biliary cirrhosis react against a ubiquitous xenobiotic-metabolizing bacterium. Hepatology. 2003;38:1250-1257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 223] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 11. | Liwinski T, Heinemann M, Schramm C. The intestinal and biliary microbiome in autoimmune liver disease-current evidence and concepts. Semin Immunopathol. 2022;44:485-507. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 33] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 12. | Tyc O, Jansen C, Schierwagen R, Uschner FE, Israelsen M, Klein S, Ortiz C, Strassburg CP, Zeuzem S, Gu W, Torres S, Praktiknjo M, Kersting S, Langheinrich M, Nattermann J, Servant F, Arumugam M, Krag A, Lelouvier B, Weismüller TJ, Trebicka J. Variation in Bile Microbiome by the Etiology of Cholestatic Liver Disease. Liver Transpl. 2020;26:1652-1657. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 13. | Hiramatsu K, Harada K, Tsuneyama K, Sasaki M, Fujita S, Hashimoto T, Kaneko S, Kobayashi K, Nakanuma Y. Amplification and sequence analysis of partial bacterial 16S ribosomal RNA gene in gallbladder bile from patients with primary biliary cirrhosis. J Hepatol. 2000;33:9-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 69] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 14. | Tedesco D, Thapa M, Chin CY, Ge Y, Gong M, Li J, Gumber S, Speck P, Elrod EJ, Burd EM, Kitchens WH, Magliocca JF, Adams AB, Weiss DS, Mohamadzadeh M, Grakoui A. Alterations in Intestinal Microbiota Lead to Production of Interleukin 17 by Intrahepatic γδ T-Cell Receptor-Positive Cells and Pathogenesis of Cholestatic Liver Disease. Gastroenterology. 2018;154:2178-2193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 105] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 15. | Sandler NG, Koh C, Roque A, Eccleston JL, Siegel RB, Demino M, Kleiner DE, Deeks SG, Liang TJ, Heller T, Douek DC. Host response to translocated microbial products predicts outcomes of patients with HBV or HCV infection. Gastroenterology. 2011;141:1220-1230, 1230.e1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 250] [Cited by in RCA: 251] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 16. | Wang L, Llorente C, Hartmann P, Yang AM, Chen P, Schnabl B. Methods to determine intestinal permeability and bacterial translocation during liver disease. J Immunol Methods. 2015;421:44-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 191] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 17. | Bozward AG, Ronca V, Osei-Bordom D, Oo YH. Gut-Liver Immune Traffic: Deciphering Immune-Pathogenesis to Underpin Translational Therapy. Front Immunol. 2021;12:711217. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 18. | Luissint AC, Parkos CA, Nusrat A. Inflammation and the Intestinal Barrier: Leukocyte-Epithelial Cell Interactions, Cell Junction Remodeling, and Mucosal Repair. Gastroenterology. 2016;151:616-632. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 393] [Cited by in RCA: 396] [Article Influence: 44.0] [Reference Citation Analysis (0)] |
| 19. | Seki E, Schnabl B. Role of innate immunity and the microbiota in liver fibrosis: crosstalk between the liver and gut. J Physiol. 2012;590:447-458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 270] [Cited by in RCA: 339] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 20. | Wexler HM. Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev. 2007;20:593-621. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1141] [Cited by in RCA: 1504] [Article Influence: 83.6] [Reference Citation Analysis (0)] |
| 21. | Floreani A, Baragiotta A, Pizzuti D, Martines D, Cecchetto A, Chiarelli S. Mucosal IgA defect in primary biliary cirrhosis. Am J Gastroenterol. 2002;97:508-510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 22. | Zhang H, Liu M, Zhong W, Zheng Y, Li Y, Guo L, Zhang Y, Ran Y, Zhao J, Zhou L, Wang B. Leaky Gut Driven by Dysbiosis Augments Activation and Accumulation of Liver Macrophages via RIP3 Signaling Pathway in Autoimmune Hepatitis. Front Immunol. 2021;12:624360. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 23. | Feld JJ, Meddings J, Heathcote EJ. Abnormal intestinal permeability in primary biliary cirrhosis. Dig Dis Sci. 2006;51:1607-1613. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 50] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 24. | Manfredo Vieira S, Hiltensperger M, Kumar V, Zegarra-Ruiz D, Dehner C, Khan N, Costa FRC, Tiniakou E, Greiling T, Ruff W, Barbieri A, Kriegel C, Mehta SS, Knight JR, Jain D, Goodman AL, Kriegel MA. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science. 2018;359:1156-1161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 576] [Cited by in RCA: 596] [Article Influence: 85.1] [Reference Citation Analysis (0)] |
| 25. | Nakamoto N, Sasaki N, Aoki R, Miyamoto K, Suda W, Teratani T, Suzuki T, Koda Y, Chu PS, Taniki N, Yamaguchi A, Kanamori M, Kamada N, Hattori M, Ashida H, Sakamoto M, Atarashi K, Narushima S, Yoshimura A, Honda K, Sato T, Kanai T. Gut pathobionts underlie intestinal barrier dysfunction and liver T helper 17 cell immune response in primary sclerosing cholangitis. Nat Microbiol. 2019;4:492-503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 306] [Article Influence: 51.0] [Reference Citation Analysis (0)] |
| 26. | Liu S, Liu M, Zhang ML, Wang CZ, Zhang YL, Zhang YJ, Du CY, Sheng SF, Wang W, Fan YT, Song JN, Huang JC, Feng YY, Qiao W, Huang JL, Li YH, Zhou L, Zhang J, Chang YS. Transcription factor Klf9 controls bile acid reabsorption and enterohepatic circulation in mice via promoting intestinal Asbt expression. Acta Pharmacol Sin. 2022;43:2362-2372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 27. | Mayo MJ. Mechanisms and molecules: What are the treatment targets for primary biliary cholangitis? Hepatology. 2022;76:518-531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 31] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 28. | Zhu Y, Liu H, Zhang M, Guo GL. Fatty liver diseases, bile acids, and FXR. Acta Pharm Sin B. 2016;6:409-412. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 93] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 29. | Fiorucci S, Distrutti E, Carino A, Zampella A, Biagioli M. Bile acids and their receptors in metabolic disorders. Prog Lipid Res. 2021;82:101094. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 157] [Article Influence: 39.3] [Reference Citation Analysis (0)] |
| 30. | Trauner M, Fuchs CD, Halilbasic E, Paumgartner G. New therapeutic concepts in bile acid transport and signaling for management of cholestasis. Hepatology. 2017;65:1393-1404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 176] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 31. | Blesl A, Stadlbauer V. The Gut-Liver Axis in Cholestatic Liver Diseases. Nutrients. 2021;13:1018. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 44] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 32. | Liao L, Schneider KM, Galvez EJC, Frissen M, Marschall HU, Su H, Hatting M, Wahlström A, Haybaeck J, Puchas P, Mohs A, Peng J, Bergheim I, Nier A, Hennings J, Reißing J, Zimmermann HW, Longerich T, Strowig T, Liedtke C, Cubero FJ, Trautwein C. Intestinal dysbiosis augments liver disease progression via NLRP3 in a murine model of primary sclerosing cholangitis. Gut. 2019;68:1477-1492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 128] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 33. | Tilg H, Cani PD, Mayer EA. Gut microbiome and liver diseases. Gut. 2016;65:2035-2044. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 348] [Article Influence: 38.7] [Reference Citation Analysis (0)] |
| 34. | Lemoinne S, Kemgang A, Ben Belkacem K, Straube M, Jegou S, Corpechot C; Saint-Antoine IBD Network, Chazouillères O, Housset C, Sokol H. Fungi participate in the dysbiosis of gut microbiota in patients with primary sclerosing cholangitis. Gut. 2020;69:92-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 148] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 35. | Lv LX, Fang DQ, Shi D, Chen DY, Yan R, Zhu YX, Chen YF, Shao L, Guo FF, Wu WR, Li A, Shi HY, Jiang XW, Jiang HY, Xiao YH, Zheng SS, Li LJ. Alterations and correlations of the gut microbiome, metabolism and immunity in patients with primary biliary cirrhosis. Environ Microbiol. 2016;18:2272-2286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 181] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 36. | Mattner J. Impact of Microbes on the Pathogenesis of Primary Biliary Cirrhosis (PBC) and Primary Sclerosing Cholangitis (PSC). Int J Mol Sci. 2016;17:1864. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 37. | Nilsson HO, Taneera J, Castedal M, Glatz E, Olsson R, Wadström T. Identification of Helicobacter pylori and other Helicobacter species by PCR, hybridization, and partial DNA sequencing in human liver samples from patients with primary sclerosing cholangitis or primary biliary cirrhosis. J Clin Microbiol. 2000;38:1072-1076. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 161] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 38. | Katt J, Schwinge D, Schoknecht T, Quaas A, Sobottka I, Burandt E, Becker C, Neurath MF, Lohse AW, Herkel J, Schramm C. Increased T helper type 17 response to pathogen stimulation in patients with primary sclerosing cholangitis. Hepatology. 2013;58:1084-1093. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 134] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 39. | Verbeke L, Farre R, Verbinnen B, Covens K, Vanuytsel T, Verhaegen J, Komuta M, Roskams T, Chatterjee S, Annaert P, Vander Elst I, Windmolders P, Trebicka J, Nevens F, Laleman W. The FXR agonist obeticholic acid prevents gut barrier dysfunction and bacterial translocation in cholestatic rats. Am J Pathol. 2015;185:409-419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 170] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 40. | Watanabe M, Fukiya S, Yokota A. Comprehensive evaluation of the bactericidal activities of free bile acids in the large intestine of humans and rodents. J Lipid Res. 2017;58:1143-1152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 95] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 41. | Jetter A, Kullak-Ublick GA. Drugs and hepatic transporters: A review. Pharmacol Res. 2020;154:104234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 106] [Article Influence: 21.2] [Reference Citation Analysis (0)] |