Published online Jan 7, 2025. doi: 10.3748/wjg.v31.i1.101198
Revised: September 28, 2024
Accepted: November 14, 2024
Published online: January 7, 2025
Processing time: 93 Days and 7.9 Hours
Hepatocellular carcinoma (HCC) has been a pervasive malignancy throughout the world with elevated mortality. Efficient therapeutic targets are beneficial to treat and predict the disease. Currently, the exact molecular mechanisms leading to the progression of HCC are still unclear. Research has shown that the microRNA-142-3p level decreases in HCC, whereas bioinformatics analysis of the cancer genome atlas database shows the ASH1L expression increased among liver tumor tissues. In this paper, we will explore the effects and mechanisms of microRNA-142-3p and ASH1L affect the prognosis of HCC patients and HCC cell bioactivity, and the association between them.
To investigate the effects and mechanisms of microRNA-142-3p and ASH1L on the HCC cell bioactivity and prognosis of HCC patients.
In this study, we grouped HCC patients according to their immunohistochemistry results of ASH1L with pathological tissues, and retrospectively analyzed the prognosis of HCC patients. Furthermore, explored the roles and mechanisms of microRNA-142-3p and ASH1L by cellular and animal experiments, which involved the following experimental methods: Immunohistochemical staining, western blot, quantitative real-time-polymerase chain reaction, flow cytometric analysis, tumor xenografts in nude mice, etc. The statistical methods involved in this study contained t-test, one-way analysis of variance, the
In this study, we found that HCC patients with high expression of ASH1L possess a more recurrence rate as well as a decreased overall survival rate. ASH1L promotes the tumorigenicity of HCC and microRNA-142-3p exhibits reduced expression in HCC tissues and interacts with ASH1L through targeting the ASH1L 3′untranslated region. Furthermore, microRNA-142-3p promotes apoptosis and inhibits proliferation, invasion, and migration of HCC cell lines in vitro via ASH1L. For the exploration mechanism, we found ASH1L may promote an immunosuppressive microenvironment in HCC and ASH1L affects the expression of the cell junction protein zonula occludens-1, which is potentially relevant to the immune system.
Loss function of microRNA-142-3p induces cancer progression and immune evasion through upregulation of ASH1L in HCC. Both microRNA-142-3p and ASH1L can feature as new biomarker for HCC in the future.
Core Tip: This article mainly focuses on the new targets which may be beneficial to treat and predict hepatocellular carcinoma. We would like to shed light on the effects of microRNA-142-3p and ASH1L on the hepatocellular carcinoma cell bioactivity and prognosis of hepatocellular carcinoma patients. Based on the experiments, loss function of microRNA-142-3p induces cancer progression and immune evasion through upregulation of ASH1L in hepatocellular carcinoma. Both microRNA-142-3p and ASH1L can feature as new biomarker for hepatocellular carcinoma in the future. In-depth comprehensive research is indispensable to completely clarity the ASH1L pathology and relevant molecular mechanisms in hepatocellular carcinoma progression.
- Citation: Yu XH, Xie Y, Yu J, Zhang KN, Guo ZB, Wang D, Li ZX, Zhang WQ, Tan YY, Zhang L, Jiang WT. Loss-of-function mutations of microRNA-142-3p promote ASH1L expression to induce immune evasion and hepatocellular carcinoma progression. World J Gastroenterol 2025; 31(1): 101198
- URL: https://www.wjgnet.com/1007-9327/full/v31/i1/101198.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i1.101198
Hepatocellular carcinoma (HCC) occupies 90% of primary liver cancers and ranks the sixth in terms of diagnoses while becoming the third main factor of cancer-associated deaths globally in 2020, including more than 906000 new cases and 830000 death cases[1,2]. Early diagnosis is crucial for HCC patients’ chances of recovery; nevertheless, owing to challenges of early diagnosis and swift development of the disease, numerous patients are initially diagnosed as terminal cancer[3]. Liver transplantation (LT) is considered the most comprehensive treatment option for HCC[4]. Nevertheless, the LT success rates are still not optimistic with 46.8% 5-year survival rate and 36.7% relapse rate[5]. As a result, there exists a pressing demand to recognize therapeutic targets and precious biomarkers to boost early diagnosis and prognosis, as well as customize treatment plans. Studies have shown remarkable individual variances in epigenetic changes and gene mutations of HCC, offering opportunities for the exploration of novel biomarkers[6].
Relevant studies have demonstrated a common connection between human cancers and altered expression of microRNA (miR)[7]. Analysis through deep RNA sequencing of the miR-142 gene on human chromosome 17 unveiled that miR-142-3p and miR-142-5p strands from the precursor miR-142, interacted with the RNA-induced silencing complex, effectively directing the RNA interference machinery towards their specific target mRNAs[8]. Notably, miR-142-3p displays abnormal expression across various tumors, being notably downregulated in colon cancer where its reduced levels related to bad decomposition and larger-size tumor[9]. Loss of miR-142-3p function has been linked to the development of leukemia[10]. Moreover, in pancreatic ductal adenocarcinoma, miR-142-3p hampers cell proliferation through the targeting of heat shock protein 70[11].
Formally known as ASH1L, the ASH1L protein functions as a histone methyltransferase. The gene produces a 10.5 kb mRNA, from which a 2962-residue protein is translated[12,13]. ASH1L comprises distinct structural domains, including four AT Hook structural domains, an suppressor of variegation, enhancer of zeste and trithorax (SET) structural domain, bromo domain, and a plant homeodomain (PHD) finger[12,14]. Its methylation of Lys36 of histone H3 (H3K36) regulates the Homeobox gene via offsetting Polycomb silencing[15]. Structural analyses have uncovered the essential nature of the ASH1L self-repressor circuit for the role of the SET structural domain catalyzes the H3K36 monomethylation while facilitating gene activation[14,16]. Research has emphasized that the important function of connecting hydrophobic plaques and DNAs was disrupted by histones H2A and H3, in H3K36 methylation[17-19]. Previous research has proven the pathology of ASH1L among various human illnesses, including its association with disparate categories of cancer[20]. As an illustration, it is enlarged in HCC cell lines and can boost tumor cell transfer via the CDK5/P35 pathway[21,22]. Analysis on genome sequence data (n = 958) from the cancer genome atlas (TCGA) database showed ASH1L gene amplification among tumor samples, with increased expression among 11 breast cancer cell lines by comparison with the control breast epithelial cell MCF10A[23]. Through fresh frozen samples, biochemical analyses exhibited five-fold higher ASH1L expression in anaplastic thyroid carcinoma (ATC) (n = 6) than papillary thyroid carcinoma (n = 5), and that the ASH1L gene knockdown lessened cell proliferation among ATC cell lines and mouse xenograft models[24]. ASH1L was also identified as a prominently amplified genes[25]. Integrated analysis on whole-genome sequencing of 300 HCC cases revealed structural variants of the ASH1L gene among some samples[26]. The oncomine platform provided evidence that ASH1L expression is markedly increased in HCC tissues in comparison to normal liver tissues[27]. Furthermore, database analysis showed significant amplification of the ASH1L gene in 34% of neuroendocrine prostate carcinomas, 13.5% of HCC, 17.9% of uterine carcinomas, 16.6% of HCCs, and 13.8% of pancreatic carcinomas[28].
Certain research has illustrated a pertinence between lower miR-142-3p expression and higher ASH1L expression in some types of tumors. Furthermore, miR-142-3p has been shown to mitigate thyroid follicular tumors via modulating ASH1L[29]. ASH1L has also been linked to the pathogenesis of leukemia[30]. Additionally, experimental data has demon
Of 111 paraffin-embedded liver slices from patients with HCC were supplied by the Department of Pathology at Tianjin First Central Hospital. The immunohistochemistry process was conducted following aforementioned approaches[35]. Initially, the paraffin-embedded slices underwent xylene deparaffinization, rehydration, ethylenediaminetetraacetic acid antigen recovery buffer treatment, and microwave heating for antigen recovery. After that, the slices were treated with 3% hydrogen peroxide, with the aim of suppressing endogenous peroxidase function. Following rinsing, they were hatched within 5% goat serum, in an effort to inhibit non-specific binding. Following this, the slices were left to incubate all night at 4 °C using rabbit anti-ASH1L antibody. After washed again, the slices were processed using goat anti-rabbit IgG secondary antibody (Sino Biological, 1:500) marked with horseradish peroxidase (HRP). Subsequently, the slices were stained using diaminobenzidine, counterstained using hematoxylin, decomposed using 1% ethanol hydrochloride solution, blued using 1% ammonia solution, dehydrated, and equipped.
The research adopted the Barnes semi-quantitative integral approach to assess the stained segments, employing the following approach: Counting five randomly selected high-magnification fields. A score of 0, 1, 2, 3, or 4 was designated on the grounds of the percentage of positive cells, corresponding to 0%-5%, 6%-25%, 51%-75%, and 76%-100%, separately. The intensity of immunohistochemical staining received scores: 0 denoted no staining, 1 denoted light-yellow staining, 2 denoted brown staining, and 3 denoted tan staining. The ultimate outcome was attained via multiplying two scores: 0 denoted negative, 1-4 denoted weakly positive, 5-8 denoted mildly positive, and 9-12 denoted strongly positive. The segments were scored independently by two readers[36]. The research involving human participators were retrospected and approved by the Ethics Committee of Tianjin First Central Hospital (protocol number: YC-BY-LC-2023-038).
In total, 111 HCC patients experiencing liver surgery between July 2016 and June 2018 at Tianjin First Central Hospital affiliated with Nankai University were included in our research. Clinicopathological staging was measured as per to the American Joint Committee on Cancer criteria. Previous approval was supplied by the institutional research ethics committee to adopt patients’ clinical data. Inclusion criteria for our research were listed below: (1) Patients aged 18 to 70 years; (2) Absence of metastasis or macrovascular invasion detected on preoperative imaging; and (3) Confirmation of HCC diagnosis by postoperative pathological examination. Exclusion criteria were listed below: (1) Presence of mixed HCC on pathological examination; (2) Presence of tumors in organs other than the liver; and (3) Missing laboratory or pathological data.
HCC cell lines were sourced from the Cell Resource Centre of Shanghai Institutes for Life Sciences, China. The cells were cultivated in Dulbecco’s Modified Eagle’s Medium (DMEM) (Corning, 10017CV). The cultures were sustained under 50 mL/L carbon dioxide (CO2) at 37 °C, as described before[37].
Total RNA was attained from the cells fostered. The RNA concentration was measured. Next, the total RNA was reverse transcribed into cDNA following the producer’s protocols. The target gene expression levels were standardized through the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Primer sequences for GAPDH and ASH1L are provided below: MiR-142-3p forward 5’-CGCGTGTAGTGTTTCCTACTT-3’, ASH1L forward 5’-CCCAAAGGAGTAAAGGAGCAAGA-3’, ASH1L reverse 5’-TCTTCAATCAAGGGTAGAGCATCA-3’, GAPDH forward 5’-GGAAGCTTGTCATCAATGGAAATC-3’, GAPDH reverse: 5’-TGATGACCCTTTTGGCTCCC-3’.
All samples were checked three times in order to ensure accuracy. The gene expression quantification was implemented through the SYBR Green quantitative real-time-polymerase chain reaction on one LightCycler 96 instrument following the producer’s guidelines. The thermal cycling profiles included an initial denaturation for 10 minutes at 95 °C, 40 amplification cycles, and melting curve analysis. The data was construed through the 2-ΔΔCt approach and viewed through the GraphPad Prism 9.0. (The above results were statistically analyzed after repeating the experiment three times independently.)
Western blot was conducted following a previously described method[38]. By and large, cells were rinsed two times with phosphatase inhibitors, then gained and centrifuged. The supernatant containing the proteins was gathered, and the density of the protein was measured through a bicinchoninic acid assay protein assay kit. Equivalent quantities (10 μg) of proteins were denatured for 5 minutes at 100 °C and loaded onto 4%-12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels. The proteins were separated and delivered onto a polyvinylidene difluoride membrane (ImmobilonP; MA, United States), followed by blocking with 5% skimmed milk for 2 hours at ordinary temperature. Following rinsed through the Tris-buffered saline (TBST) containing 0.1% Tween20 detergent, the membranes were exposed to primary antibody all night at 4 °C, rinsed using TBST, and subsequently treated using a secondary antibody conjugated to HRP for 2 hours at normal temperature. The signals were detected through an improved chemiluminescence kit (China). Primary antibodies utilized for western blot were listed below: ASH1 antibody and GAPDH antibody. Secondary antibody incubations were carried out using goat anti-rabbit IgG HRP and anti-mouse IgG HRP (the above results were statistically analyzed by after repeating the experiment three times independently).
The lentivirus overexpression (OE) system was constructed by OBiO and GeneChem Inc (Shanghai, China). Transfected cells were selected using puromycin hydrochloride (China) and geneticin (G-418) (China). The silencing lentivirus was generated through Gene Wiz Inc (China). Transfected cells were filtered via puromycin hydrochloride and labeled with green fluorescent protein for confirmation (Supplementary Table 1).
In light of the producer’s guidelines, target cells were nurtured in 6-well plates at a concentration of 1 × 105 cells/mL for 24 hours to achieve 50%-60% confluence in each well prior to viral infection. The transfer solution was prepared by mixing the old cell medium containing lentiviral auxiliary reagent at a ratio of 24:1. The target lentivirus was mixed with 1 mL transfer solution and supplemented to each well. Following 16 hours of transfection, the medium was displaced by fresh culture solution. After 48 hours, cell lines overexpressing genes were screened with puromycin hydrochloride (3 μg/mL) and G-418 (500 μg/mL), although silencing cell lines were screened with puromycin hydrochloride (3 μg/mL). Subsequently, steady transfected cell lines were produced.
Cells were exposed into a 96-well plate with a density of 20000 cells/mL; besides, 100 μL cell suspension was added to each well. Next, 10 μL cell counting kit 8 (CCK-8) reagent was supplemented to each well. Then, those plates were hatched for 0, 24, 48, 72, 96 and 120 hours pursuant to the manufacturer’s protocols, with an additional 2-hour incubation period. Optical density readings were determined at 450 nm. The assays were implemented in triplicate; additionally, the data was construed and plotted through GraphPad Prism 9.0.
The upper chamber of the Transwell insert (China) was coated with Matrigel in light of the manufacturer’s guidelines for invasion test, while it was left uncoated for the migration test. A 200 μL cell suspension (1 × 105 cells/mL) was seeded into serum-free DMEM, and then 650 μL of DMEM comprising 10% fetal bovine serum (FBS) was placed into the lower chamber as one chemotaxis inducer. Following 40-hour incubation under 5% CO2 at 37 °C, transmembrane cells were immobilized and stained using 0.1% crystal violet (Maclean’s, Shanghai, China). Subsequently, five randomly selected fields of view were imaged and counted through one light microscope. Each assay was redone at lowest three times.
Cells were fostered within 6-well plates with a concentration of 500-800 cells each well in DMEM added with 10% FBS. Following 10 days, the cells were immobilized using 4% paraformaldehyde and stained using 0.1% crystal violet.
Each sample was rinsed two times using cold phosphate buffer saline (PBS) and resuspended within 1 × binding buffer at a density of approximately 1 × 106 cells/mL. To each sample, 5 μL Annexin V-allophycocyanin conjugate (APC) was supplemented. Cells were mildly vortexed and hatched for 15 minutes at ordinary temperature in dark to allow binding of Annexin V-APC to phosphatidylserine residues externally displayed on apoptotic cells. Following the incubation, 5 μL propidium iodide (PI) was supplemented to each tube to stain the cells that had lost membrane integrity, indicative of late apoptosis or necrosis. After gently vortexing the tubes, the cells were brooded for extra 5 minutes at normal temperature in dark. Following staining, 400 μL 1 × binding buffer was supplied to every tube. Samples were construed on one flowing cytometer equipped with a 633 nm red laser for APC and a 488 nm blue laser for PI detection. Over 10000 cases were written down for all samples.
The immunofluorescence staining and confocal microscopy processes followed prior approaches[37]. In brief, cells were cultivated on coverslips in 24-well plates, rinsed using PBS, and then immobilized within one/fixation buffer at 4 °C all night. Subsequently, they were delivered to cold PBS and hatched at 4 °C all night. In terms of staining, sections were washed three times using PBS and then incubated using blocking buffer (5% bovine albumin, 0.1% Tween in TBS) for half an hour at ordinary temperature. The primary antibodies were attenuate in blocking buffer and used in the cells, before overnight hatching at 4 °C. Following three washes using TBST, the secondary antibody was utilized and brooded in the dark for 2 hours at ordinary temperature. After three additional washing using TBST, the sections were graphed through one Leica TCS SP5 confocal microscope with consistent imaging parameters at the central laboratory of the School of Life Sciences, Nankai University. Primary antibodies adopted were listed below: ASH1 antibody (1:1000, United Kingdom) and zonula occludens-1 (ZO1) antibody (1:1000, United States). Secondary antibodies were adopted as follows: Anti-mouse IgG Alexa Fluor 594 (115-585-044), anti-mouse IgG Alexa Fluor 488 (715-545-150), anti-rabbit IgG Alexa Fluor 594 (111-585-144), and anti-rabbit IgG Alexa Fluor 488 (711-545-152), supplied by Jackson ImmunoResearch Laboratories at dilutions of 1:500-1:1000.
The animal protocol was designed to minimize pain or discomfort to the animals. C-nude/BALB male mice aged 4-6 weeks were supplied by the Vital River Laboratory Animal Centre of China. Each group comprised 5 nude mice, with each mouse being subcutaneously injected with 5 × 106 target cells. The mice were housed within one sterile animal device; besides, tumor growth was supervised every day. Tumor volume was assessed weekly through vernier calipers and figured out through the formula V = length × width2 × 0.25. Following 4 weeks, the mice were euthanized; besides, the tumors were excised and explored histologically. The animal experiment was approved by the Animal Experiment Ethics Committee of the Institute of Radiation Medicine, Chinese Academy of Medical Sciences and Peking Union Medical College, Tianjin (protocol number: IRM/2-IACUC-2408-027).
The interaction between miR-142-3p and the 3’ untranslated region (UTR) of ASH1L was confirmed in 97H HCC cells using a group transfection approach with a lentivirus overexpressing miR-142-3p, targeting the double luciferase reporter construct containing the ASH1L 3’ UTR sequence from nucleotides 955 to 961 (refer to Supplementary Figure 1). Fluorescent probes were designed based on the miR-142-3p and ASH1L 3’ UTR sequences. These tissue slices were subjected to dual staining through fluorescence in situ hybridization and subsequently captured using laser confocal microscopy. The hsa-miR-142-3p probe was labeled with (5’HEX) in red, while the ASH1L probe was labeled with (5’6-FAM) in green. The sequences of the probes were designed as follows: ASH1L probe, ACCGTGCCCTTCTGACCCCA-5’6 -FAM; Hsa-miR-142-3p probe, TCCATAAAGTAGGAAACACTACA -5’HEX.
We obtained segments each kilobase per million mapped fragments (FPKM) and count values originating in TCGA database through the genomic data commons. The FPKM values were subsequently changed to transcripts each kilobase million for further analysis. Additionally, gene expression and clinical data for HCC patients were found in the international cancer genome consortium (ICGC) database, yielding a final cohort of 212 samples. For the GEO datasets, GSE76427 and GSE14520 were downloaded from the GEO website using “GEOquery” R package. The comparative abundance of 28 immune cell categories in HCC across different datasets were then figured out through the single-sample gene set enrichment analysis algorithm on the grounds of gene sets identified in Charoentong’s study[39].
Analysis of differential gene expression was executed through an empirical Bayesian approach with the “limma” R package. Genes were considered differentially expressed if they had an absolute log2 fold change over 2.5 and a regulated P value below 0.05. Gene ontology, and Kyoto encyclopedia of genes and genomes gene sets were supplied by MSigDB for subsequent gene set enrichment analysis.
Statistical analyses were conducted through statistical product and service solutions 26.0 software. A two-tailed P value below 0.05 held statistical significance. The relationship between ASH1L expression and clinicopathological parameters was assessed through the χ2 test. Survival analysis was implemented through the Kaplan-Meier approach; besides, contrasts were made through the log-rank test. The relationship between ASH1L expression and the biological activity of HCC cell lines was evaluated by one-way analysis of variance and t test.
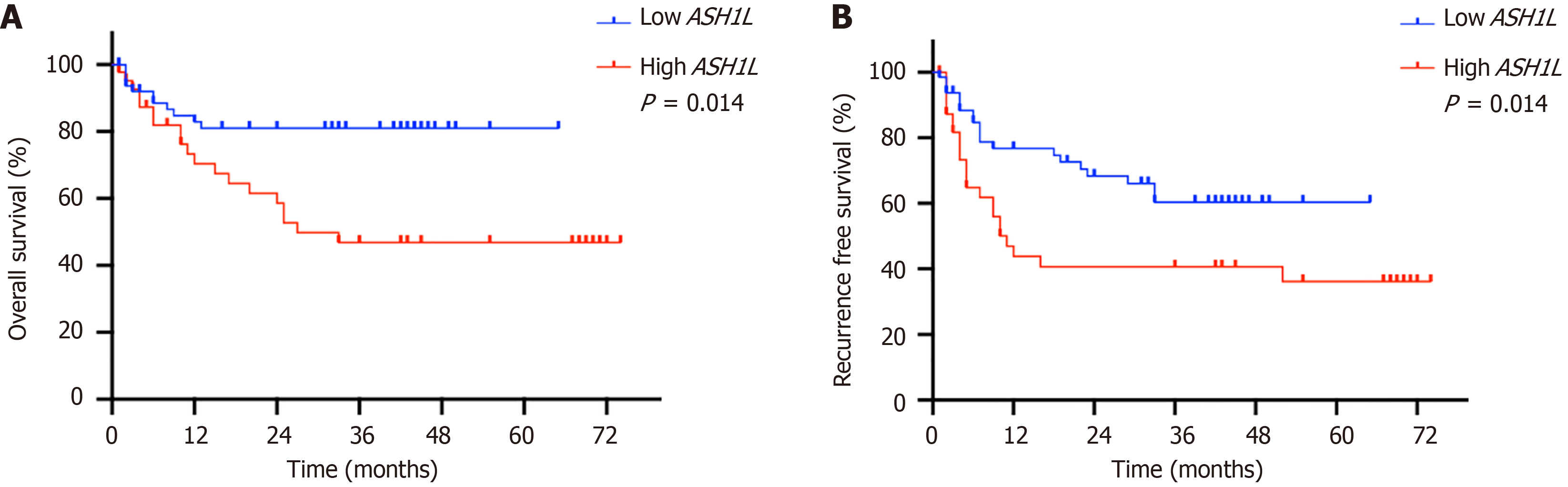
Of 111 human tissue samples were gathered with the expression levels of ASH1L analyzed through immunohistochemistry (IHC) (Figure 1A-C and Supplementary Figure 2). Contrasting the expression levels between healthy and cancer tissues from the identical participator, stronger ASH1L staining intensity emerged in tumor samples than that in controls (Figure 1A). Furthermore, fresh tumor tissues were obtained with liver tissues from 7 HCC patients for protein and RNA extraction, which confirmed the high expression of ASH1L in tumor tissues at both the protein and RNA levels (cP < 0.001, dP < 0.0001 t-tests) (Figure 1D-F). We categorized all samples into the low group (44 cases) and the high group (67 cases) on the grounds of signal intensity and analyzed their clinicopathological features (Table 1). Notably, the ASH1L high-expression group exhibited elevated blood levels of well-known liver function markers, including total bilirubin and aspartate aminotransferase, suggesting a greater liver function burden (Table 2). Moreover, the high group related to a more advanced clinicopathological stage than the low group, which indicates a more serious disease among patients (Table 3). Additionally, patients in the high group possess a more recurrence rate as well as a decreased overall survival rate (Figure 2 and Table 3).
| Characteristics | Number of cases | Percentage of cases (%) | |
| Age (year) | < 60 | 78 | 70.3 |
| ≥ 60 | 33 | 29.7 | |
| Gender | Male | 93 | 83.8 |
| Female | 18 | 16.2 | |
| Hepatitis | HBV | 89 | 80.2 |
| HCV | 13 | 11.7 | |
| None | 9 | 8.1 | |
| Cirrhosis | Y | 109 | 98.2 |
| N | 2 | 1.8 | |
| AFP (ug/L) | < 400 | 71 | 64 |
| ≥ 400 | 40 | 36 | |
| Meld score | ≤ 14 | 108 | 97.3 |
| 15-18 | 3 | 2.7 | |
| > 18 | 0 | 0 | |
| Clinicopathological stag | I | 27 | 24.3 |
| II | 34 | 30.6 | |
| III | 50 | 45.1 | |
| IV | 0 | 0 | |
| MVI | Y | 28 | 25.2 |
| N | 83 | 74.8 | |
| Pathological differentiation | Well | 1 | 0.9 |
| Moderate | 69 | 62.1 | |
| Poor | 33 | 29.7 | |
| Undifferentiated | 8 | 7.2 | |
| Number of tumors | Single | 48 | 43.2 |
| Multiple | 63 | 56.8 | |
| Satellite nodules | N | 88 | 79.3 |
| Y | 23 | 20.7 | |
| Relapse states (at follow-up) | N | 69 | 62.2 |
| Y | 42 | 37.8 | |
| Vital states (at follow-up) | Alive | 81 | 73 |
| Dead | 30 | 27 | |
| Expression of ASH1L | Low expression | 67 | 60.4 |
| High expression | 44 | 39.6 | |
| Variable | High expression (n = 44) | Low expression (n = 67) | P value |
| ALT (U/L) | 36.30 (24.40-56.35) | 34.50 (20.60-47.90) | 0.228 |
| AST (U/L) | 60.85 (42.20-104.13) | 41.20 (27.30-64.20) | 0.003 |
| TB (μmol/L) | 36.49 (17.53-75.96) | 21.19 (13.70-37.71) | 0.021 |
| Scr (μmol/L) | 65.05 (58.03-80.00) | 66.00 (53.00-77.00) | 0.424 |
| ALB (g/L), mean ± SD | 34.94 ± 5.98 | 35.20 ± 6.02 | 0.878 |
| AFP (ng/mL) | 215.90 (10.45-1705.50) | 76.68 (6.34-1210.00) | 0.187 |
| WBC (× 109/L) | 4.23 (2.80-5.77) | 4.51 (2.98-6.14) | 0.387 |
| HGB (g/L), mean ± SD | 120.84 ± 24.98 | 125.09 ± 22.44 | 0.458 |
| PLT (× 109/L) | 108.00 (55.25-145.75) | 106.0 (64.00-163.00) | 0.623 |
| INR | 1.19 (1.06-1.44) | 1.21 (1.09-1.36) | 0.594 |
| PT (second) | 12.95 (11.53-14.98) | 13.60 (12.20-15.30) | 0.197 |
| Variable | High expression (n = 44) | Low expression (n = 67) | P value |
| Gender (M/F) | 37/7 | 56/11 | 0.944 |
| Age (year) | 54.73 ± 8.56 | 54.63 ± 9.67 | 0.814 |
| Hepatitis (HBV/HCV/none) | 36/4/4 | 52/9/5 | 0.487 |
| Cirrhosis (Y/N) | 32/12 | 64/2 | 0.25 |
| MELD score | 7.67 (5.57-9.65) | 6.62 (5.43-8.09) | 0.071 |
| Number of tumors (multiple/single) | 30/14 | 33/34 | 0.05 |
| Maximum diameter of tumor (cm) | 3.50 (2.00-6.00) | 3.00 (2.00-6.00) | 0.954 |
| Satellite cooker (Y/N) | 12/32 | 11/56 | 0.169 |
| MVI (Y/N) | 16/28 | 12/55 | 0.029 |
| Tumor differentiation (well/moderate/poor/undifferentiation) | 0/28/15/1 | 1/40/19/7 | 0.649 |
| Clinicopathological stage (I/II/III/IV) | 7/12/25/0 | 20/22/25/0 | 0.032 |
| Relapse states (N/Y) | 22/22 | 47/22 | 0.002 |
| Vital states (alive/dead) | 25/19 | 56/11 | 0.033 |
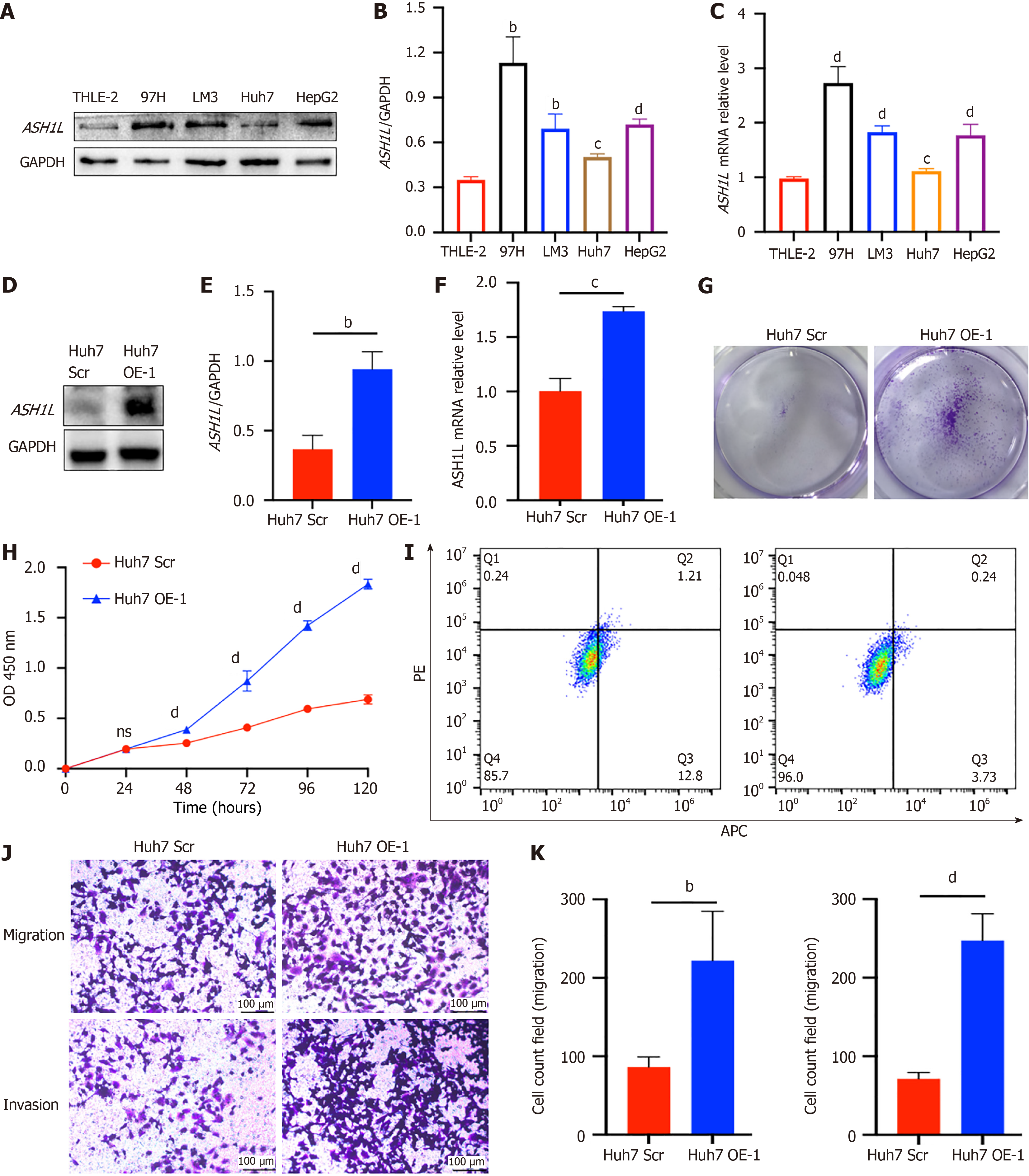
Expression levels of ASH1L in the cell lines were compared via Western blot analysis (Figure 3A and B), revealing high expression in 97H cells and the lowest Huh7 expression (aP < 0.05, bP < 0.01, cP < 0.001) (one-way analysis of variance and t-tests). Data was deeply validated via assessing mRNA levels through quantitative real-time polymerase chain reaction (RT-qPCR) (Figure 3C) (dP < 0.0001) (one-way analysis of variance and t-tests). Subsequently, ASH1L OE and knockdown (KD) cell lines were generated through the utilization of lentivirus. The Huh7 cell line, known for its comparatively lower expression level in contrast to other cell lines, was employed for this purpose. Subsequently, stable ASH1L OE was successfully established in Huh7 cells, as validated through RT-qPCR and western blot analysis (Figure 3D-F) (bP < 0.01, cP < 0.001) (t-tests). Proliferation experiments utilizing plate colony formation (Figure 3G) and CCK-8 (Figure 3H) manifested a prominent enhancement in HCC cell proliferation, on account of the OE of ASH1L (dP < 0.0001) (one-way analysis of variance and t-tests). Next, flow cytometry was adopted to show that the HCC cell increased in the second and fourth quadrants and these cells were apoptotic and necrotic cells which were significantly reduced for ASH1L OE (Figure 3I). Additionally, transwell assay also demonstrated that ASH1L OE facilitated the transfer and aggression of HCC cells (Figure 3J-K) (bP < 0.01, dP < 0.0001) (t-tests). To further validate these results, HepG2 scrambled (Scr) and HepG2 OE cell lines were employed (Supplementary Figure 3) (cP < 0.001, dP < 0.0001) (one-way analysis of variance and t-tests). Cumulatively, data powerfully indicate that ASH1L boosts the HCC cell growth, transfer, and invasion in vitro.
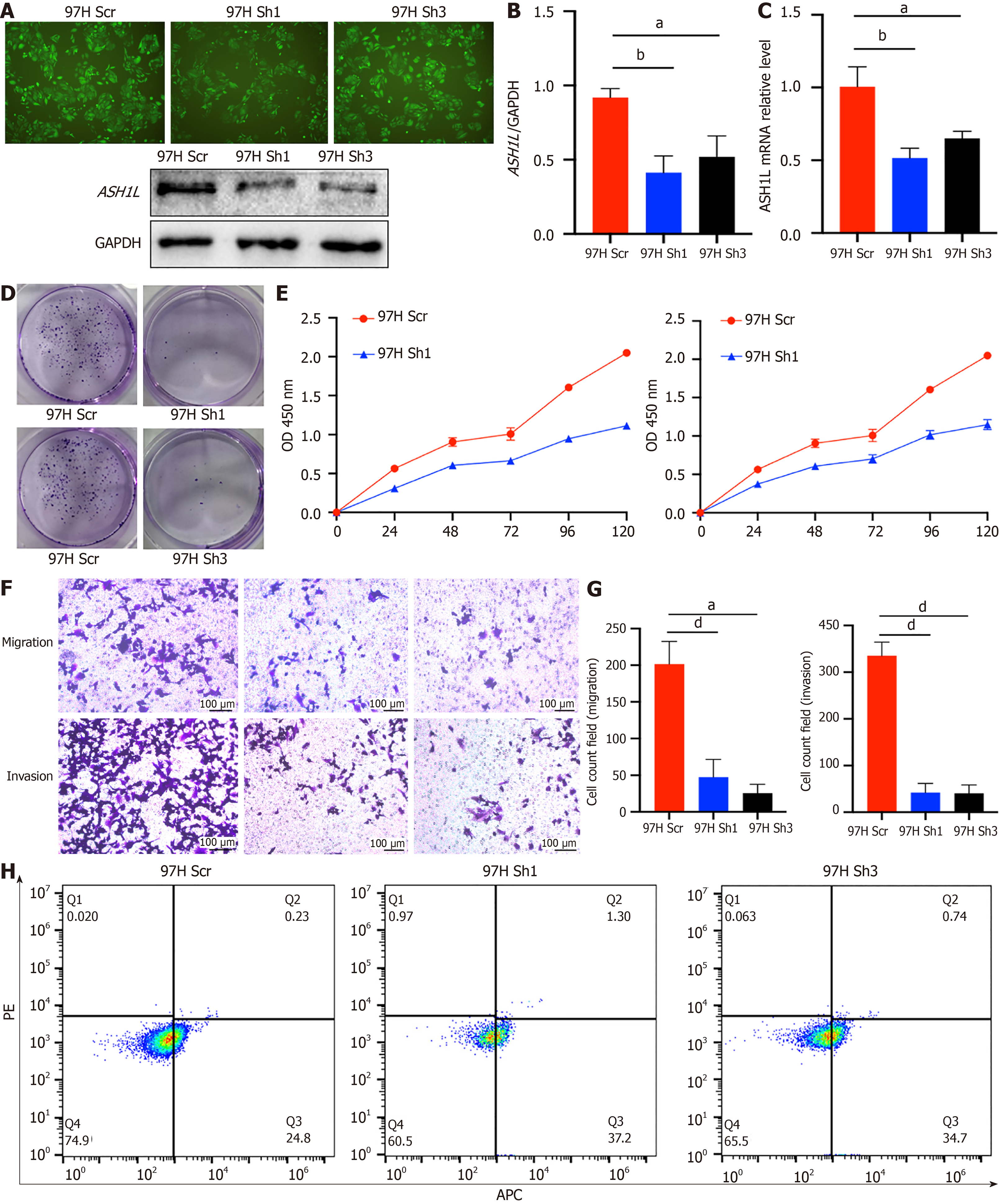
To mitigate potential side effects of OE, we implemented KD ASH1L. At first, we smoothly executed ASH1L KD in 97H cell line and confirmed stable passaging of this cell line through Western blotting (Figure 4A and B) and RT-qPCR (Figure 4C) (aP < 0.05, bP < 0.01, dP < 0.0001) (one-way analysis of variance and t-tests). Proliferation experiments utilizing plate colony formation (Figure 4D) and CCK-8 (Figure 4E) clearly displayed that ASH1L depletion hindered the proliferative capacity of HCC cells (bP < 0.01, dP < 0.0001) (one-way analysis of variance and t-tests). Given the close association of invasion and migration with tumor progression, our research deeply investigated the abilities through Transwell assays. Experimentally, it was indicated that a prominent decrease in HCC cell transfer and invasion abilities after ASH1L knocking down (Figure 4F and G) (dP < 0.0001) (one-way analysis of variance and t-tests). Next, flow cytometry was used to show that the HCC cell increased in the second and fourth quadrants which meant that apoptosis was significantly increased due to ASH1L knock-out (Figure 4H). Moreover, all findings supplied by the 97H cell line were authenticated in the HepG2 KD and HepG2 Scr cell lines (Supplementary Figure 4) (aP< 0.05, bP < 0.01, cP < 0.001, dP < 0.0001) (one-way analysis of variance and t-tests).
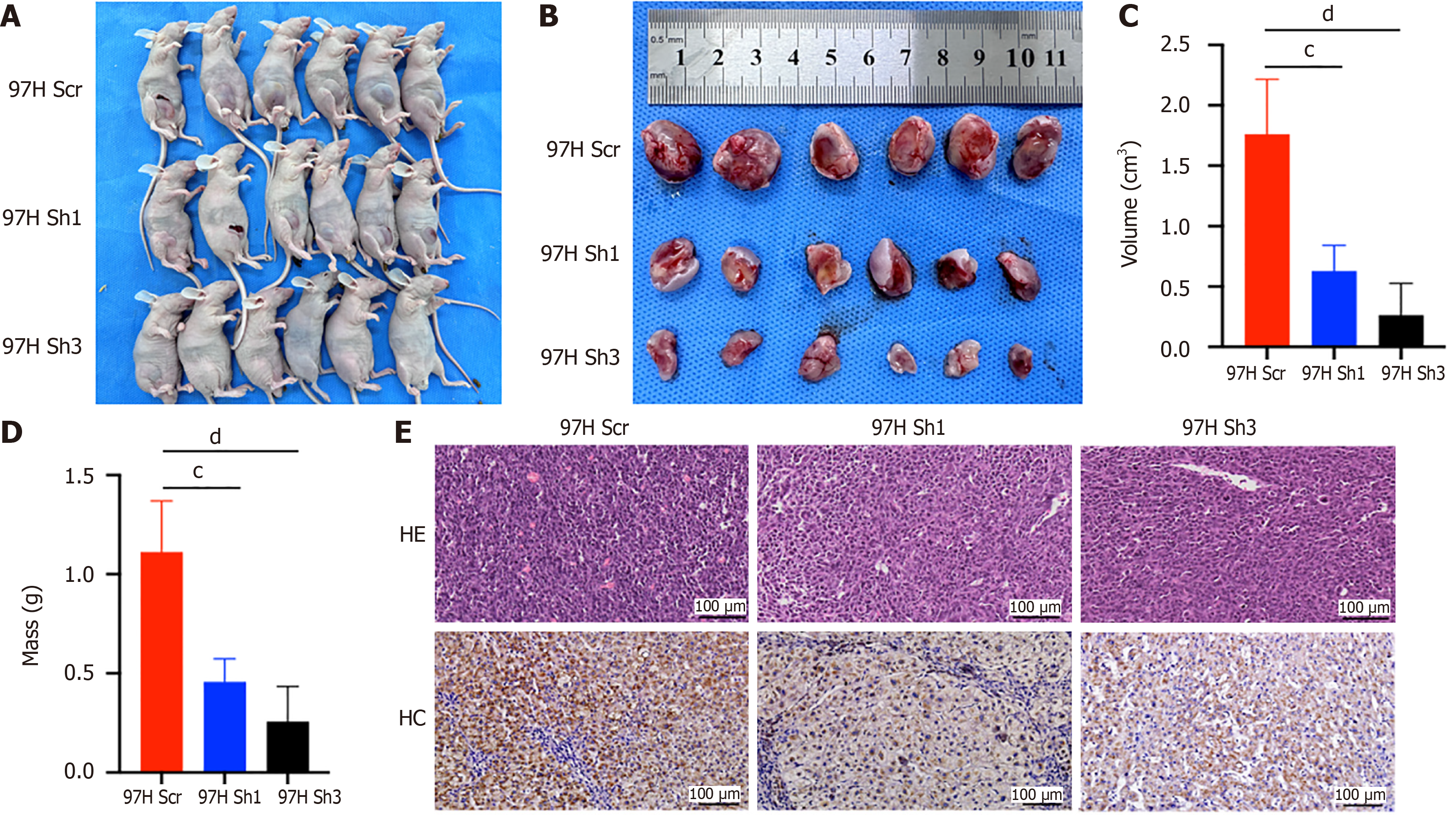
To rate the effect of ASH1L on HCC cell tumorigenic capability in vivo, we utilized three distinct cell lines-97H Scr, 97H Sh1, and 97HSh 3-to establish a subcutaneous tumorigenic model in right inguinal region of BALB/C-nude mice (Figure 5A and B). Following 2 weeks, the subcutaneous tumors were harvested for quantitative evaluation of volume and mass, as well as for tissue sectioning, hematoxylin and eosin staining, and IHC staining. The findings revealed that the volume (Figure 5C) and mass (Figure 5D) of subcutaneous tumors in BALB/C-nude mice in the ASH1L KD group were significantly smaller (cP < 0.001, dP < 0.0001) (one-way analysis of variance and t-tests). IHC staining indicated that the intensity of staining in tumors from the control group was stronger than the KD groups (Figure 5E). Experimentally, it is indicated that suppressing the ASH1L expression impacts the HCC cell tumorigenicity in vivo.
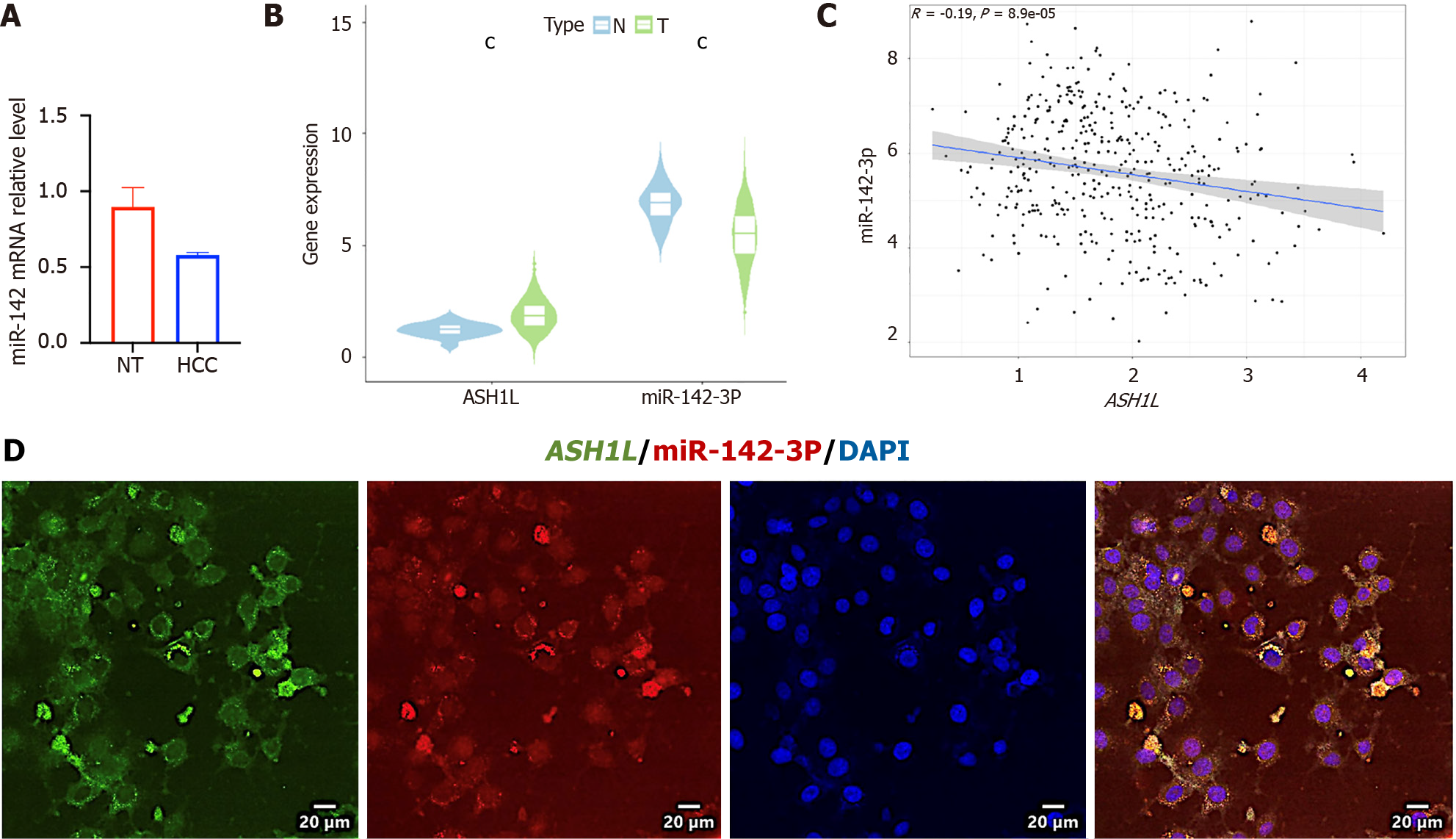
Fresh cancer tissues and liver tissues were gathered from 7 HCC patients for RNA extraction. The low miR-142-3p expression among tumor tissues and cell lines was verified at the RNA level (Figure 6A and Supplementary Figure 1) (aP < 0.05) (t-tests). To further explore the expression variances between the two molecules in cancer and paracancerous tissues, as well as their expression correlations, the TCGA database was utilized for preliminary analyses using the Wilcox Test (Figure 6B) and spearman test (Figure 6C) (cP < 0.001) (t-tests). The findings indicate that the low miR-142-3p expression occurred among HCC tissues, whilst the ASH1L expression was high (Figure 6B). Moreover, miR-142-3p exhibited a negative correlation with ASH1L (Figure 6C). To validate the directness of the negative pertinence between miR-142-3p and ASH1L, a Fluorescence in situ hybridization co-localization experiment of miR-142-3p and ASH1L 3’ UTR among 97H cells was performed, demonstrating that miR-142-3p and ASH1L were primarily expressed in the cytoplasm with binding presence (Figure 6D).
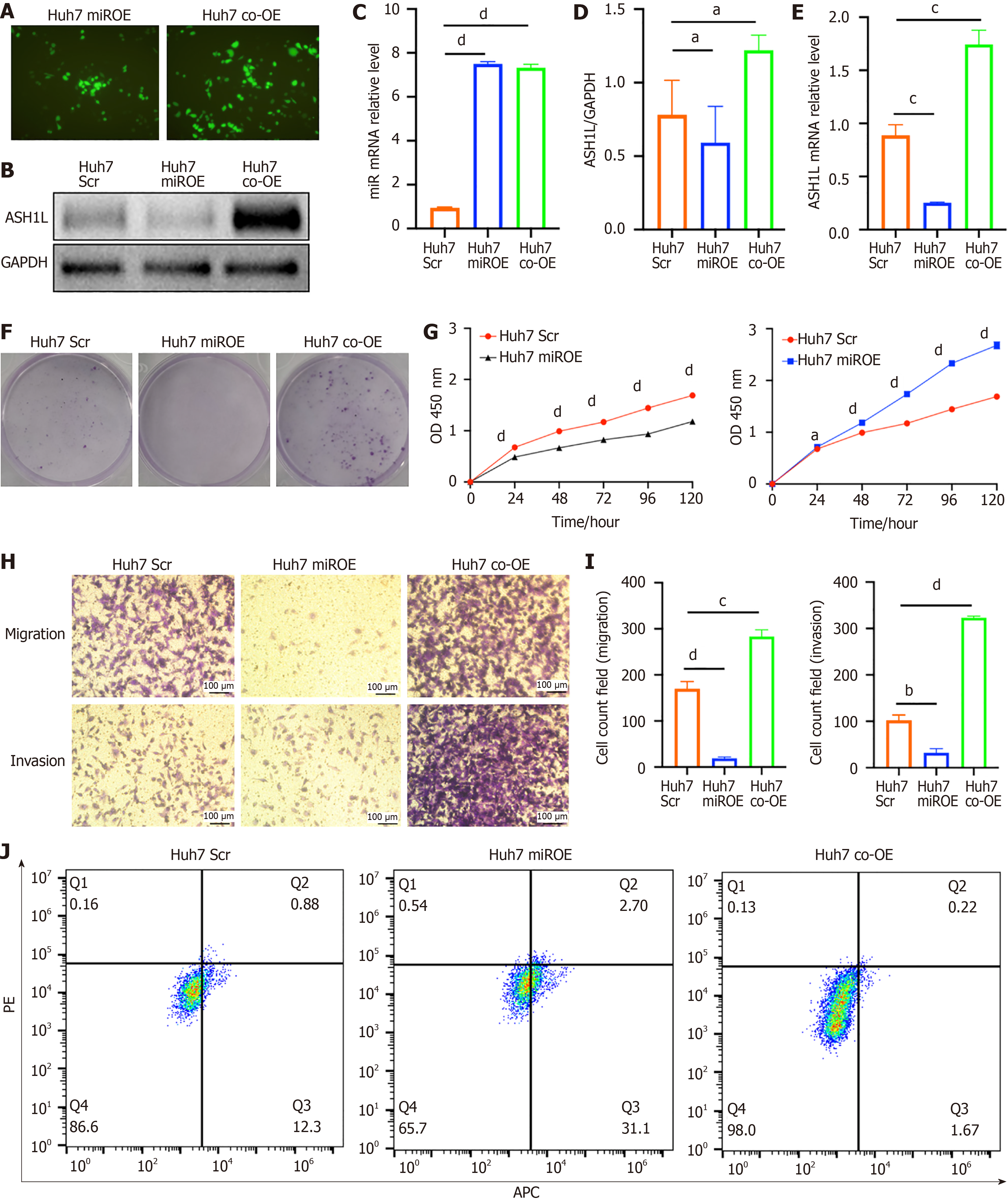
Next, we generated miR-142-3p OE and ASH1L OE cell lines via lentivirus. Specifically, our study utilized the Huh7 cell line and successfully established stable miR-142-3p and ASH1L OE among Huh7 cells, which was validated through fluorescence imaging and RT-qPCR analysis (Figure 7A-E) (aP < 0.05, cP < 0.001, dP < 0.0001) (one-way analysis of variance and t-tests). Including plate colony formation (Figure 7F) and CCK-8 assays (Figure 7G) (aP < 0.05, dP < 0.0001) (one-way analysis of variance and t-tests), proliferation assays revealed that miR-142-3p OE suppressed the HCC cell proliferation, while ASH1L OE significantly enhanced proliferation. Transwell assays demonstrated that miR-142-3p OE suppressed the invasion and transfer of HCC cells, whereas ASH1L OE promoted these processes (Figure 7H and I) (aP < 0.05, cP < 0.001, dP < 0.0001) (one-way analysis of variance and t-tests). Flow cytometry was adopted to show that the HCC cell increased in the second and fourth quadrants which meant that apoptosis was significantly increased due to miR-142-3p OE (Figure 7J). Furthermore, we corroborated these findings using 97H Scr, miR, and KD cell lines (Supplementary Figure 5) (bP < 0.01, cP < 0.001, dP < 0.0001) (one-way analysis of variance and t-tests). In summary, miR-142-3p suppresses the HCC cell growth, invasion, and metastasis in vitro by targeting ASH1L.
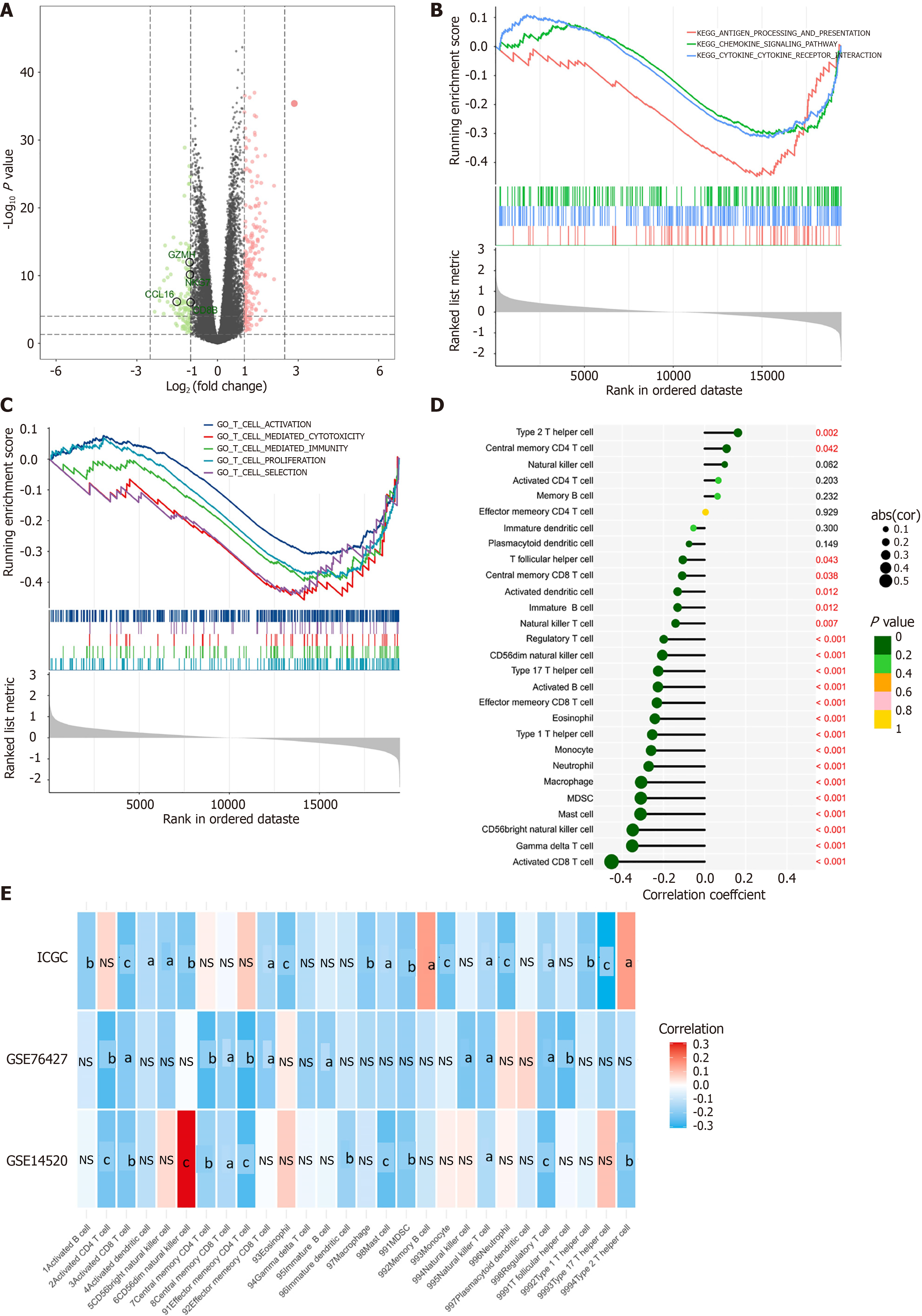
We identified ASH1L as a crucial factor in the development and progression of HCC. This led us to investigate its potential role in immune evasion. Using RNA-seq data from TCGA-liver hepatocellular carcinoma, we categorized patients into low and high ASH1L expression groups on the grounds of the median level. Interestingly, patients with high ASH1L expression exhibited significantly lower levels of GZMH, NKG7, CCL16, and CD8B (Figure 8A), all of which are key players in the immune response’s cancer-killing process. Furthermore, pathway enrichment analysis proved that elevated ASH1L expression related to significant inhibition of antigen processing, chemokine/cytokine-related pathways (Figure 8B), and T-cell-related pathways (Figure 8C). Immune infiltration analysis showed a powerful negative relationship between ASH1L expression and infiltration levels of nearly all immune cells (Figure 8D and E). These findings were further validated using the ICGC, GSE76427, and GSE14520 datasets, collectively suggesting that ASH1L contributes to an immunosuppressive microenvironment in HCC.
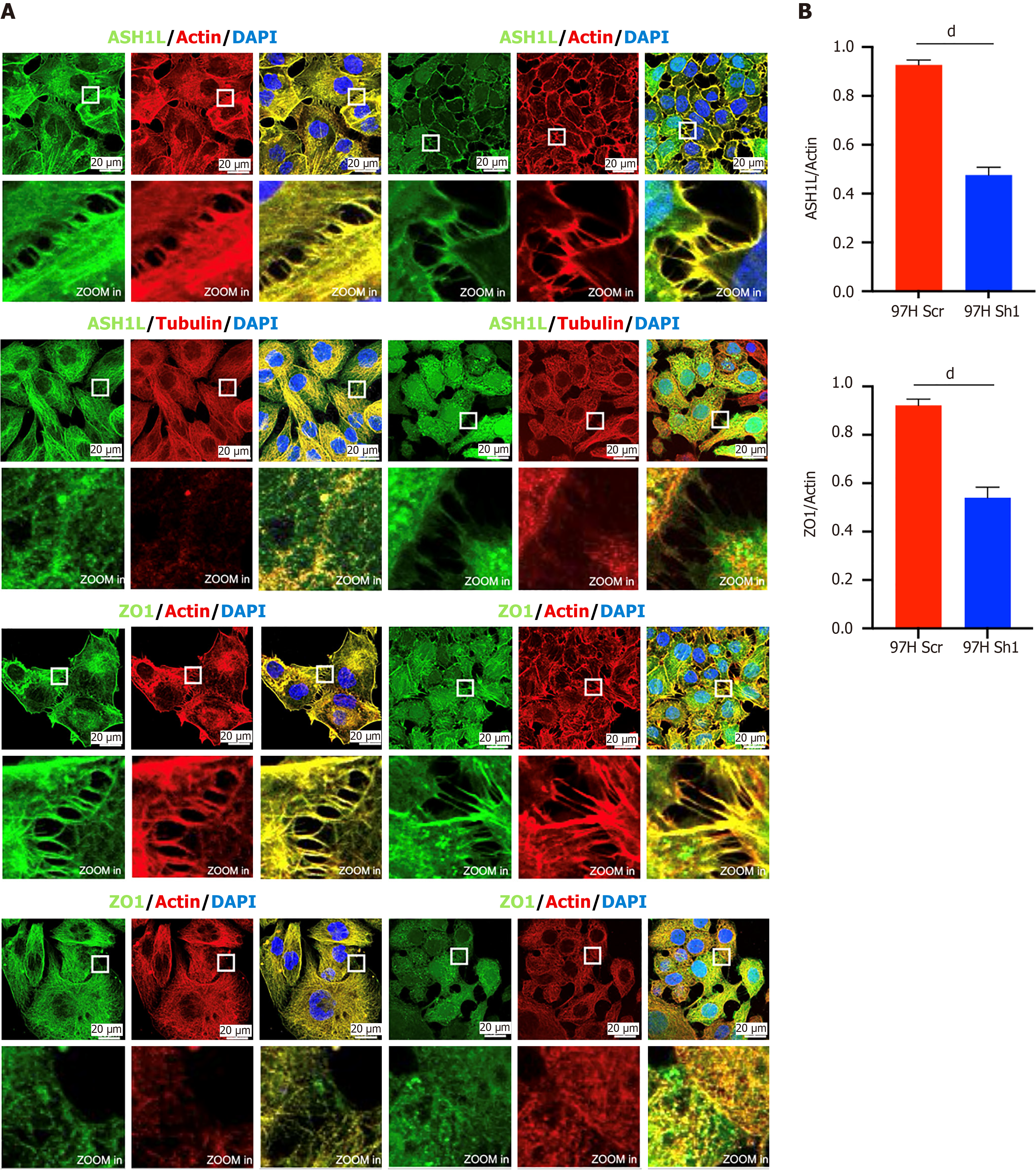
In our investigation of the effect of ASH1L on cell invasion and transfer, we observed that ASH1L is expressed at cell junctions. Subsequent reduction of ASH1L expression through lentiviral treatment led to a decrease in ASH1L presence at cell junctions (Figure 9). Notably, a concurrent decrease was noted in the levels of the classical cell junction protein ZO1 with the decrease in ASH1L expression (Figure 9) (dP < 0.0001) (t-tests). These findings were consistent across experiments conducted on the HepG2 cell line (Supplementary Figure 6) (dP < 0.0001) (t-tests). Furthermore, several studies have shown that ZO1 is associated with immunity[40,41] and we validated ASH1L could contributes to an immunosuppressive microenvironment through affecting the expression of ZO1.
ASH1L is a member of the trithorax group, consisting of transcriptional activating proteins regulating gene expression. ASH1L genes undergo transcription, and produce an mRNA of around 10.5 kb, translated into a 2962-residue protein[12,13]. It comprises an AT Hook domain, an SET domain, a bromodomain, and a PHD finger. ASH1L is predominantly found in numerous small nuclear speckles and cell-cell junctions[12,14]. Prior research has investigated the pathology implications of ASH1L among various human illnesses through structural and functional analyses[37]. Nevertheless, limited data exist concerning the ASH1L role in HCC development. This study aims to address this gap by exploring the possible role and the ASH1L mechanisms in the progress of HCC.
Experimental results highly suggest ASH1L’s engagement in HCC cell lines apoptosis and tumor progression, illustrating its potential for a new prognostic biomarker for patients suffering from liver cancer. Human tissue staining unveiled improved ASH1L expression among tumor tissues than that among normal tissues. More than 100 HCC patient tissues were gathered. Survival curves also revealed a less overall survival rate of patients in the high group. Addi
In our research findings, ASH1L was shown to effectively inhibit apoptosis in HCC cell lines. Apoptosis is an important player in sustaining the health and stability of tissues by eliminating damaged or unnecessary cells. When the process of apoptosis is disrupted, such as by inhibition through cancer-related genes, it can lead to unregulated cell survival, thereby promoting tumor formation and growth. Through a series of cellular and molecular biology experiments, we observed that HCC cells overexpressing ASH1L exhibited a significantly reduced rate of apoptosis. Flow cytometry analysis using APC/PI staining confirmed a lower percentage of apoptotic cells in ASH1L overexpressing HCC cell lines compared to control groups. The discoveries underscore the vital function of ASH1L in adjusting apoptosis pathways and its potential impact on the aggressiveness and progression of HCC.
We conducted a deeper investigation into miR-142-3p, an upstream regulator of ASH1L, whose altered expression has frequently been linked to various human cancers in the literature[7], including colon cancer[9], leukemia[10], and pancreatic ductal adenocarcinoma[11]. Analysis of the TCGA database exhibited a relationship between ASH1L and miR-142-3p. Functional miR-142-3p readouts were obtained by establishing the miR 97H and miR Huh7 cell lines, and conducting in vitro apoptosis, growth, transfer, and invasion experiments. These experiments denoted that miR-142-3p inhibited ASH1L expression, cell lines apoptosis, and the bioactivity of HCC cell lines.
Our results showed the impact of ASH1L KD on 97H cell lines in immunofluorescence (IF) analysis; besides, our RNAseq data revealed that the ASH1L influenced genes engaging in cellular metabolism. As a histone methyltransferase, ASH1L was classified as a set of transcriptional activating proteins. While the ASH1L pathology among various types of carcinomas have been documented, our present RNAseq analysis offers valuable perceptions on discerning the genes regulated by ASH1L in HCC. In-depth exploration to elucidate the specific ASH1L pathology and its relevant molecular mechanisms in HCC progression is guaranteed.
Current studies have even recognized a new ASH1L suppressor and proven its efficiency in curing mixed lineage leukemia leukemia[42]. ASH1L comprises a catalytic SET field and three chromatin reader fields: PHD, bromo adjacent homology, and bromodomain[43]. These researches specifically focused on the SET field of ASH1L, Alaska airlines 99 (AS-99). They acknowledged the SET field as one valuable chemical probe for investigating the ASH1L roles[43]. Data presents new chances of developing ASH1L suppressors as a promising therapy for HCC. The next phase of my research will further investigate the safety and efficacy of AS-99, an ASH1L inhibitor, in HCC.
Many HCC biomarkers are extensively examined, with alpha-fetoprotein (AFP) as the highest rated biomarker experiencing all five stages of biomarker rating[44]. The efficiency of other alternative biomarkers is commonly evaluated via contrasting them with AFP. As an illustration, as a liposomal enzyme, α-L-fucosidase has exhibited a prominent rise among HCC patients in comparison to benign tumor patients[45,46]. Moreover, it has been observed that Golgi protein 73 (GP73) is highly expressed among HCC patients[47], therefore empowering its alternatives as a novel biomarker. Our findings contribute these biomarkers, suggesting that the ASH1L can feature as a new HCC biomarker. In order to validate this assumption, serum samples from numerous studies will be indispensable, just like the research implemented on GP73.
To sum up, this research put forward convincing proof ASH1L’s engagement in HCC progression. On the grounds of the current proof, we present that ASH1L can become both an underlying therapeutic target and a precious prognostic biomarker for HCC. Additionally, our discoveries illustrate the effect of ASH1L expression on cell metastasis, invasion, and proliferation, highlighting its function in HCC development. Though RNAseq and IF, we initiated the exploration of the molecular mechanisms leading to the ASH1L stimulation effect on HCC progression. In-depth comprehensive research is indispensable to completely clarity the ASH1L pathology and relevant molecular mechanisms in HCC progression. And, we will also apply ASH1L’s small molecule inhibitor AS-99 for further study.
Loss function of miR-142-3p induces cancer progression and immune evasion through upregulation of ASH1L in HCC. Both miR-142-3p and ASH1L can feature as new biomarker for HCC in the future, and we will further explore the mechanism of action of miR-142-3p and ASH1L.
The authors would like to acknowledge Yoshida Sei (Nankai University, Tianjin, China) for experimental technical assistance.
| 1. | Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S, Lencioni R, Koike K, Zucman-Rossi J, Finn RS. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7:6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4432] [Cited by in RCA: 3887] [Article Influence: 971.8] [Reference Citation Analysis (3)] |
| 2. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64702] [Article Influence: 16175.5] [Reference Citation Analysis (177)] |
| 3. | Fu J, Wang H. Precision diagnosis and treatment of liver cancer in China. Cancer Lett. 2018;412:283-288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 224] [Article Influence: 32.0] [Reference Citation Analysis (1)] |
| 4. | Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16:589-604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2184] [Cited by in RCA: 2908] [Article Influence: 484.7] [Reference Citation Analysis (17)] |
| 5. | Xu X, Chen J, Wei Q, Liu ZK, Yang Z, Zhang M, Wang GY, Gao J, Yang ZX, Guo WY, Xing TH, Shao Z, Xie QF, Zheng SS. Clinical practice guidelines on liver transplantation for hepatocellular carcinoma in China (2018 edition). Hepatobiliary Pancreat Dis Int. 2019;18:307-312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (1)] |
| 6. | Castelli G, Pelosi E, Testa U. Liver Cancer: Molecular Characterization, Clonal Evolution and Cancer Stem Cells. Cancers (Basel). 2017;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 117] [Article Influence: 14.6] [Reference Citation Analysis (1)] |
| 7. | Tsang FH, Au SL, Wei L, Fan DN, Lee JM, Wong CC, Ng IO, Wong CM. MicroRNA-142-3p and microRNA-142-5p are downregulated in hepatocellular carcinoma and exhibit synergistic effects on cell motility. Front Med. 2015;9:331-343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 41] [Article Influence: 4.1] [Reference Citation Analysis (1)] |
| 8. | Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foà R, Schliwka J, Fuchs U, Novosel A, Müller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401-1414. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3103] [Cited by in RCA: 2974] [Article Influence: 165.2] [Reference Citation Analysis (1)] |
| 9. | Shen WW, Zeng Z, Zhu WX, Fu GH. MiR-142-3p functions as a tumor suppressor by targeting CD133, ABCG2, and Lgr5 in colon cancer cells. J Mol Med (Berl). 2013;91:989-1000. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 113] [Article Influence: 9.4] [Reference Citation Analysis (1)] |
| 10. | Trissal MC, Wong TN, Yao JC, Ramaswamy R, Kuo I, Baty J, Sun Y, Jih G, Parikh N, Berrien-Elliott MM, Fehniger TA, Ley TJ, Maillard I, Reddy PR, Link DC. MIR142 Loss-of-Function Mutations Derepress ASH1L to Increase HOXA Gene Expression and Promote Leukemogenesis. Cancer Res. 2018;78:3510-3521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 36] [Article Influence: 5.1] [Reference Citation Analysis (1)] |
| 11. | MacKenzie TN, Mujumdar N, Banerjee S, Sangwan V, Sarver A, Vickers S, Subramanian S, Saluja AK. Triptolide induces the expression of miR-142-3p: a negative regulator of heat shock protein 70 and pancreatic cancer cell proliferation. Mol Cancer Ther. 2013;12:1266-1275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 115] [Article Influence: 9.6] [Reference Citation Analysis (1)] |
| 12. | Nakamura T, Blechman J, Tada S, Rozovskaia T, Itoyama T, Bullrich F, Mazo A, Croce CM, Geiger B, Canaani E. huASH1 protein, a putative transcription factor encoded by a human homologue of the Drosophila ash1 gene, localizes to both nuclei and cell-cell tight junctions. Proc Natl Acad Sci U S A. 2000;97:7284-7289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 89] [Article Influence: 3.6] [Reference Citation Analysis (1)] |
| 13. | Gregory GD, Vakoc CR, Rozovskaia T, Zheng X, Patel S, Nakamura T, Canaani E, Blobel GA. Mammalian ASH1L is a histone methyltransferase that occupies the transcribed region of active genes. Mol Cell Biol. 2007;27:8466-8479. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 174] [Article Influence: 9.7] [Reference Citation Analysis (1)] |
| 14. | An S, Yeo KJ, Jeon YH, Song JJ. Crystal structure of the human histone methyltransferase ASH1L catalytic domain and its implications for the regulatory mechanism. J Biol Chem. 2011;286:8369-8374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 108] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 15. | Miyazaki H, Higashimoto K, Yada Y, Endo TA, Sharif J, Komori T, Matsuda M, Koseki Y, Nakayama M, Soejima H, Handa H, Koseki H, Hirose S, Nishioka K. Ash1l methylates Lys36 of histone H3 independently of transcriptional elongation to counteract polycomb silencing. PLoS Genet. 2013;9:e1003897. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 73] [Article Influence: 6.1] [Reference Citation Analysis (1)] |
| 16. | Rogawski DS, Ndoj J, Cho HJ, Maillard I, Grembecka J, Cierpicki T. Two Loops Undergoing Concerted Dynamics Regulate the Activity of the ASH1L Histone Methyltransferase. Biochemistry. 2015;54:5401-5413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 26] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 17. | Zheng W, Ibáñez G, Wu H, Blum G, Zeng H, Dong A, Li F, Hajian T, Allali-Hassani A, Amaya MF, Siarheyeva A, Yu W, Brown PJ, Schapira M, Vedadi M, Min J, Luo M. Sinefungin derivatives as inhibitors and structure probes of protein lysine methyltransferase SETD2. J Am Chem Soc. 2012;134:18004-18014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 116] [Article Influence: 8.9] [Reference Citation Analysis (1)] |
| 18. | Yang S, Zheng X, Lu C, Li GM, Allis CD, Li H. Molecular basis for oncohistone H3 recognition by SETD2 methyltransferase. Genes Dev. 2016;30:1611-1616. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 111] [Article Influence: 13.9] [Reference Citation Analysis (1)] |
| 19. | Lam UTF, Tan BKY, Poh JJX, Chen ES. Structural and functional specificity of H3K36 methylation. Epigenetics Chromatin. 2022;15:17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 36] [Article Influence: 12.0] [Reference Citation Analysis (1)] |
| 20. | Balbach ST, Orkin SH. An Achilles' Heel for MLL-Rearranged Leukemias: Writers and Readers of H3 Lysine 36 Dimethylation. Cancer Discov. 2016;6:700-702. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.6] [Reference Citation Analysis (1)] |
| 21. | Liu J, Lee W, Jiang Z, Chen Z, Jhunjhunwala S, Haverty PM, Gnad F, Guan Y, Gilbert HN, Stinson J, Klijn C, Guillory J, Bhatt D, Vartanian S, Walter K, Chan J, Holcomb T, Dijkgraaf P, Johnson S, Koeman J, Minna JD, Gazdar AF, Stern HM, Hoeflich KP, Wu TD, Settleman J, de Sauvage FJ, Gentleman RC, Neve RM, Stokoe D, Modrusan Z, Seshagiri S, Shames DS, Zhang Z. Genome and transcriptome sequencing of lung cancers reveal diverse mutational and splicing events. Genome Res. 2012;22:2315-2327. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 155] [Cited by in RCA: 164] [Article Influence: 12.6] [Reference Citation Analysis (1)] |
| 22. | Demelash A, Rudrabhatla P, Pant HC, Wang X, Amin ND, McWhite CD, Naizhen X, Linnoila RI. Achaete-scute homologue-1 (ASH1) stimulates migration of lung cancer cells through Cdk5/p35 pathway. Mol Biol Cell. 2012;23:2856-2866. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 71] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 23. | Liu L, Kimball S, Liu H, Holowatyj A, Yang ZQ. Genetic alterations of histone lysine methyltransferases and their significance in breast cancer. Oncotarget. 2015;6:2466-2482. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 127] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 24. | Xu B, Qin T, Yu J, Giordano TJ, Sartor MA, Koenig RJ. Novel role of ASH1L histone methyltransferase in anaplastic thyroid carcinoma. J Biol Chem. 2020;295:8834-8845. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 25. | Shahrisa A, Tahmasebi-Birgani M, Ansari H, Mohammadi Z, Carloni V, Mohammadi Asl J. The pattern of gene copy number alteration (CNAs) in hepatocellular carcinoma: an in silico analysis. Mol Cytogenet. 2021;14:33. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 26. | Fujimoto A, Furuta M, Totoki Y, Tsunoda T, Kato M, Shiraishi Y, Tanaka H, Taniguchi H, Kawakami Y, Ueno M, Gotoh K, Ariizumi S, Wardell CP, Hayami S, Nakamura T, Aikata H, Arihiro K, Boroevich KA, Abe T, Nakano K, Maejima K, Sasaki-Oku A, Ohsawa A, Shibuya T, Nakamura H, Hama N, Hosoda F, Arai Y, Ohashi S, Urushidate T, Nagae G, Yamamoto S, Ueda H, Tatsuno K, Ojima H, Hiraoka N, Okusaka T, Kubo M, Marubashi S, Yamada T, Hirano S, Yamamoto M, Ohdan H, Shimada K, Ishikawa O, Yamaue H, Chayama K, Miyano S, Aburatani H, Shibata T, Nakagawa H. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet. 2016;48:500-509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 465] [Cited by in RCA: 538] [Article Influence: 59.8] [Reference Citation Analysis (0)] |
| 27. | Zheng Y, Tang L, Chen G, Liu Z. Comprehensive Bioinformatics Analysis of Key Methyltransferases and Demethylases for Histone Lysines in Hepatocellular Carcinoma. Technol Cancer Res Treat. 2020;19:1533033820983284. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 28. | Panaccione A, Zhang Y, Mi Y, Mitani Y, Yan G, Prasad ML, McDonald WH, El-Naggar AK, Yarbrough WG, Ivanov SV. Chromosomal abnormalities and molecular landscape of metastasizing mucinous salivary adenocarcinoma. Oral Oncol. 2017;66:38-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 29. | Colamaio M, Puca F, Ragozzino E, Gemei M, Decaussin-Petrucci M, Aiello C, Bastos AU, Federico A, Chiappetta G, Del Vecchio L, Torregrossa L, Battista S, Fusco A. miR-142-3p down-regulation contributes to thyroid follicular tumorigenesis by targeting ASH1L and MLL1. J Clin Endocrinol Metab. 2015;100:E59-E69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 49] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 30. | Zhu L, Li Q, Wong SH, Huang M, Klein BJ, Shen J, Ikenouye L, Onishi M, Schneidawind D, Buechele C, Hansen L, Duque-Afonso J, Zhu F, Martin GM, Gozani O, Majeti R, Kutateladze TG, Cleary ML. ASH1L Links Histone H3 Lysine 36 Dimethylation to MLL Leukemia. Cancer Discov. 2016;6:770-783. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 125] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 31. | Tanaka Y, Kawahashi K, Katagiri Z, Nakayama Y, Mahajan M, Kioussis D. Dual function of histone H3 lysine 36 methyltransferase ASH1 in regulation of Hox gene expression. PLoS One. 2011;6:e28171. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 38] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 32. | Wong CM, Kai AK, Tsang FH, Ng IO. Regulation of hepatocarcinogenesis by microRNAs. Front Biosci (Elite Ed). 2013;5:49-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 33. | Elsemman IE, Mardinoglu A, Shoaie S, Soliman TH, Nielsen J. Systems biology analysis of hepatitis C virus infection reveals the role of copy number increases in regions of chromosome 1q in hepatocellular carcinoma metabolism. Mol Biosyst. 2016;12:1496-1506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 34. | Li GM, Wang YG, Pan Q, Wang J, Fan JG, Sun C. RNAi screening with shRNAs against histone methylation-related genes reveals determinants of sorafenib sensitivity in hepatocellular carcinoma cells. Int J Clin Exp Pathol. 2014;7:1085-1092. [PubMed] |
| 35. | Collett K, Eide GE, Arnes J, Stefansson IM, Eide J, Braaten A, Aas T, Otte AP, Akslen LA. Expression of enhancer of zeste homologue 2 is significantly associated with increased tumor cell proliferation and is a marker of aggressive breast cancer. Clin Cancer Res. 2006;12:1168-1174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 214] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 36. | Deng L, Chen T, Xu H, Li Y, Deng M, Mo D, Tian H, Ren Y. The Expression of Snail, Galectin-3, and IGF1R in the Differential Diagnosis of Benign and Malignant Pheochromocytoma and Paraganglioma. Biomed Res Int. 2020;2020:4150735. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 37. | Yoshida S, Wei X, Zhang G, O'Connor CL, Torres M, Zhou Z, Lin L, Menon R, Xu X, Zheng W, Xiong Y, Otto E, Tang CA, Hua R, Verma R, Mori H, Zhang Y, Hu CA, Liu M, Garg P, Hodgin JB, Sun S, Bitzer M, Qi L. Endoplasmic reticulum-associated degradation is required for nephrin maturation and kidney glomerular filtration function. J Clin Invest. 2021;131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 45] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 38. | Sun X, Liu Y, Zhou S, Wang L, Wei J, Hua R, Shen Z, Yoshida S. Circular dorsal ruffles disturb the growth factor-induced PI3K-AKT pathway in hepatocellular carcinoma Hep3B cells. Cell Commun Signal. 2022;20:102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 39. | Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, Hackl H, Trajanoski Z. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017;18:248-262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1446] [Cited by in RCA: 3190] [Article Influence: 398.8] [Reference Citation Analysis (0)] |
| 40. | Huang ZQ, Zhou XM, Yuan T, Liu J, Ong HH, Sun LY, Tu JH, Li MY, Thong KTM, Ye J, Shi L, Wang DY, Xu Y. Epithelial Tight Junction Anomalies in Nasal Inverted Papilloma. Laryngoscope. 2024;134:552-561. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 41. | Zhou S, Liu C, Wang J, Ye J, Lian Q, Gan L, Deng S, Xu T, Guo Y, Li W, Zhang Z, Yang GY, Tang Y. CCL5 mediated astrocyte-T cell interaction disrupts blood-brain barrier in mice after hemorrhagic stroke. J Cereb Blood Flow Metab. 2024;44:367-383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 11] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 42. | Rogawski DS, Deng J, Li H, Miao H, Borkin D, Purohit T, Song J, Chase J, Li S, Ndoj J, Klossowski S, Kim E, Mao F, Zhou B, Ropa J, Krotoska MZ, Jin Z, Ernst P, Feng X, Huang G, Nishioka K, Kelly S, He M, Wen B, Sun D, Muntean A, Dou Y, Maillard I, Cierpicki T, Grembecka J. Discovery of first-in-class inhibitors of ASH1L histone methyltransferase with anti-leukemic activity. Nat Commun. 2021;12:2792. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 43. | Hou P, Huang C, Liu CP, Yang N, Yu T, Yin Y, Zhu B, Xu RM. Structural Insights into Stimulation of Ash1L's H3K36 Methyltransferase Activity through Mrg15 Binding. Structure. 2019;27:837-845.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 44. | Nathani P, Singal AG. Imaging and Biomarker Approaches to HCC Surveillance. Clin Liver Dis (Hoboken). 2021;17:401-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 45. | El-Shayeb AF, El-Habachi NM, Mansour AR, Zaghloul MS. Serum midkine is a more sensitive predictor for hepatocellular carcinoma than Dickkopf-1 and alpha-L-fucosidase in cirrhotic HCV patients. Medicine (Baltimore). 2021;100:e25112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 46. | Zhang W, Chen Z, Xue C, Zhang Y, Wu L, Zhu J, Xuan S, Tian J, Pang Z. The Applicability of ADA, AFU, and LAC in the Early Diagnosis and Disease Risk Assessment of Hepatitis B-Associated Liver Cirrhosis and Hepatocellular Carcinoma. Front Med (Lausanne). 2021;8:740029. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 47. | Liu Y, Wang J, Yang R, Cheng Y, Zhou Y, Li H, Jiang W, Zhang X. GP73-mediated secretion of AFP and GP73 promotes proliferation and metastasis of hepatocellular carcinoma cells. Oncogenesis. 2021;10:69. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |