Published online Feb 28, 2024. doi: 10.3748/wjg.v30.i8.969
Peer-review started: October 25, 2023
First decision: December 21, 2023
Revised: January 2, 2024
Accepted: February 1, 2024
Article in press: February 1, 2024
Published online: February 28, 2024
Processing time: 124 Days and 2.4 Hours
Three-dimensional organoid culture systems have been established as a robust tool for elucidating mechanisms and performing drug efficacy testing. The use of gastric organoid models holds significant promise for advancing personalized medicine research. However, a comprehensive bibliometric review of this bur-geoning field has not yet been published.
To analyze and understand the development, impact, and direction of gastric organoid research using bibliometric methods using data from the Web of Science Core Collection (WoSCC) database.
This analysis encompassed literature pertaining to gastric organoids published between 2010 and 2023, as indexed in the WoSCC. CiteSpace and VOSviewer were used to depict network maps illustrating collaborations among authors, institutions and keywords related to gastric organoid. Citation, co-citation, and burst analysis methodologies were applied to assess the impact and progress of research.
A total of 656 relevant studies were evaluated. The majority of research was published in gastroenterology-focused journals. Globally, Yana Zavros, Hans Clevers, James M Wells, Sina Bartfeld, and Chen Zheng were the 5 most productive authors, while Hans Clevers, Huch Meritxell, Johan H van Es, Marc Van de Wetering, and Sato Toshiro were the foremost influential scientists in this area. Institutions from the University Medical Center Utrecht, Netherlands Institute for Developmental Biology (Utrecht), and University of Cincinnati (Cincinnati, OH, United States) made the most significant contributions. Currently, gastric organoids are used mainly in studies investigating gastric cancer (GC), Helicobacter pylori-infective gastritis, with a focus on the mechanisms of GC, and drug screening tests.
Key focus areas of research using gastric organoids include unraveling disease mechanisms and enhancing drug screening techniques. Major contributions from renowned academic institutions highlight this field’s dynamic growth.
Core Tip: This study highlights the pivotal role of organoid technology in gastric disease research, emphasizing its growth in publications and citations. Key contributions from leading researchers and institutions, particularly in understanding gastric cancer and Helicobacter pylori-infective gastritis, mark advances in the field. Focused on deciphering cancer mechanisms and improving drug screening, this area of exploration provides crucial insights for future gastroenterology research.
- Citation: Jiang KL, Jia YB, Liu XJ, Jia QL, Guo LK, Wang XX, Yang KM, Wu CH, Liang BB, Ling JH. Bibliometrics analysis based on the Web of Science: Current trends and perspective of gastric organoid during 2010-2023. World J Gastroenterol 2024; 30(8): 969-983
- URL: https://www.wjgnet.com/1007-9327/full/v30/i8/969.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i8.969
The term “organoid” was first introduced as an anatomical diagnosis of tumors in a study of tumor mechanisms in 1946[1]. Organoid generally refers to tissues or structures that are similar to organs used in vitro biology. A gastric organoid is a specific cluster of cells derived from gastric or pluripotent stem cells[2,3]. Gastric organoids are highly similar to gastric epithelial tissue in terms of cellular components, tissue architecture, and specific functions, and have corresponding functional characteristics, which enable the replication of gastric epithelial tissues in vitro[4]. Gastrointestinal organoid technology represents a technical revolution compared with conventional cell biology methods. In conventional cell culture, a single-cell clone is cultured in a two-dimensional environment. However, in gastric organoid culture, many cell clones from the stomach can be cultured in a three-dimensional (3D) environment exhibiting a higher degree of similarity to the in vivo microenvironment, thus providing a more accurate and physiologically pertinent model for study[5,6]. Gastric organoids cultured in vitro, especially those derived from human tissues, provide a new platform for the study of human gastric physiology and diseases[7]. Since Barkerfirst described a protocol using mouse leucine-rich repeat-containing G-protein coupled receptor 5+ (Lgr5+) gastric stem cells to construct gastric organoids in 2010, gastric organoids have attracted increasing attention in the field of digestive tract research[8].
In recent years, numerous living biobanks of tumor organoids have been established. For instance, a human gastric cancer (GC) organoid biobank encompasses 34 patients and 63 organoid lines, capturing the heterogeneity of tumor subtypes and enabling therapeutic screening[9]. These established organoid biobanks serve as biomaterial for human cancer research and personalized medicine. They have been widely used in fundamental research investigating gastric physiology and pathology, clinical areas, such as diagnostic processes, pharmacological screening and development, as well as gene therapy applications[10,11]. Hans Clevers from Utrecht University in the Netherlands is a pioneer in the field, having made significant contributions to the technology[12].
The Web of Science Core Collection (WoSCC) database houses many published academic studies. Because these publications are fragmented, strong conclusions from studies investigating gastric organoids should coherently bring research findings together. In the present study, we used bibliometric analysis to generate a network map of author(s), institution(s), keywords, and co-cited references derived from publications addressing gastric organoids. We then systematically analyzed the main topics based on the results of this analysis to build an in-depth and comprehensive understanding of research investigating gastric organoids.
Literature sources included in this study were retrieved from the WoSCC database. The database was searched by combining the terms “organoid”, “gastroid”, “spheroid”, “gastric”, “gastritis”, and “stomach”. Studies published from 2010 up to September 2023 were searched to form the basis of data processing and analysis.
Data extracted from the studies retrieved in the literature search of the WoSCC database included author information, title, keywords, abstract, and references, and underwent preliminary screening. Quantitative variables were analyzed using spreadsheet software (Excel 2019, Microsoft Corporation, Redmond, WA, United States). Citespace version 6.1.R3, developed by Chen Chaomei, and VOSviewer version 1.68, developed by Nees Jan van Eck and Ludo Waltman, were used for scientific knowledge mapping and analysis[13,14]. VOSviewer 1.68 was used to calculate citations and publications of authors and institutions, and to depict the keyword network graph[15,16]. Citespace 6.1.R3 was used to calculate co-cite and citation frequencies, centrality, keyword bursts, references, as well as to depict a network graph of authors, institutions, and a timeline graph of keywords[17].
Citation analysis is a statistical method of illustrating the quantity of cited studies to identify patterns and measurement characteristics. Generally, the more often a study is cited, the greater its impact. Co-citation is when 2 studies are cited by ≥ 1 study at the same time, which indicates how closely the 2 studies are related[18,19]. Moreover, the greater the number of studies cited simultaneously, the closer the relationship between the two. Centrality is an indicator of the importance of a node in a network, and documents with high centrality are often the key hubs connecting two different domains. Evolution of a field of knowledge can be indicated by references with citation bursts. A burst is a keyword/citation reference that has received scholarly attention in a particular field. The timeline is presented as a blue bar, while the interval, when a topic is found to have a burst, is presented as a red section indicating the year it started, the year it ended, and the time of the burst. Higher intensity indicates a higher citation frequency.
Each circle on the network map represents an “individual”, and the size of the circle represents the active degree and the number of publications of the keyword. The closer the distance between an “individual” and an “individual”, the closer the relationship between them. The increased number of connected lines indicates the increased number of co-citations between the “individual”[14].
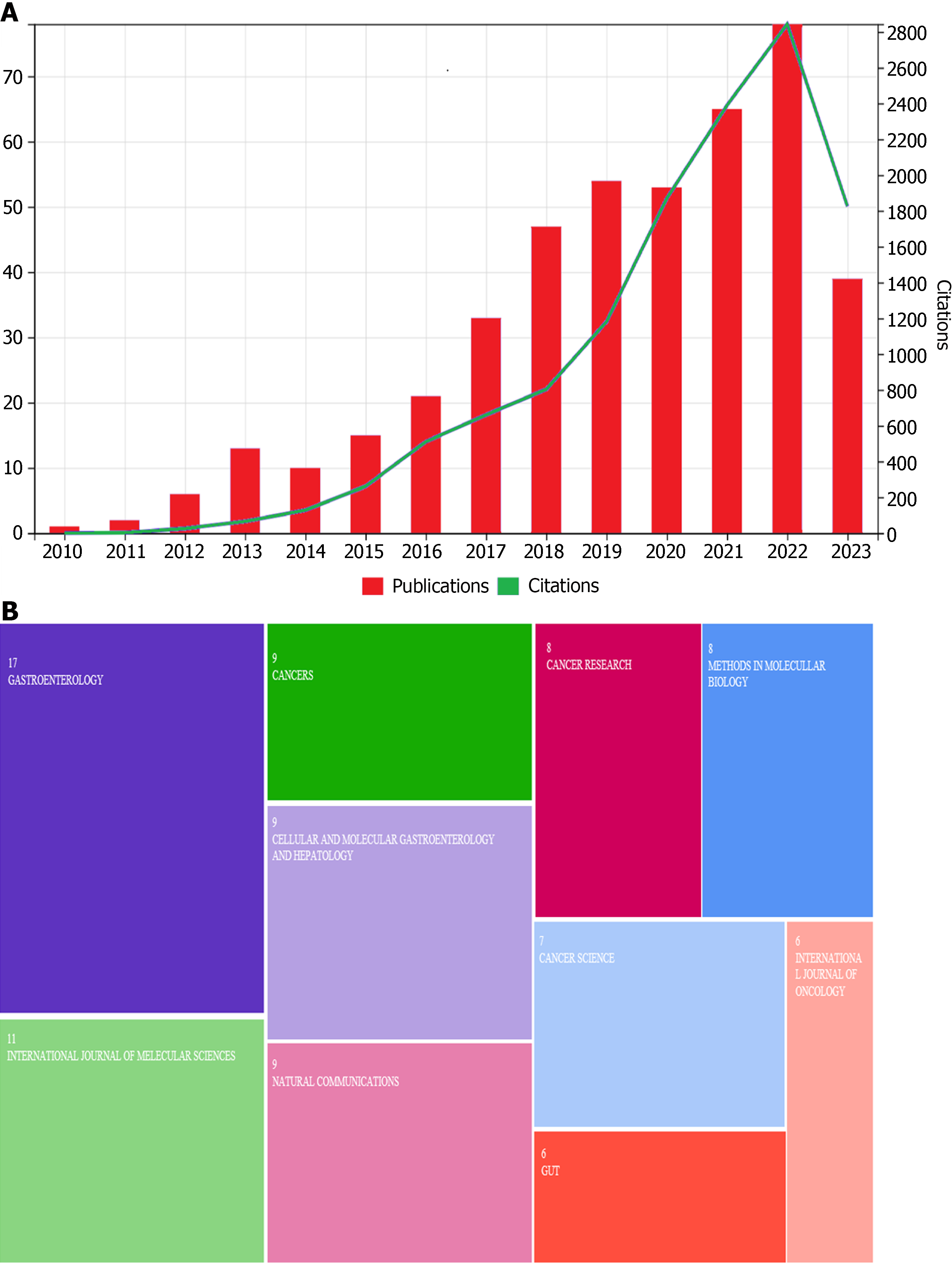
A total of 656 articles were retrieved in the literature search. The successful culture of gastric-like organs was first reported in 2010, and relevant research developed slowly and steadily in the ensuing years[8]. The number of publications and citations exhibited stable growth from 2010 to 2016 and then a sharp increase after 2017 (Figure 1A). Researchers discovered the great advantage of the organoid model in the past 5 years; therefore, relevant publications and citations have increased rapidly. Most articles have been high quality and were published in Gastroenterology, International Journal of Molecular Science, Cancers, Cellular and Molecular Gastroenterology and Hepatology, Nature Communications, Cancer Research, Cancer Science, and Gut (Figure 1B). These journals focus mainly on cancer and gastroenterological diseases.
A total of 474 investigators published research in this area, with 32 publishing > 5 studies. Yana Zavros, Hans Clevers, James M Wells, Sato Toshiro, and Koo Bon-kyoung were the 5 most productive authors. Hans Clevers, Huch Meritxell, Johan H van Es, Marc Van de Wetering, and Sato Toshiro were the 5 most popular scientists. Hans Clevers, Yana Zavros, and Richard Peek demonstrated high centrality (0.04). Most of the productive and influential investigators were from The Netherlands and the United States, mainly from the University Medical Center Utrecht (Utrecht, Netherlands) and the University of Cincinnati (Cincinnati, OH, United States), respectively. Table 1 summarizes the top 15 most cited researchers and their respective countries.
| Rank | Author | Publications | Citations | Country | Centrality |
| 1 | Clevers, Hans | 30 | 5144 | Netherland | 0.08 |
| 2 | Huch, Meritxell | 10 | 3987 | German | 0.04 |
| 3 | Van es, Johan | 7 | 3244 | Netherland | 0.00 |
| 4 | Van de Wetering, Marc | 6 | 3175 | Netherland | 0.00 |
| 5 | Sato, Toshiro | 14 | 3141 | Japan | 0.08 |
| 6 | Stange, Daniel | 12 | 1764 | German | 0.01 |
| 7 | Barker, Nick | 13 | 1707 | Netherland | 0.03 |
| 8 | Koo, Bon-kyoung | 15 | 1618 | Netherland | 0.25 |
| 9 | Bartfeld, Sina | 13 | 1405 | German | 0.03 |
| 10 | Zavros, Yana | 46 | 1312 | United States | 0.04 |
| 11 | Wells, James | 22 | 1040 | United States | 0.01 |
| 12 | Spence, Jason | 7 | 973 | United States | 0.00 |
| 13 | Peek, Richard | 5 | 715 | United States | 0.05 |
| 14 | Schumacher, Michael | 6 | 657 | Japan | 0.00 |
| 15 | Chen, Zheng | 12 | 615 | United States | 0.03 |
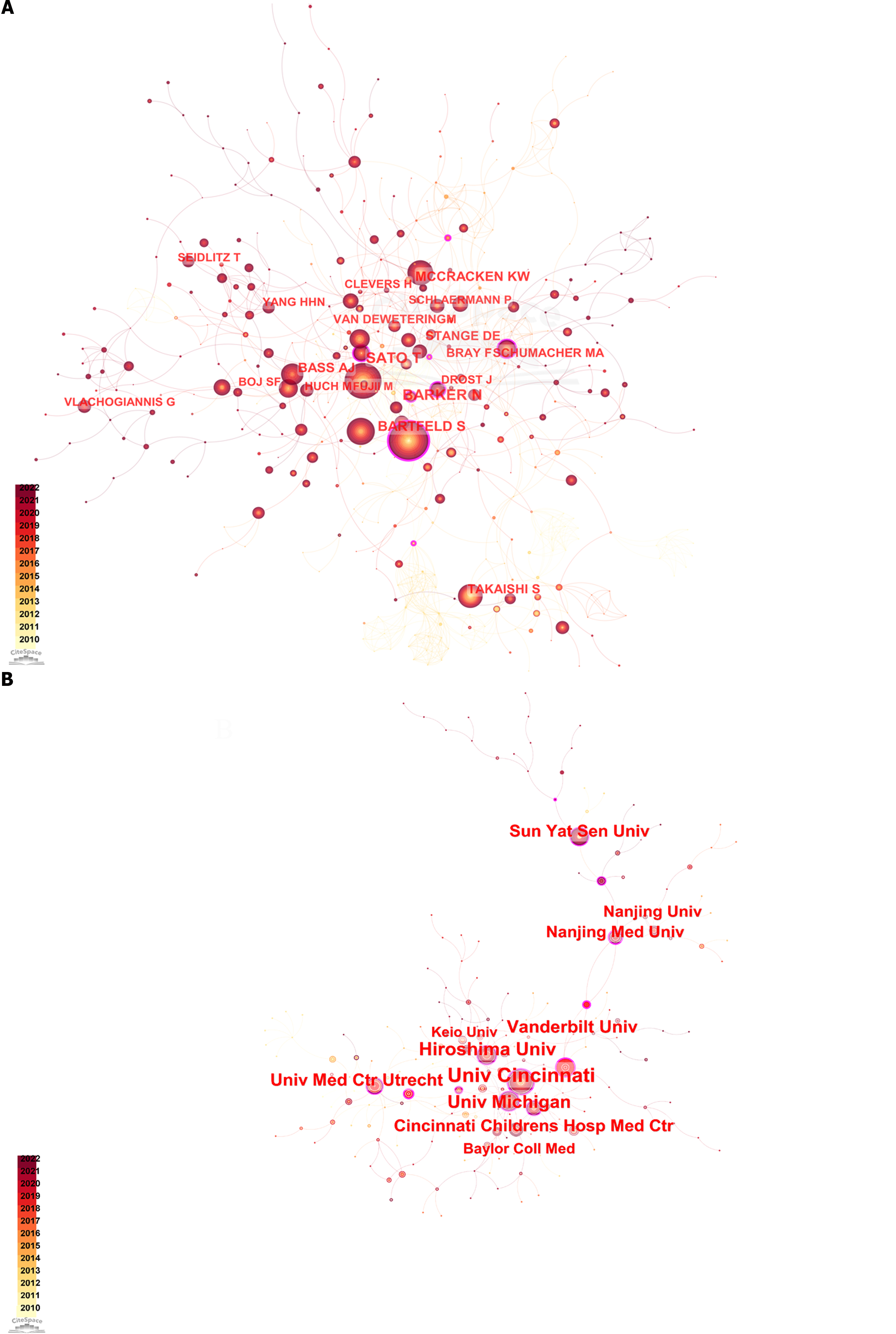
Hans Clevers, Sato Toshiro, Sina Bartfeld, N Barker, Huch Meritxel, and Marc van de Wetering established a close and vital cooperative network. HHN Yang worked closely with T Seldlitz, G Vlachogiannis worked closely with L Broutier, and another network was formed between S Takaishi and A Jemal. The co-authorship network diagram is presented in Figure 2A.
A total of 314 institutions published research on this topic, and 54 published > 5 studies. The top 5 most-productive institutions in this context were the University of Cincinnati, Hiroshima University, University of Michigan (Ann Arbor, MI, United States), Vanderbilt University (Nashville, TN, United States), and the University Medical Center Utrecht. The top 5 most cited institutions were the University Medical Center Utrecht, the Netherlands Institute for Developmental Biology Utrecht, University of Cincinnati, University of Hong Kong, and the University of Michigan. The top 15 most-cited institutions, which were mostly from the Netherlands, United States, China, and Japan, are summarized in Table 2. The University of Cincinnati had the highest centrality (0.25), followed by the University Medical Center Utrecht (0.18).
| Rank | Institution | Country | Publications | Citations | Centrality |
| 1 | University Medical Center Utrecht | Netherland | 19 | 5083 | 0.18 |
| 2 | Netherlands Institute for Developmental Biology Utrecht | Netherland | 8 | 3064 | 0.00 |
| 3 | University of Cincinnati | United States | 33 | 1451 | 0.25 |
| 4 | University of Hong Kong | Hongkong, China | 6 | 1148 | 0.08 |
| 5 | University of Michigan | United States | 22 | 1104 | 0.04 |
| 6 | Cincinnati Children’s Hospital Medical Center | United States | 16 | 999 | 0.07 |
| 7 | Columbia University | United States | 11 | 986 | 0.05 |
| 8 | Vanderbilt University | United States | 22 | 941 | 0.25 |
| 9 | Stanford University | United States | 11 | 881 | 0.02 |
| 10 | Cambridge University | United Kingdom | 7 | 758 | 0.08 |
| 11 | Harvard University | United States | 12 | 731 | 0.08 |
| 12 | Shinshu University | Japan | 17 | 625 | 0.07 |
| 13 | Keio University | Japan | 13 | 602 | 0.13 |
| 14 | Nanjing med University | China | 14 | 600 | 0.20 |
| 15 | Miami University | United States | 7 | 556 | 0.00 |
The United States had the most extensive cooperative network. The University of Cincinnati, Vanderbilt University, University of Michigan, and Cincinnati Children’s Hospital Medicine Center formed the centers of each United States-collaborating institution. Chinese and Japanese scientists contributed the most to research from Asian institutions. Nanjing University (Nanjing, Jiangsu Province, China), Sun Yat-sen University (Guangzhou, Guangdong Province, China), and Fudan University (Shanghai, China) played central roles in the Chinese institutional network of collaborations, radiating out to other institutions. Japanese institutions were dominated, in this context, by Hiroshima University (Hiroshima, Japan), with more collaborations with the University of Cincinnati and other Japanese and Korean institutions.
The University Medical Center Utrecht in the Netherlands has a network of collaborations with European institutions. A network diagram illustrating institutional collaboration is presented in Figure 2B.
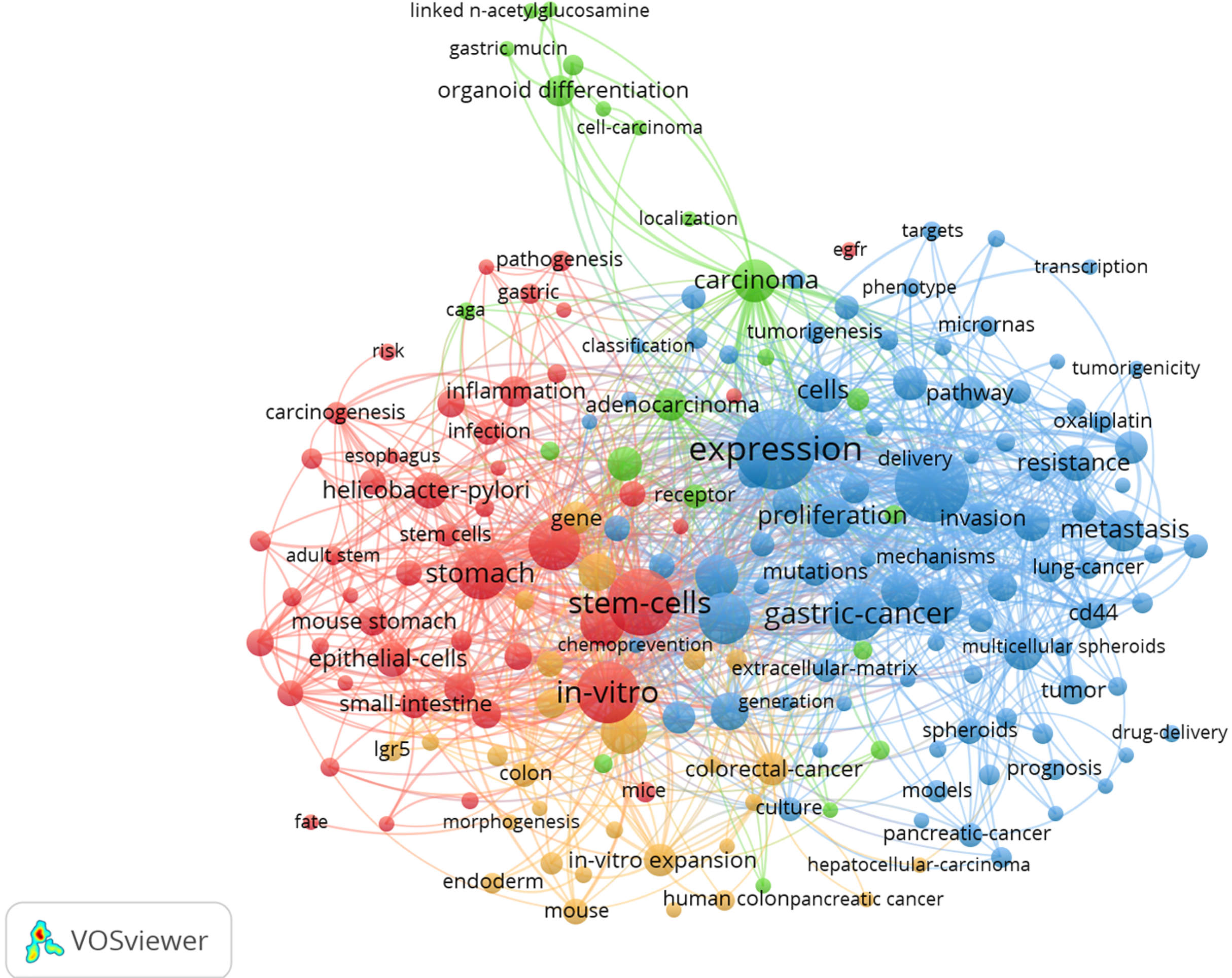
VOSviewer was used to construct a keyword network graph. Based on the network analysis the high-frequency keywords of published articles were clustered into 4 categories. The orange area (cluster #1, “Gastric organoid formation”) included the main terms related to gastric organoid construction such as stem cells, gastric epithelium, pyloric gland, and fundic gland. The green area (cluster #2, “Gastric cancer organoid formation”) explored gastric organoid construction such as tumor stem cells, tumor proliferation and differentiation, and immunohistochemistry. The blue area (cluster #3, “Applications of gastric cancer organoids”) occupied a large panel and described the application of organoids in GC therapeutic research, which indicated the study of tumor drug resistance, gene mutations, signaling pathways, and immunohistochemistry. It contains proteins such as WNT, kif11, and pi3k. Another interesting finding in these network diagrams is the red cluster, which is related to the application of organoids in gastritis (cluster #4, “Application of organoid in gastritis research”), including Helicobacter pylori (H. pylori) infection, inflammatory pathway activation, and cagA proteins (Figure 3). Many early studies investigating organoids focused on the formation and differentiation of GC organoids, whereas more recent studies focused on the treatment and mechanism of gastritis and GC.
After filtering out non-relevant and repetitive keywords, such as “expression” and “identification”, the 20 most popular keywords in the field of gastric organoids were shortlisted and are summarized in Table 3. GC and H. pylori-related gastritis were identified as the most prevalent diseases. Stem cells, cancer stem cells, epithelial cells, progenitor cells, and chief cells were identified as the main cellular “hotpot”. Cellular events related to cancer development and progression, such as metastasis, proliferation, apoptosis, migration, epithelial-mesenchymal transition, and mutation have received more attention.
| Keyword | Freq | Centrality |
| Gastric cancer | 177 | 0.06 |
| Stem cell | 110 | 0.04 |
| Cancer stem cell | 45 | 0.10 |
| Differentiation | 41 | 0.06 |
| Helicobacter pylori | 32 | 0.01 |
| Metastasis | 30 | 0.19 |
| Proliferation | 30 | 0.04 |
| Resistance | 29 | 0.06 |
| Epithelial cell | 27 | 0.16 |
| Self-renewal | 26 | 0.11 |
| Apoptosis | 22 | 0.07 |
| Progression | 22 | 0.04 |
| Protein | 20 | 0.03 |
| Progenitor cell | 18 | 0.04 |
| Gene expression | 18 | 0.05 |
| Chief cell | 17 | 0.04 |
| Migration | 16 | 0.01 |
| Chemotherapy | 16 | 0.07 |
| Epithelial mesenchymal transition | 14 | 0.02 |
| Mutation | 15 | 0.03 |
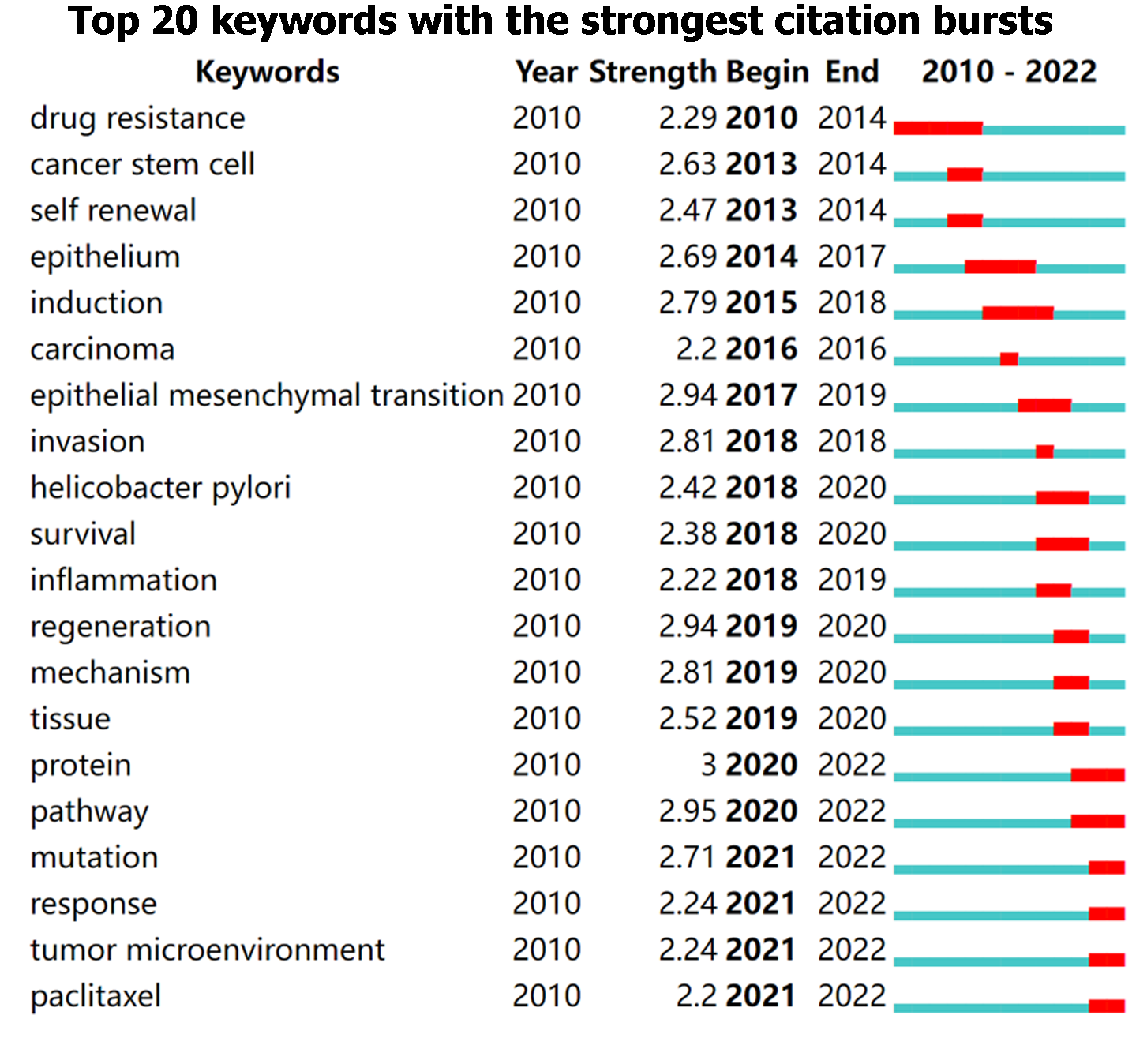
In addition, burst keywords were considered to be the indicators of emerging trends. Overall, it can be classified into 3 phases, beginning from 2010 to 2014 with high intensity in drug resistance, cancer stem cell, and self-renewal, followed by that from 2015 to 2020 with epithelium, carcinoma, epithelial-mesenchymal transition, H. pylori, inflammation, and regeneration year by year, and the third phase from 2020 until present with mutation, response, tumor microenvironment, and paclitaxel. Figure 4 depicts the keywords with the strongest citation bursts in this field.
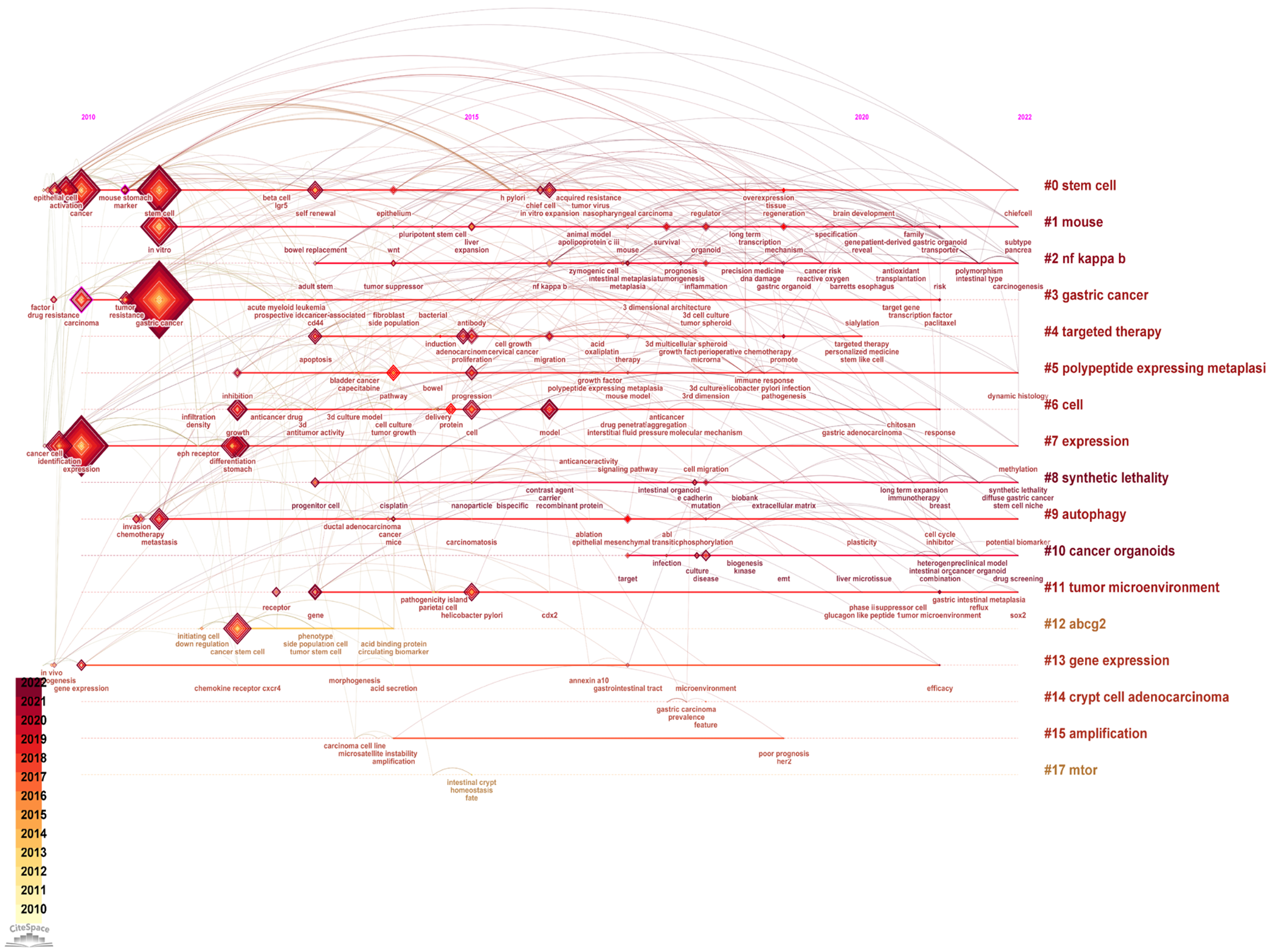
The timeline view of the keywords in gastric organoid is presented in Figure 5. There were a total of 17 main research clusters incorporating gastric organoid applications. In addition, the typical labels in each cluster are reported in Table 4.
| No. | Cluster label | Year | Label |
| 0 | Stem cell | 2015 | Stem cell; cancer; differentiation; model; acid gastric cancer; signet ring carcinoma; matrix metalloproteinase; spdef; transcription factor; stem cell; regeneration; gastric cancer; progenitor; in vitro expansion |
| 1 | Mouse | 2018 | Gastric cancer; transcription factor; regeneration; tumor suppressor genes; identification in vitro; expansion; liver; leptin deficiency; en-y gastric bypass; mouse; liver; infection; bariatric surgery; gastrointestinal cancer |
| 2 | Noncanonical nuclear factor-kappa B | 2019 | Gastric cancer; her2; capecitabine; open label; oxaliplatin drug screening; cancer organoids; precision medicine; Helicobacter pylori; living biobanks; cancer organoids; organoid; drug screenin; tptep1; secretion |
| 3 | Gastric cancer | 2013 | Gene; differentiation; receptor; lgr5; r spondin, gastric cancer; morphogenetic protein; epidermal growth factor receptor; Helicobacter pylori; tissue engineering; tumor microenvironment; receptor; gastric intestinal metaplasia; gene; csc markers (6.04, 0.05) |
| 4 | Targeted therapy | 2016 | Gastric cancer; cancer stem cell; endothelial growth; mammary stem cell; gastric cancers, cancer stem cells; translational research; tumor suppressor genes; gastric organoids; cancer microenvironment; abcg2; cancer stem cell; lgr4; acbp-3; cycle arrest |
| 5 | Polypeptide expression | 2017 | Cancer; progression; microenvironment; mmp 9; angiogenesis, gene expression; endoderm; gastrointestinal tract; mouse small intestine; gata4; gene expression; angiogenesis; epstein barr virus; toripalimab; mmp 9 |
| 6 | Cell | 2015 | Goblet cell carcinoid; crypt cell adenocarcinoma; appendix; mucinous; neuroendocrine; amphophilic; crypt cell adenocarcinoma; appendix; amphophilic; mucinous; neuroendocrine |
| 7 | Expression | 2012 | Survival; tumor suppressor; her2; poor prognosis; amplification, organoids; stomach; chemotherapy; gastroids; proliferation; amplification; gastroids; myc; inactivation; conditional mouse model |
| 8 | Synthetic lethality | 2019 | Gastric cancer; gastric organoids; gland fission; gastric stem cells; mtor; mtor; gland fission; gastric stem cells; gastric organoids; gastric cancer |
| 9 | Autophagy | 2014 | Gastric cancer; Helicobacter pylori; base excision repair; stomach neoplasms; gastric stem cells, nf kappa; trefoil protein; Helicobacter pylori; robust model; atrophic gastritis; NF kappa b; Helicobacter pylori; inflammation; antral epithelium; dynamic histology |
| 10 | Cancer organoids | 2020 | Gastric cancer; resistance; cancer; cetuximab; heterogeneity, cancer stem cell; lauren classification; mucin phenotype expression; cluster; transcription factor; gastric cancer; cd44; stomach; cancer stem cell; chemoresistance |
| 11 | Tumor microenvironment | 2017 | Gastric cancer; t-cell infiltration; recombinant protein; precision oncology; cancer model, stem cell; stomach; promote; cancer; gene expression; targeted therapy; self-renewa; promote; anti-egfr single domain antibody; cancer evolution |
| 12 | abcg 2 | 2012 | Gastric cancer; cancer stem cells; Helicobacter pylori; beta catenin; wound repair identification; stem cell; expression; inhibition; stomach; polypeptide expressing metaplasia; contribute; network; immune response; inhibition |
| 13 | Gene expression | 2013 | Gastric cancer; drug sensitivity; shaker; albumin-bound paclitaxel; stomach, breast cancer; model; growth; drug; identification; cell; breast cancer; growth; recombinant protein; irgd |
| 14 | Crypt cell adenocarcinoma | 2018 | Expression; stomach; stem cell; identification; progenitor cell, gastric cancer; stem cells; cancer biology; beta-galactoside alpha; sialic acid; expression; identification; pathogenesis; metaplasia; progenitor cell |
| 15 | Amplification | 2015 | Gastric cancer; cell adhesion; cell matrix interaction; magnetic resonance imaging; hereditary diffuse gastric cancer, synthetic lethality; discoidin domain receptor; diffuse gastric cancer; magnetic resonance imaging; hereditary diffuse gastric cancer; synthetic lethality; hdgc; e cadherin; e-cadherin; chemoprevention |
| 16 | mTOR | 2015 | Gastric cancer; stem-like subtype; adult mouse; cluster; cathepsin c ovarian; shaker; dynamic; fluid; peritoneal; autophagy; hypoxia inducible factor 1 alpha; cancer; metastasis; fibroblasts |
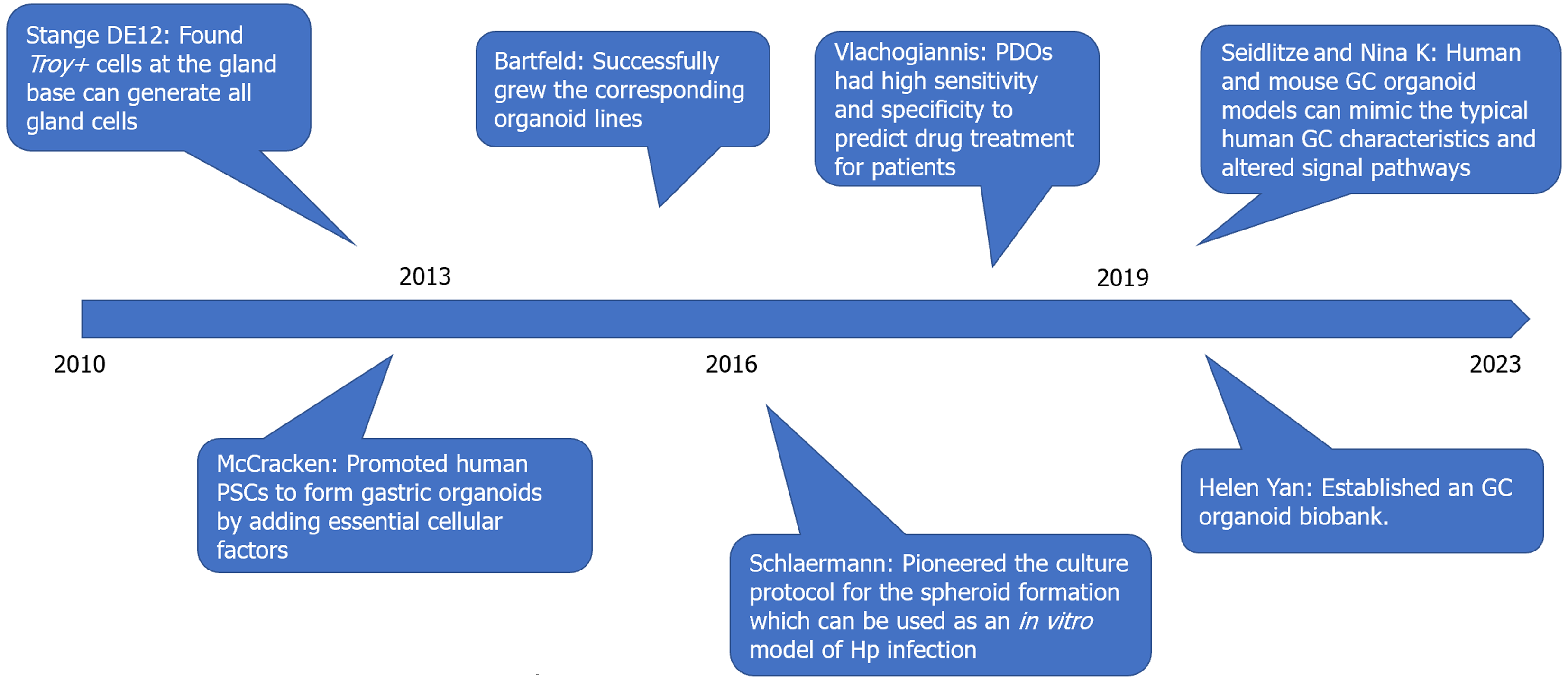
A total of 492 references were co-cited, 11 of which were co-cited > 30 times. Table 5 summarizes the top 10 co-cited studies, and Figure 6 illustrates the main elements of these important studies investigating gastric organoids and the significant findings. McCracken et al[20] used human pluripotent stem cells to form gastric organoids by adding essential cellular factors [such as EGF, BMP4, WNT5a, and fibroblast growth factors (e.g., FGF10)]. Bartfeld et al[6] used flow cytometry to sort 4 specific cell lines, pit mucous cells, gland mucous cells, chief cells, and enteroendocrine cells, and successfully cultured the corresponding organoid lines. Schlaermann et al[7] pioneered a culture protocol for the spheroid formation of the gastric corpus and body, which can be used as an in vitro model of H. pylori infection. Vlachogiannis et al[21] compared the responses of 21 patients with gastrointestinal tumors and patient-derived organoids (PDOs) to targeted drugs or chemotherapy and found that sensitivity and specificity to predict drug treatment for patients were 100% and 93%. Yan et al[9] established an organoid biobank and recorded differentially expressed genes between tumor organoids and paired tumor tissues. Seidlitz et al[22] and Nanki et al[23] demonstrated that human and mouse GC organoid models can mimic the typical human GC characteristics and altered signal pathways, demonstrating their potential role as biomarkers of treatment response.
| Ref. | Title | Journal | Freq | Centrality |
| Bartfeld et al[6], 2015 | In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection | Gastroenterology | 50 | 0.10 |
| Bray et al[24], 2018 | Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries | CA Cancer J Clin | 49 | 0.00 |
| Yan et al[9], 2018 | A Comprehensive Human Gastric Cancer Organoid Biobank Captures Tumor Subtype Heterogeneity and Enables Therapeutic Screening | Cell Stem Cell | 45 | 0.05 |
| Vlachogiannis et al[21], 2018 | Patient-derived organoids model treatment response of metastatic gastrointestinal cancers | Science | 43 | 0.06 |
| Seidlitz et al[22], 2019 | Human gastric cancer modelling using organoids | GUT | 43 | 0.02 |
| Schlaermann et al[7], 2016 | A novel human gastric primary cell culture system for modelling Helicobacter pylori infection in vitro | GUT | 37 | 0.09 |
| Mccracken et al[20], 2014 | Modelling human development and disease in pluripotent stem-cell-derived gastric organoids | Nature | 35 | 0.05 |
| Nanki et al[23], 2018 | Divergent Routes toward Wnt and R-spondin Niche Independency during Human Gastric Carcinogenesis | Cell | 32 | 0.07 |
| Cancer Genome Atlas Research Network[26], 2014 | Comprehensive molecular characterization of gastric adenocarcinoma | Nature | 31 | 0.05 |
| Stange et al[19], 2013 | Differentiated Troy+ chief cells act as reserve stem cells to generate all lineages of the stomach epithelium | Cell | 28 | 0.12 |
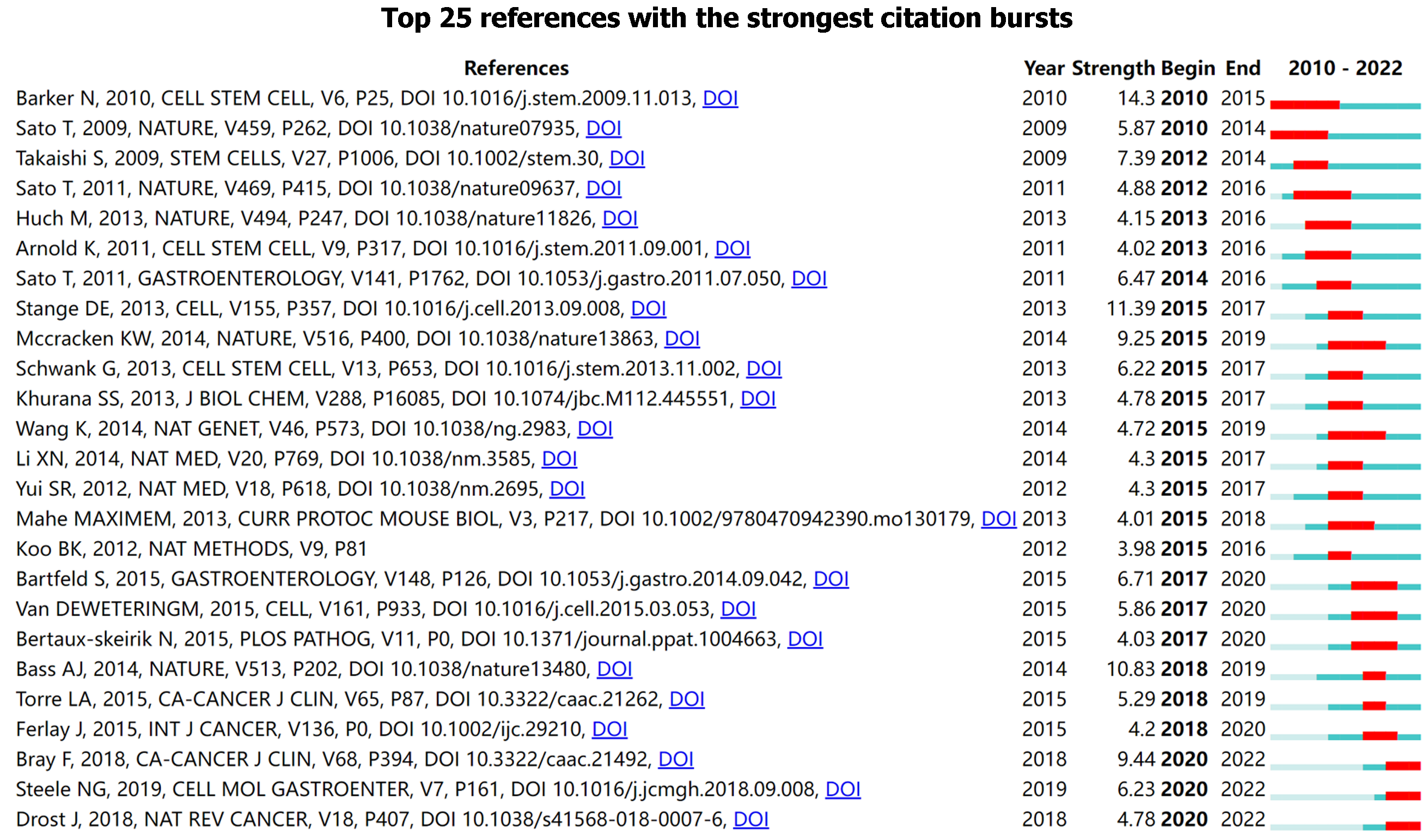
Burst analysis displayed the minimum duration of the bursts as 1 year, although the longest was 5 years. Of these citations, 28% (7/25) of the bursts occurred before 2015, with 36% (9/25) of literature bursts in 2015 and 36% (9/25) after 2017[24]. Twelve percent (3/25) of the citation outbreaks occurred in the past 2 years. Of the top 25 references, the strongest citation burst (14.3) was for studies by Barker et al[25], with the longest citation burst. Barker et al[25] reported that single Lgr5+ stem cells generated gastric-like organs in vitro and that the WNT pathway promoted Lgr5+ stem cell transformation, thereby promoting gastric adenoma formation. The second-highest citation burst appeared in a study by Stange et al[19], with a burst strength of 11.39 from 2015 to 2017. Stange et al[19] reported that Troy+ cells at the gland base can generate all gland cells, which represents remarkable plasticity ability in the field of epithelial stem cell biology. Molecular characterization of gastric adenocarcinoma also demonstrated a citation burst from 2018 to 2019, with a strength of 10.83[26]. In the past 2 years, Steel et al[27] reported that patient tissues exhibited a response to combination drug therapy similar to PDOs and PDOs, which resembled the original patient’s tissue gene mutation, which made a citation burst. Another review from Drost et al[28], which commented on current cancer organoid protocols and the method as a reliable model for cancer research also presented a burst. Figure 7 depicts the citation bursts for the 25 most-cited references.
Professor Hans Clevers, a prolific scientist in the field of adult stem cells and organoid technology, identified Lgr5 as a marker of intestinal stem cells and established the first in vitro 3D organoid culture system for intestinal stem cells with his team. This pioneered the area of organoid research as a disease model for diagnostics, basic science, and preclinical testing. Sato Toshiro exhibited the highest centrality among Asian researchers. He was a postdoctoral researcher in Hans Clevers’ laboratory and was involved in the initial establishment of “mini-guts” in culture, after which he returned to Keio University (Tokyo, Japan) to focus on digestive tract organoids. The most predominant researchers were from the Clevers’ laboratories, including Sina Bartfeld, Johan H Van es, Marc Van de Wetering, and Huch Meritxell[29-31].
The main institutions were those of the more influential authors, such as the University Medical Center Utrecht, University of Cincinnati, University of Michigan, and Keio University. The first research investigating gastrointestinal organoids establishment from the University of Cincinnati was in collaboration with Keio University, where Toshiro Sato worked[32]. Institutions in Europe, the United States, and Japan were more willing to form collaborative networks. Notably, Chinese institutions formed closer collaborations within the country than with external agencies. Chinese researchers are characterized by a lack of influence, which may be attributed to the lack of external collaboration(s), thus necessitating encouragement to form partnerships with institutions around the world[33,34].
The network diagram of keywords revealed that the study of gastric organoids is currently being applied to 2 diseases: H. pylori-infective gastritis and GC. The timeline and burst analysis of keywords reflect past research hotspots and possible future research trends. Based on changes in keywords in this field in this decade, research investigating gastric organoids has gradually shifted from the exploration of model building in the early years to basic research and clinical applications, with the main focus on the mechanism of GC and drug sensitivity testing. In addition, research investigating H. pylori has received attention. The gastric organoid is expected to become a stable and powerful preclinical model in the future.
The results of co-cited literature in this decade suggest a shift from the earliest attempts to successfully construct gastric-like organs to a greater focus on the establishment of gastric organ biobanks. Work by Barker et al[8,25] was the initiating study in this field, laying the foundation for culturing gastric glands in culture dishes and the culture protocol by Bartfeld et al[6], which has become the reference standard for most studies[22,23,35,36]. In clinical applications, isolating GC tissues from patients to establish an organoid sample bank requires high technical expertise[9]. In addition, current technology cannot acquire pure cancer organoids because PDOs are always mixed with healthy tissues[23]. These limitations are offset by many advantages, such as their 3D physiology and the ability to test patient tumor tissues over time to facilitate clinical decision making. Several studies have compared organoid and patient responses to chemotherapeutics and achieved encouraging results[9,22,23,27]. Studies investigating larger-population cohorts are needed to confirm the accuracy of the predictive value of organoids to antitumor drugs. Drug screening for tumor organoids is more likely to be used as a predictive model similar to antibiotic susceptibility testing. Until then, larger samples require diagnostic tests to determine their sensitivity and specificity. To achieve effective clinical translation, clinical research investigating tumor organoids should focus on improving the efficiency and accuracy of drug screening and reappearing the tumor characteristics of patients to the greatest extent. Although these challenges need to be addressed, the overall outlook for predicting clinical outcomes is promising[38,39].
There were some limitations to the present study. Because gastric organoid research is an emerging field, the literature base is still in its infancy; as such, results of bibliometric analyses are inevitably limited. Second, there are currently some frontier technologies in gastric organoids; however, bibliometrics cannot highlight them because they are too innovative and have not been cited enough. These include organoid-on-chips[40] and clustered regularly interspaced palindromic repeats (i.e., “CRISPR”)-associated protein 9-mediated base editing organoids. Jeong developed an innovative human stomach microphysiological system-on-a-chip (hsMPS), which exhibits a significantly improved gastric mucosal barrier. This advancement results from integrating MPS technology with organoid structures. In this hsMPS model, epithelial cells obtained from human antral organoids are co-cultivated with primary gastric mesenchymal stromal cells, isolated from gastric tissues, in an environment with regulated fluid dynamics[41]. Chen et al[42] utilized CRISPR technology in GCOs to reveal that SOX9-marked gastric stem cells play a pivotal role in the malignant transformation process, with biased symmetric cell division being crucial for their malignancy, particularly in regulating symmetric cell division and influencing the expansion of GC cells. Finally, the latest culture method described by Eicher et al[43], who differentiated human pluripotent stem cells into three germ layers, neuroglial, mesenchymal, and epithelial precursors, and subsequently cultured organoids to overcome the limitation of the lack of non-epithelial cells in organoids.
Our study revealed a research area in continuous growth, highlighting a clear trend of increasing focus on GC and H. pylori-infective gastritis. The analysis demonstrated that gastric organoids, as a research platform, has made significant strides in decoding disease mechanisms, optimizing drug screening processes, and developing new therapeutic methods. By evaluating the increase in related publications, the rise in citation rates, and the shifts in keywords, we confirmed that gastric organoid research is an active and evolving branch within the field of gastroenterology. Significant academic contributions have come from top research institutions worldwide, particularly those in the Netherlands and the United States, which have led the way in advancing basic research and clinical applications. Additionally, the influence of individual researchers, such as Hans Clevers and Sato Toshiro, has highlighted the importance of personal contributions in driving scientific progress. Gastric organoid research has demonstrated its importance on multiple levels within gastroenterology and the broader field of biomedical science. It not only offers a platform that more accurately simulates the human physiological state but also opens new avenues for personalized medicine in the future. With technological advancements and methodological improvements, gastric organoid research is expected to continue as a focal point of biomedical innovation, particularly in understanding complex disease mechanisms and developing new treatment strategies.
This study conducts a comprehensive bibliometric analysis of gastric organoid research from 2010 to 2023, shedding light on its evolution and emerging trends.
To systematically map the progress and key developments in gastric organoid research, the study highlights the field’s significance.
To analyze and understand the development, impact, and direction of gastric organoid research using bibliometric methods.
Employed a combination of bibliometric tools and analytical techniques to assess publications and trends in gastric organoid research.
Identified key contributors and institutions in the field, highlighted the application of gastric organoids in studying gastric cancer and Helicobacter pylori-related gastritis.
The study underscores the critical role of gastric organoids in advancing our understanding of disease mechanisms and drug screening processes.
Future research should further explore the potential of gastric organoids in personalized medicine and enhance our comprehension of gastric diseases.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Mijwil MM, Iraq S-Editor: Chen YL L-Editor: A P-Editor: Cai YX
| 1. | Smith E, Cochrane WJ. CYSTIC ORGANOID TERATOMA: (Report of a Case). Can Med Assoc J. 1946;55:151-152. [PubMed] |
| 2. | Pompaiah M, Bartfeld S. Gastric Organoids: An Emerging Model System to Study Helicobacter pylori Pathogenesis. Curr Top Microbiol Immunol. 2017;400:149-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 3. | Cheng L, Xie M, Qiao W, Song Y, Zhang Y, Geng Y, Xu W, Wang L, Wang Z, Huang K, Dong N, Sun Y. Generation and characterization of cardiac valve endothelial-like cells from human pluripotent stem cells. Commun Biol. 2021;4:1039. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 4. | Merker SR, Weitz J, Stange DE. Gastrointestinal organoids: How they gut it out. Dev Biol. 2016;420:239-250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 61] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 5. | Schumacher MA, Aihara E, Feng R, Engevik A, Shroyer NF, Ottemann KM, Worrell RT, Montrose MH, Shivdasani RA, Zavros Y. The use of murine-derived fundic organoids in studies of gastric physiology. J Physiol. 2015;593:1809-1827. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 96] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 6. | Bartfeld S, Clevers H. Organoids as Model for Infectious Diseases: Culture of Human and Murine Stomach Organoids and Microinjection of Helicobacter Pylori. J Vis Exp. 2015;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 83] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 7. | Schlaermann P, Toelle B, Berger H, Schmidt SC, Glanemann M, Ordemann J, Bartfeld S, Mollenkopf HJ, Meyer TF. A novel human gastric primary cell culture system for modelling Helicobacter pylori infection in vitro. Gut. 2016;65:202-213. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 171] [Cited by in RCA: 177] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 8. | Barker N, Clevers H. Leucine-rich repeat-containing G-protein-coupled receptors as markers of adult stem cells. Gastroenterology. 2010;138:1681-1696. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 241] [Cited by in RCA: 275] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 9. | Yan HHN, Siu HC, Law S, Ho SL, Yue SSK, Tsui WY, Chan D, Chan AS, Ma S, Lam KO, Bartfeld S, Man AHY, Lee BCH, Chan ASY, Wong JWH, Cheng PSW, Chan AKW, Zhang J, Shi J, Fan X, Kwong DLW, Mak TW, Yuen ST, Clevers H, Leung SY. A Comprehensive Human Gastric Cancer Organoid Biobank Captures Tumor Subtype Heterogeneity and Enables Therapeutic Screening. Cell Stem Cell. 2018;23:882-897.e11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 278] [Cited by in RCA: 486] [Article Influence: 69.4] [Reference Citation Analysis (0)] |
| 10. | Ren X, Chen W, Yang Q, Li X, Xu L. Patient-derived cancer organoids for drug screening: Basic technology and clinical application. J Gastroenterol Hepatol. 2022;37:1446-1454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 31] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 11. | Pang MJ, Burclaff JR, Jin R, Adkins-Threats M, Osaki LH, Han Y, Mills JC, Miao ZF, Wang ZN. Gastric Organoids: Progress and Remaining Challenges. Cell Mol Gastroenterol Hepatol. 2022;13:19-33. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 12. | Puschhof J, Pleguezuelos-Manzano C, Clevers H. Organoids and organs-on-chips: Insights into human gut-microbe interactions. Cell Host Microbe. 2021;29:867-878. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 117] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 13. | Pan X, Yan E, Cui M, Hua W. Examining the usage, citation, and diffusion patterns of bibliometric mapping software: a comparative study of three tools. J Informetr. 2018;12:481-493. [RCA] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 177] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 14. | van Eck NJ, Waltman L. Software survey: VOSviewer, a computer program for bibliometric mapping. Scientometrics. 2010;84:523-538. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4505] [Cited by in RCA: 5155] [Article Influence: 322.2] [Reference Citation Analysis (1)] |
| 15. | Xie P. Study of international anticancer research trends via co-word and document co-citation visualization analysis. Scientometrics. 2015;105:611-622. [RCA] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 74] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 16. | Gao Y, Shi S, Ma W, Chen J, Cai Y, Ge L, Li L, Wu J, Tian J. Bibliometric analysis of global research on PD-1 and PD-L1 in the field of cancer. Int Immunopharmacol. 2019;72:374-384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 104] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 17. | Chen C. CiteSpace II: Detecting and Visualizing Emerging Trends and Transient Patterns in Scientific Literature. J Am Soc Inf Sci. 2006;57:359-377. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2009] [Cited by in RCA: 2057] [Article Influence: 108.3] [Reference Citation Analysis (0)] |
| 18. | Chen C. Searching for intellectual turning points: progressive knowledge domain visualization. Proc Natl Acad Sci U S A. 2004;101 Suppl 1:5303-5310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1484] [Cited by in RCA: 1315] [Article Influence: 62.6] [Reference Citation Analysis (1)] |
| 19. | Stange DE, Koo BK, Huch M, Sibbel G, Basak O, Lyubimova A, Kujala P, Bartfeld S, Koster J, Geahlen JH, Peters PJ, van Es JH, van de Wetering M, Mills JC, Clevers H. Differentiated Troy+ chief cells act as reserve stem cells to generate all lineages of the stomach epithelium. Cell. 2013;155:357-368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 424] [Cited by in RCA: 420] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 20. | McCracken KW, Catá EM, Crawford CM, Sinagoga KL, Schumacher M, Rockich BE, Tsai YH, Mayhew CN, Spence JR, Zavros Y, Wells JM. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature. 2014;516:400-404. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 649] [Cited by in RCA: 718] [Article Influence: 65.3] [Reference Citation Analysis (0)] |
| 21. | Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernández-Mateos J, Khan K, Lampis A, Eason K, Huntingford I, Burke R, Rata M, Koh DM, Tunariu N, Collins D, Hulkki-Wilson S, Ragulan C, Spiteri I, Moorcraft SY, Chau I, Rao S, Watkins D, Fotiadis N, Bali M, Darvish-Damavandi M, Lote H, Eltahir Z, Smyth EC, Begum R, Clarke PA, Hahne JC, Dowsett M, de Bono J, Workman P, Sadanandam A, Fassan M, Sansom OJ, Eccles S, Starling N, Braconi C, Sottoriva A, Robinson SP, Cunningham D, Valeri N. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359:920-926. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1436] [Cited by in RCA: 1330] [Article Influence: 190.0] [Reference Citation Analysis (0)] |
| 22. | Seidlitz T, Merker SR, Rothe A, Zakrzewski F, von Neubeck C, Grützmann K, Sommer U, Schweitzer C, Schölch S, Uhlemann H, Gaebler AM, Werner K, Krause M, Baretton GB, Welsch T, Koo BK, Aust DE, Klink B, Weitz J, Stange DE. Human gastric cancer modelling using organoids. Gut. 2019;68:207-217. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 150] [Cited by in RCA: 234] [Article Influence: 39.0] [Reference Citation Analysis (0)] |
| 23. | Nanki K, Toshimitsu K, Takano A, Fujii M, Shimokawa M, Ohta Y, Matano M, Seino T, Nishikori S, Ishikawa K, Kawasaki K, Togasaki K, Takahashi S, Sukawa Y, Ishida H, Sugimoto S, Kawakubo H, Kim J, Kitagawa Y, Sekine S, Koo BK, Kanai T, Sato T. Divergent Routes toward Wnt and R-spondin Niche Independency during Human Gastric Carcinogenesis. Cell. 2018;174:856-869.e17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 226] [Article Influence: 37.7] [Reference Citation Analysis (0)] |
| 24. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55853] [Article Influence: 7979.0] [Reference Citation Analysis (132)] |
| 25. | Barker N, Huch M, Kujala P, van de Wetering M, Snippert HJ, van Es JH, Sato T, Stange DE, Begthel H, van den Born M, Danenberg E, van den Brink S, Korving J, Abo A, Peters PJ, Wright N, Poulsom R, Clevers H. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6:25-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1056] [Cited by in RCA: 1198] [Article Influence: 79.9] [Reference Citation Analysis (0)] |
| 26. | Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202-209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5015] [Cited by in RCA: 4859] [Article Influence: 441.7] [Reference Citation Analysis (2)] |
| 27. | Steele NG, Chakrabarti J, Wang J, Biesiada J, Holokai L, Chang J, Nowacki LM, Hawkins J, Mahe M, Sundaram N, Shroyer N, Medvedovic M, Helmrath M, Ahmad S, Zavros Y. An Organoid-Based Preclinical Model of Human Gastric Cancer. Cell Mol Gastroenterol Hepatol. 2019;7:161-184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 104] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 28. | Drost J, Clevers H. Organoids in cancer research. Nat Rev Cancer. 2018;18:407-418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 813] [Cited by in RCA: 1091] [Article Influence: 155.9] [Reference Citation Analysis (0)] |
| 29. | Bartfeld S. Realizing the potential of organoids-an interview with Hans Clevers. J Mol Med (Berl). 2021;99:443-447. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 30. | Lin L, DeMartino J, Wang D, van Son GJF, van der Linden R, Begthel H, Korving J, Andersson-Rolf A, van den Brink S, Lopez-Iglesias C, van de Wetering WJ, Balwierz A, Margaritis T, van de Wetering M, Peters PJ, Drost J, van Es JH, Clevers H. Unbiased transcription factor CRISPR screen identifies ZNF800 as master repressor of enteroendocrine differentiation. Science. 2023;382:451-458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 21] [Reference Citation Analysis (0)] |
| 31. | Huch M, Bonfanti P, Boj SF, Sato T, Loomans CJ, van de Wetering M, Sojoodi M, Li VS, Schuijers J, Gracanin A, Ringnalda F, Begthel H, Hamer K, Mulder J, van Es JH, de Koning E, Vries RG, Heimberg H, Clevers H. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 2013;32:2708-2721. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 460] [Cited by in RCA: 538] [Article Influence: 44.8] [Reference Citation Analysis (0)] |
| 32. | Mahe MM, Aihara E, Schumacher MA, Zavros Y, Montrose MH, Helmrath MA, Sato T, Shroyer NF. Establishment of Gastrointestinal Epithelial Organoids. Curr Protoc Mouse Biol. 2013;3:217-240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 246] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 33. | Chen Z, Li Z, Soutto M, Wang W, Piazuelo MB, Zhu S, Guo Y, Maturana MJ, Corvalan AH, Chen X, Xu Z, El-Rifai WM. Integrated Analysis of Mouse and Human Gastric Neoplasms Identifies Conserved microRNA Networks in Gastric Carcinogenesis. Gastroenterology. 2019;156:1127-1139.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 34. | Chen W, Zhang J, Fu H, Hou X, Su Q, He Y, Yang D. KLF5 Is Activated by Gene Amplification in Gastric Cancer and Is Essential for Gastric Cell Proliferation. Cells. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 35. | Manieri E, Tie G, Malagola E, Seruggia D, Madha S, Maglieri A, Huang K, Fujiwara Y, Zhang K, Orkin SH, Wang TC, He R, McCarthy N, Shivdasani RA. Role of PDGFRA(+) cells and a CD55(+) PDGFRA(Lo) fraction in the gastric mesenchymal niche. Nat Commun. 2023;14:7978. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 36. | Kawasaki K, Toshimitsu K, Matano M, Fujita M, Fujii M, Togasaki K, Ebisudani T, Shimokawa M, Takano A, Takahashi S, Ohta Y, Nanki K, Igarashi R, Ishimaru K, Ishida H, Sukawa Y, Sugimoto S, Saito Y, Maejima K, Sasagawa S, Lee H, Kim HG, Ha K, Hamamoto J, Fukunaga K, Maekawa A, Tanabe M, Ishihara S, Hamamoto Y, Yasuda H, Sekine S, Kudo A, Kitagawa Y, Kanai T, Nakagawa H, Sato T. An Organoid Biobank of Neuroendocrine Neoplasms Enables Genotype-Phenotype Mapping. Cell. 2020;183:1420-1435.e21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 142] [Article Influence: 28.4] [Reference Citation Analysis (0)] |
| 37. | Kishimoto K, Iwasawa K, Sorel A, Ferran-Heredia C, Han L, Morimoto M, Wells JM, Takebe T, Zorn AM. Directed differentiation of human pluripotent stem cells into diverse organ-specific mesenchyme of the digestive and respiratory systems. Nat Protoc. 2022;17:2699-2719. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 38. | Tuveson D, Clevers H. Cancer modeling meets human organoid technology. Science. 2019;364:952-955. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 358] [Cited by in RCA: 367] [Article Influence: 61.2] [Reference Citation Analysis (0)] |
| 39. | Lau HCH, Kranenburg O, Xiao H, Yu J. Organoid models of gastrointestinal cancers in basic and translational research. Nat Rev Gastroenterol Hepatol. 2020;17:203-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 126] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 40. | Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE. Reconstituting organ-level lung functions on a chip. Science. 2010;328:1662-1668. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3032] [Cited by in RCA: 2677] [Article Influence: 178.5] [Reference Citation Analysis (0)] |
| 41. | Carvalho MR, Yan LP, Li B, Zhang CH, He YL, Reis RL, Oliveira JM. Gastrointestinal organs and organoids-on-a-chip: advances and translation into the clinics. Biofabrication. 2023;15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 24] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 42. | Chen Q, Weng K, Lin M, Jiang M, Fang Y, Chung SSW, Huang X, Zhong Q, Liu Z, Huang Z, Lin J, Li P, El-Rifai W, Zaika A, Li H, Rustgi AK, Nakagawa H, Abrams JA, Wang TC, Lu C, Huang C, Que J. SOX9 Modulates the Transformation of Gastric Stem Cells Through Biased Symmetric Cell Division. Gastroenterology. 2023;164:1119-1136.e12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 43. | Eicher AK, Kechele DO, Sundaram N, Berns HM, Poling HM, Haines LE, Sanchez JG, Kishimoto K, Krishnamurthy M, Han L, Zorn AM, Helmrath MA, Wells JM. Functional human gastrointestinal organoids can be engineered from three primary germ layers derived separately from pluripotent stem cells. Cell Stem Cell. 2022;29:36-51.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 83] [Article Influence: 27.7] [Reference Citation Analysis (0)] |