Published online Mar 7, 2024. doi: 10.3748/wjg.v30.i9.994
Peer-review started: November 29, 2023
First decision: December 26, 2023
Revised: January 3, 2024
Accepted: February 18, 2024
Article in press: February 18, 2024
Published online: March 7, 2024
Processing time: 98 Days and 0.5 Hours
In this editorial, we comment on the article by Lyu et al published in the recent issue of the World Journal of Gastroenterology (2023; 2219-2840). Hepatocellular carcinoma (HCC) is a frequently encountered and highly aggressive primary liver cancer, which remains the third-commonest cause of cancer-related death despite the current therapeutic modalities. There is urgency in developing novel thera
Core Tip: This paper sheds light on the role of exosomal circular RNAs as microRNA sponges and their potential targeting for suppressing hepatocellular carcinoma growth and progression.
- Citation: Papadopoulos N, Trifylli EM. Role of exosomal circular RNAs as microRNA sponges and potential targeting for suppressing hepatocellular carcinoma growth and progression. World J Gastroenterol 2024; 30(9): 994-998
- URL: https://www.wjgnet.com/1007-9327/full/v30/i9/994.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i9.994
Hepatocellular carcinoma (HCC) is a frequently encountered and highly aggressive primary liver cancer, characterized by a high number of cancer-related deaths worldwide, constituting the third most common cause of cancer-related death[1]. Despite the novelties in the diagnostic and therapeutic strategies, including immunochemotherapy, targeted and local treatments, radiotherapy, and surgical approaches, the overall survival of HCC patients remains low due to chemoresistance and tumor recurrence[2]. There is a call for the discovery and development of novel therapeutic approaches, with the understanding of the molecular complexity of this malignancy being pivotal. The role of extracellular vesicles (EVs) and non-coding RNA molecules and their implications in HCC development and progression are in the spotlight of their ongoing studies, aiming to discover novel therapeutic targets and strategies[3].
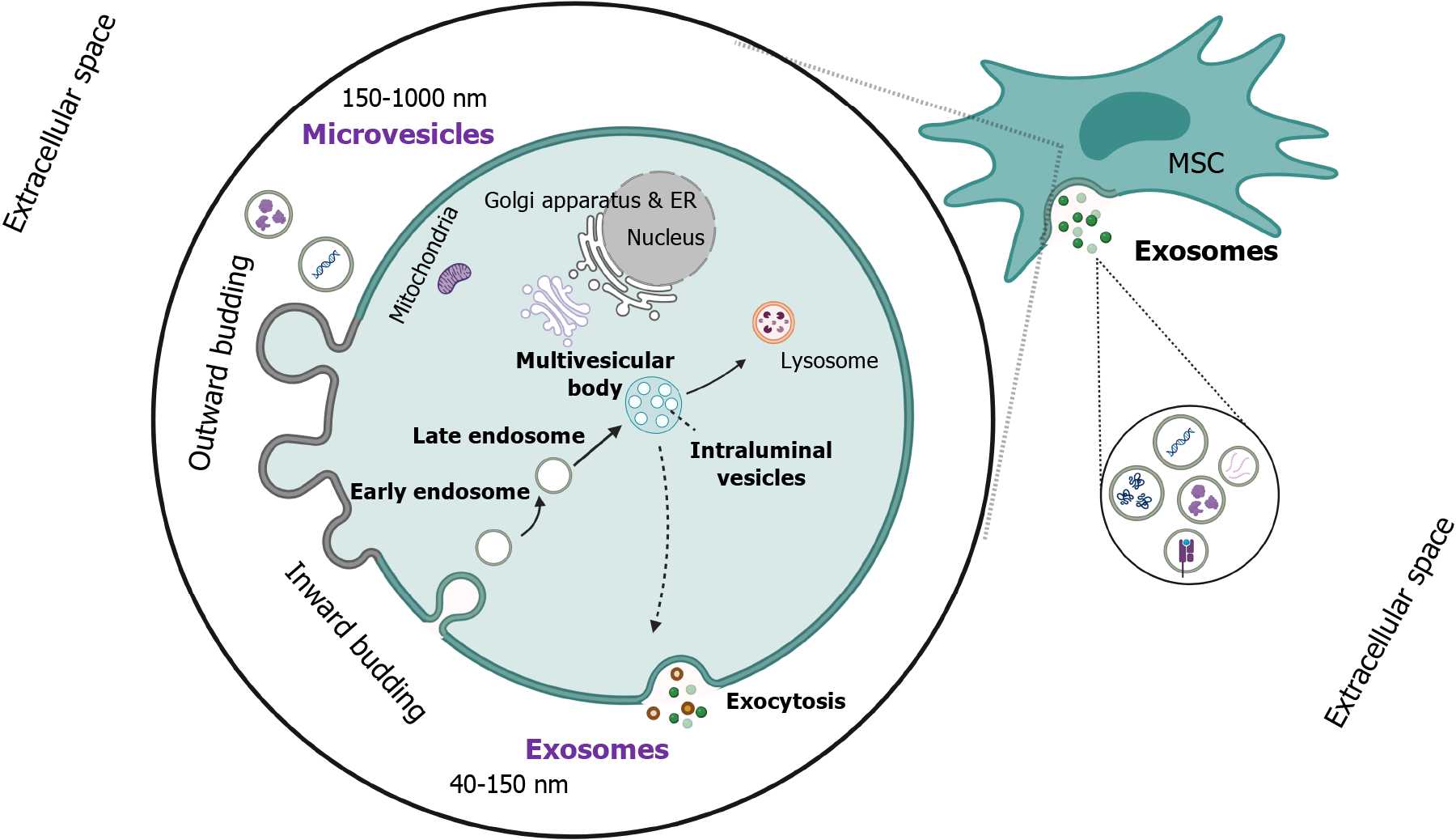
EVs constitute a highly heterogeneous population of nanostructures composed of a lipid membrane (lipid bilayer) with several enclosed cargoes[4]. Their heterogeneity is attributed to their various sizes, biogenetic mechanisms, and the diversity of their cargoes, including coding or non-coding RNA molecules, proteins, lipids, several ligands, DNA molecules, and autophagosomes. A wide variety of cells can produce these vesicles via different mechanisms, including the inward or outward budding of the cell membrane or apoptosis, which lead to exosome, microvesicle, and apoptotic body generation, respectively[5]. The main classification of EVs is based on their size, including: (1) Exosomes; (2) microvesicles; and (3) apoptotic bodies, being the smallest (40-150 nm), medium-sized (150-1000 nm), and largest (> 1000 nm) subclass, respectively.
However, there is also another classification based on their biogenetic mechanism, including exosomes and ectosomes, with the former being produced by the integration of the multivesicular body (MVB) with the plasma membrane and the exocytosis of exosomes, while the latter via plasma membrane budding[6].
Focusing on the mechanism of exosome generation, the inward plasma membrane budding is primarily required, accompanied by the internalization of transmembrane proteins and the formation of vesicles. This process is followed by several distinct steps for the final generation and release of exosomes from the parental cell, including the formation of early endosomes and the maturation of early endosomes into late endosomes, which either lead to intraluminal vesicles (ILVs) via the invagination of the membrane of the latter or they are retransferred to the cell membrane. Afterward, the incorporation of ILVs will lead to MVBs, which are either destructed in lysosomes or fused with the cell membrane to release exosomes[7]. All the aforementioned biogenetic pathways are strictly orchestrated under the influence of endosomal sorting (ESCRT) complexes 0-III to modify and remodel the membrane and generate ILVs and, eventually, MVBs. Moreover, soluble NSF attachment protein receptor (SNARE) proteins are required for the MVB fusion with plasma membrane release of exosomes in the extracellular space (Figure 1)[8]. These nanostructures have a pivotal role in cell-to-cell communication, as they can modify the functional and transcriptional level of the recipient cells via the uptake of their cargoes. Meanwhile, their high biocompatibility, low immunogenicity, and their role in intercellular com
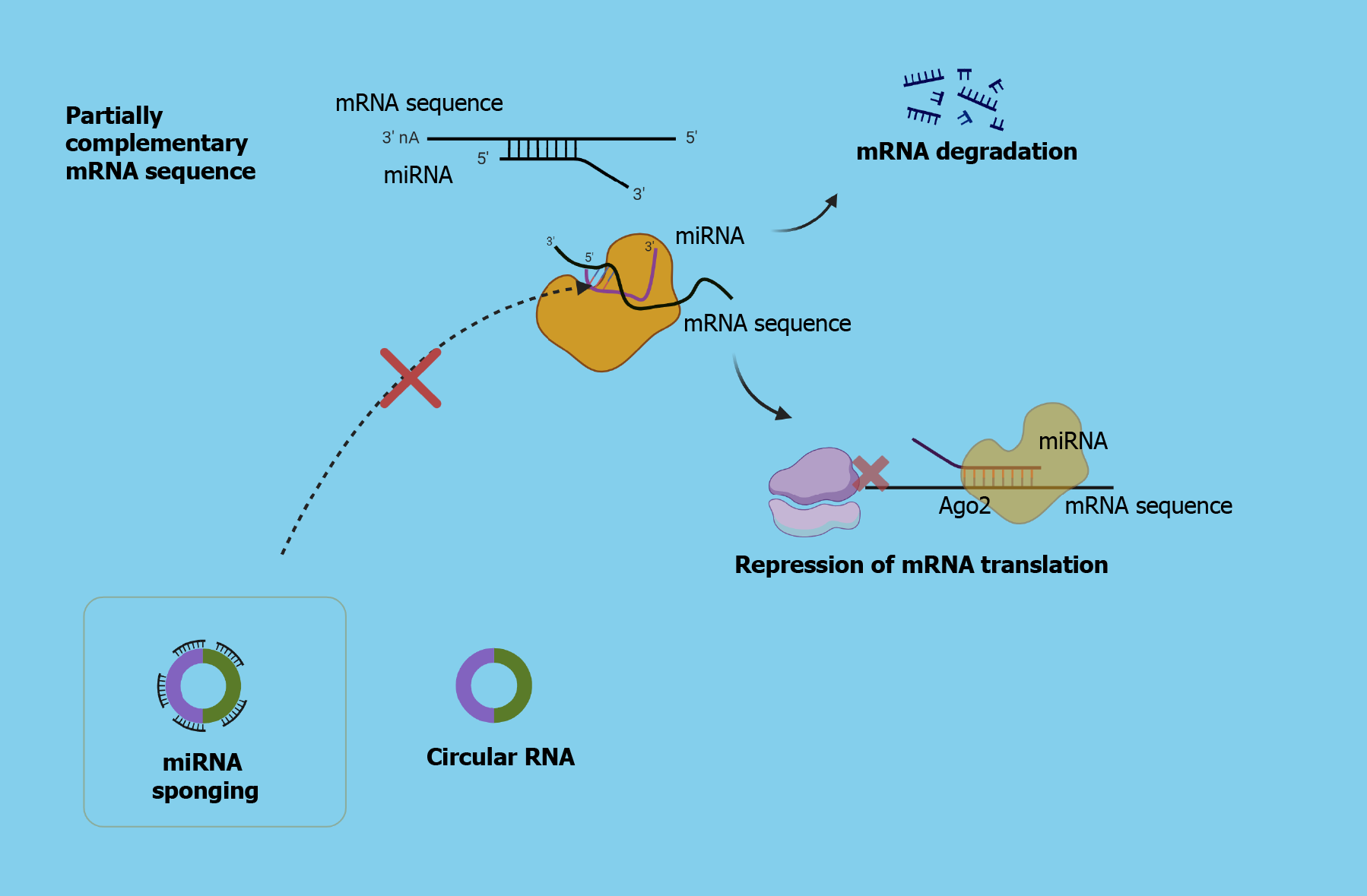
Several aberrations are observed in the circRNA expression levels in HCC tissue compared to physiological ones. The mechanisms on how circRNAs are implicated in HCC are still not adequately clear; however, their significant contribution cannot be doubted[15]. Taking advantage of their functions will open new horizons in the development of circRNA-centered therapeutic and diagnostic perspectives.
It is demonstrated that the role of circRNAs is dual in HCC, as they can either promote or suppress tumor progression via interacting with oncogenic or tumor-suppressive miRNAs. The aforementioned phenomenon is attributed to their role as a “sponge” for oncogenic miRNAs or tumor suppressive miRNAs, which can lead to tumor inhibition or promotion, respectively[16]. There are several aberrations in the expression levels of circRNAs, which may lead to tumor growth and progression and generally poor prognosis or they can induce suppression of HCC development. The study by Lyu et al[17], which was published in the recent issue of World Journal of Gastroenterology, demonstrated the role of mesenchymal stem cells (MSCs)-derived exosomal hsa_circ_0000563 (circ-563) as a sponge for miR-148a-3p leading to tumor progression, whereas the silencing of circ-563 suppressed the HCC growth and development[17].
Exploring earlier studies, in the study by Zhang et al[18], tumor-suppressive miR-148a-3p was found to be downregulated in HCC tissue, compared to physiological hepatic tissue, which was associated with aggressive tumor behavior and worrisome prognosis[18]. Additionally, they observed that hepatic stellate cell (HSC)-derived exosomes, in which miR-148a-3p was depleted, led to HCC progression via the ITGA5/PI3K/Akt axis, which is involved in cell proliferation, migration, and survival, as well as in cancer development and drug resistance. However, the increased expression of exosomal miR-148-3p in HCC tissue leads to tumor suppression[18].
Another study about the role of miR-148a-3p in HCC by Lyu et al[19] has demonstrated the interplay between the aforementioned miRNA and metal-regulatory transcription factor-1 (MTF-1) in HCC progression[19]. More specifically, MTF-1 is closely implicated in metal homeostasis, while its overexpression leads to hepatocarcinogenesis, tumor proliferation, and metastatic dissemination, as was demonstrated in conditions like copper exposure. Enhanced miR-148a-3p expression successfully suppressed HCC growth and progression that were induced via MTF-1. However, it was observed that exosomal miR-148a-3p was notably downregulated in HCC patients, whereas its enhanced expression led to suppression of MTF-1 and HCC inhibition[19].
Eventually, the development of RNA sequencing, the identification of circRNAs, and the development of the competitive endogenous RNA (ceRNA) theory expanded the research approaches for HCC pathogenesis and therapeutic targets. The ceRNA theory suggests that several RNA molecules like long non-coding RNA, circRNA, and mRNA can competitively share miRNA binding sites. In this recent study of Lyu et al[17], they also focused on the significant role of the tumor microenvironment (TME) in HCC, focusing on the implication of MSCs via releasing exosomes. MSCs are recruited in HCC TME, exerting various effects, including suppression of anti-tumor immunity, and promoting neoangiogenesis that favors tumor growth and progression. Additionally, they differentiate into stromal cells, enhancing the tumor stroma. They can also induce several signaling pathways via secreting cytokines and EVs, like exosomes. The manipulation of MSC-derived molecules like exosomes for modifying the functionality of the recipient cells through the delivery of anti-HCC agents or genetic modulatory molecules can potentially widen the therapeutic perspectives. In the aforementioned study, they utilized labeled isolated MSC-derived exosomes co-cultured with HCC. As demonstrated in the previous study by Lyu et al[19], overexpression of exosomal miR-148a-3p, which targets MTF-1, notably decreased HCC progression, whereas, in the present study, they reported that among the various circRNAs from the databases, has_circ563 had the most partially complementary sequence for miR-148a-3p[17]. In addition, they demonstrated a correlation between MTF-1 and circ563 overexpression, as well as a correlation between circ563 upregulation and decreased miR-148-3p expression levels, implying its potential role as an “miRNA sponge” and eventually as an HCC promoter. On the other hand, silencing of circ-563 led to tumor suppression, suggesting its potential use as an anti-HCC therapeutic strategy[17].
New opportunities for HCC management could be opened up via the deep understanding of ceRNA theory, suggesting the role of circRNAs as “miRNA sponges”, accompanied by the utilization of exosomes as delivery vectors.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Greece
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Zheng Y, China S-Editor: Chen YL L-Editor: Wang TQ P-Editor: Yuan YY
| 1. | Rumgay H, Arnold M, Ferlay J, Lesi O, Cabasag CJ, Vignat J, Laversanne M, McGlynn KA, Soerjomataram I. Global burden of primary liver cancer in 2020 and predictions to 2040. J Hepatol. 2022;77:1598-1606. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 1058] [Article Influence: 352.7] [Reference Citation Analysis (0)] |
| 2. | Reig M, Forner A, Rimola J, Ferrer-Fàbrega J, Burrel M, Garcia-Criado Á, Kelley RK, Galle PR, Mazzaferro V, Salem R, Sangro B, Singal AG, Vogel A, Fuster J, Ayuso C, Bruix J. BCLC strategy for prognosis prediction and treatment recommendation: The 2022 update. J Hepatol. 2022;76:681-693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1904] [Cited by in RCA: 2593] [Article Influence: 864.3] [Reference Citation Analysis (59)] |
| 3. | Koustas E, Trifylli EM, Sarantis P, Papadopoulos N, Papanikolopoulos K, Aloizos G, Damaskos C, Garmpis N, Garmpi A, Matthaios D, Karamouzis MV. An Insight into the Arising Role of MicroRNAs in Hepatocellular Carcinoma: Future Diagnostic and Therapeutic Approaches. Int J Mol Sci. 2023;24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 4] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 4. | Akers JC, Gonda D, Kim R, Carter BS, Chen CC. Biogenesis of extracellular vesicles (EV): exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J Neurooncol. 2013;113:1-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 760] [Cited by in RCA: 1055] [Article Influence: 87.9] [Reference Citation Analysis (0)] |
| 5. | Willms E, Cabañas C, Mäger I, Wood MJA, Vader P. Extracellular Vesicle Heterogeneity: Subpopulations, Isolation Techniques, and Diverse Functions in Cancer Progression. Front Immunol. 2018;9:738. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 387] [Cited by in RCA: 676] [Article Influence: 96.6] [Reference Citation Analysis (0)] |
| 6. | Cocucci E, Meldolesi J. Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015;25:364-372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 786] [Cited by in RCA: 1081] [Article Influence: 108.1] [Reference Citation Analysis (0)] |
| 7. | Han QF, Li WJ, Hu KS, Gao J, Zhai WL, Yang JH, Zhang SJ. Exosome biogenesis: machinery, regulation, and therapeutic implications in cancer. Mol Cancer. 2022;21:207. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 335] [Article Influence: 111.7] [Reference Citation Analysis (0)] |
| 8. | Arya SB, Collie SP, Parent CA. The ins-and-outs of exosome biogenesis, secretion, and internalization. Trends Cell Biol. 2023;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 154] [Article Influence: 154.0] [Reference Citation Analysis (0)] |
| 9. | Berumen Sánchez G, Bunn KE, Pua HH, Rafat M. Extracellular vesicles: mediators of intercellular communication in tissue injury and disease. Cell Commun Signal. 2021;19:104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 150] [Article Influence: 37.5] [Reference Citation Analysis (0)] |
| 10. | Meng W, He C, Hao Y, Wang L, Li L, Zhu G. Prospects and challenges of extracellular vesicle-based drug delivery system: considering cell source. Drug Deliv. 2020;27:585-598. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 387] [Article Influence: 96.8] [Reference Citation Analysis (0)] |
| 11. | Meng H, Niu R, Huang C, Li J. Circular RNA as a Novel Biomarker and Therapeutic Target for HCC. Cells. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 38] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 12. | Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Wu YM, Dhanasekaran SM, Engelke CG, Cao X, Robinson DR, Nesvizhskii AI, Chinnaiyan AM. The Landscape of Circular RNA in Cancer. Cell. 2019;176:869-881.e13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1086] [Cited by in RCA: 1184] [Article Influence: 197.3] [Reference Citation Analysis (0)] |
| 13. | Jarlstad Olesen MT, S Kristensen L. Circular RNAs as microRNA sponges: evidence and controversies. Essays Biochem. 2021;65:685-696. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 56] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 14. | Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675-691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3273] [Cited by in RCA: 3145] [Article Influence: 524.2] [Reference Citation Analysis (0)] |
| 15. | Qiu L, Wang T, Ge Q, Xu H, Wu Y, Tang Q, Chen K. Circular RNA Signature in Hepatocellular Carcinoma. J Cancer. 2019;10:3361-3372. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 59] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 16. | Yao R, Zou H, Liao W. Prospect of Circular RNA in Hepatocellular Carcinoma: A Novel Potential Biomarker and Therapeutic Target. Front Oncol. 2018;8:332. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 55] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 17. | Lyu ZZ, Li M, Yang MY, Han MH, Yang Z. Exosome-mediated transfer of circRNA563 promoting hepatocellular carcinoma by targeting the microRNA148a-3p/metal-regulatory transcription factor-1 pathway. World J Gastroenterol. 2023;29:6060-6075. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (2)] |
| 18. | Zhang X, Chen F, Huang P, Wang X, Zhou K, Zhou C, Yu L, Peng Y, Fan J, Zhou J, Lu Z, Hu J, Wang Z. Exosome-depleted MiR-148a-3p derived from Hepatic Stellate Cells Promotes Tumor Progression via ITGA5/PI3K/Akt Axis in Hepatocellular Carcinoma. Int J Biol Sci. 2022;18:2249-2260. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 44] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 19. | Lyu Z, Yang M, Yang T, Ma M, Yang Z. Metal-Regulatory Transcription Factor-1 Targeted by miR-148a-3p Is Implicated in Human Hepatocellular Carcinoma Progression. Front Oncol. 2021;11:700649. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 20. | BioRENDER. Agreement number PL2696KYM8 and LA2696N8DC. [cited 29 November 2023]. Available from: https://www.biorender.com/. |