Published online Dec 14, 2024. doi: 10.3748/wjg.v30.i46.4880
Revised: September 4, 2024
Accepted: October 16, 2024
Published online: December 14, 2024
Processing time: 259 Days and 0.7 Hours
Human leukocyte antigen (HLA) class II molecules are cell surface receptor proteins found on antigen-presenting cells. Polymorphisms and mutations in the HLA gene can affect the immune system and the progression of hepatitis B.
To study the relation between rs2856718 of HLA-DQ, rs3077, and rs9277535 of HLA-DP, hepatitis B virus (HBV)-related cirrhosis, and hepatocellular carcinoma (HCC).
In this case-control study, the genotypes of these single nucleotide polymor
The physical distance separating these SNPs was 29816 kB with the disequilibrium (D’) values ranging from 0.07 to 0.34. The close linkage between rs3077 and rs9277535 was attributed to a distance of 21 kB. The D’ value decreased from moderate in the healthy control group (D’ = 0.50, P < 0.05) to weak in the hepatic disease group (D’ < 0.3, P < 0.05). In a combination of the three variants rs2856718, rs3077, and rs9277535, the A allele decreased hepatic disease risk [A-A-A haplotype, risk ratio (RR) = 0.44 (0.14; 1.37), P < 0.05]. The G allele had the opposite effect [G-A/G-G haplotype, RR = 1.12 (1.02; 1.23), P < 0.05]. In liver cancer cases, the A-A-A/G haplotype increased the risk of HCC by 1.58 (P < 0.05).
Rs9277535 affects liver fibrosis progression due to HBV infection, while rs3077 is associated with a risk of HBV-related HCC. The link between rs2856718, rs3077, and rs9277535 and disease risk was determined using a multi-clustering analysis.
Core Tip: A significant correlation was observed between Human leukocyte antigen (HLA)-DP-DQ polymorphisms and the risk of hepatitis B virus (HBV)-related liver cirrhosis (LC) and hepatocellular carcinoma (HCC). In this study, individuals with the HLA-DP and HLA-DQ genotypes were found to have a higher prevalence of LC and HCC. The hypothesis that HLA polymorphisms play a crucial role in the progression of HBV-related liver diseases has been confirmed. Specific HLA-DP and HLA-DQ alleles were associated with the risk of HBV-related LC and HCC. By delving into the genetic underpinnings of these diseases, we highlight the potential for developing personalized medical approaches and targeted therapies for affected individuals.
- Citation: Nguyen TT, Ho TC, Bui HTT, Tran VK, Nguyen TT. Multi-clustering study on the association between human leukocyte antigen-DP-DQ and hepatitis B virus-related hepatocellular carcinoma and cirrhosis in Viet Nam. World J Gastroenterol 2024; 30(46): 4880-4903
- URL: https://www.wjgnet.com/1007-9327/full/v30/i46/4880.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i46.4880
Hepatitis B virus (HBV) is the primary cause of viral hepatitis morbidity. According to estimates from the World Health Organization (2022), there were 254 million chronic hepatitis B infections, with 1.2 million new infections each year. In 2022, approximately 1.1 million deaths worldwide will be caused by HBV, mostly from liver cirrhosis (LC) and hepatocellular carcinoma (HCC)[1]. Viet Nam has been identified as one of 20 countries responsible for 75% of the global burden of viral hepatitis[2], with a high HBV prevalence in adults of approximately 10.5%[3].
Several studies have focused on identifying sensitive loci in human leukocyte antigens (HLA). HLA plays an important role in presenting viral antigens to immune cells that are responsible for killing virus-infected and cancerous cells. Genome-wide association studies (GWAS) of the HLA gene have recently shown that three single nucleotide polymor
Treatment with nucleotide analogs or interferon-alpha inhibits HBV replication in chronically infected patients and slows disease progression to HCC. Losing HBsAg is the goal of HBV treatment but is rarely achieved in these patients (≤ 1% per year), and lifelong treatment is often required. Chronic HBV infection is a heterogeneous disease divided into different phases based on various clinical parameters such as HBsAg, hepatitis B envelope antigen (HBeAg), HBV deoxyribonucleic acid (HBV DNA), and alanine aminotransferase (ALT) levels. The immune system is essential for controlling HBV infection. However, the parameters and combinations associated with achieving a functional cure is still not fully understood. Thus, a deeper understanding of clinical and immune parameters is required[11]. HLA genes have been identified as critical genetic factors for HBV persistence by GWAS through the regulation of adaptive immunity by presenting processed antigens for T cell recognition. Several 3’-untranslated region (3’-UTR) variants in HLA-B, HLA-G, and HLA-DQA1 have been associated with spontaneous HBsAg seroclearance after adjustment for host factors such as age, gender, ethnicity, and viral factors such as HBV genotype and viral load[12].
Machine learning (ML) techniques have demonstrated a significant potential for capturing intricate patterns. When harnessed, these patterns can greatly enhance our understanding of various diseases, leading to more precise advancements in diagnosis, personalized treatment recommendations, and accurate outcome predictions. ML models have demonstrated notable accuracy in addressing HBV-related issues, such as early detection, risk assessment for HBV, and prediction of HBsAg seroclearance[11].
In this study, we focused on multivariate statistics using several algorithms of the hierarchical clustering analysis (HCA) matrix and Pearson’s correlation in R (version 4.1.0)[13]. Chi-square statistics were computed using Yates’ correction for the continuity and generation Pyates. The bias-corrected correlation coefficient (Puncor) and Fisher’s r-to-z transformed correlation coefficient (Pfisher) are two alternative criteria. HCA was used to generate a dendrogram hierarchy of clusters. In combination with [principal component analysis (PCA), CA, MCA, FAMD, and MFA], hierarchical clustering, and partitioning clustering, particularly the K-means method, we studied the complex correlation between genetic polymorphisms of HLA-DP and HLA-DQ and the risk of HBV-related cirrhosis, and HCC in Viet Nam.
This case-control study enrolled healthy controls and patients with chronic hepatitis B, HBV-related LC, and HBV-related HCC patients between 2020 and 2023. 315 healthy controls were recruited from among subjects who underwent routine physical examinations at Hanoi Medical University Hospital. No healthy controls had a history of hepatitis virus infection or other liver diseases. 251 HBV-infected HCC patients positive for HBsAg were obtained from the Viet Nam National Cancer Hospital. In total, 721 patients with persistent HBV infection were enrolled from the National Hospital of Tropical Diseases, Thanh Nhan Hospital, and Thai Nguyen Hospital. Of these, 471 patients had active chronic hepatitis B infection and 250 had LC. Persistent HBV infection was defined as positivity for at least six months for both HBsAg and antibodies against the hepatitis B core antigen (anti-HBc) with or without HBeAg positivity.
Genomic DNA was extracted from peripheral venous blood using the Wizard® Genomic DNA Purification Kit (A1125; Promega, United States). DNA was isolated from whole blood samples using the manual protocol provided by the manufacturer. The isolated DNA was stored at –20 °C until use.
Three SNPs (rs2856718, rs3077, and rs9277535) were genotyped using the TaqMan-Allelic discrimination method with a QuantStudio 3 real-time PCR system (Applied Biosystems, United States), and the results were analyzed using allelic discrimination software (Applied Biosystems, United States). The PCR reactions were performed in 20 µL of reaction mixture containing 50 ng of genomic DNA, 10 µL of 2X Taqman™ Fast Advanced Master Mix (Applied Biosystems, United States), and 0.5 µL of 40X SNP-specific primer/probe. The primers/probes included HLA-DP rs3077 (C_11916951_10), HLA-DP rs9277535 (C_29715274_20), and HLA-DQ rs2856718 (C_27015374_30) (Applied Biosystems). The thermal cycle conditions were followed as: hold at 95 °C for 10 minutes, then amplify for 40 cycles at 95 °C for 15 seconds and 60 °C for 1 minute.
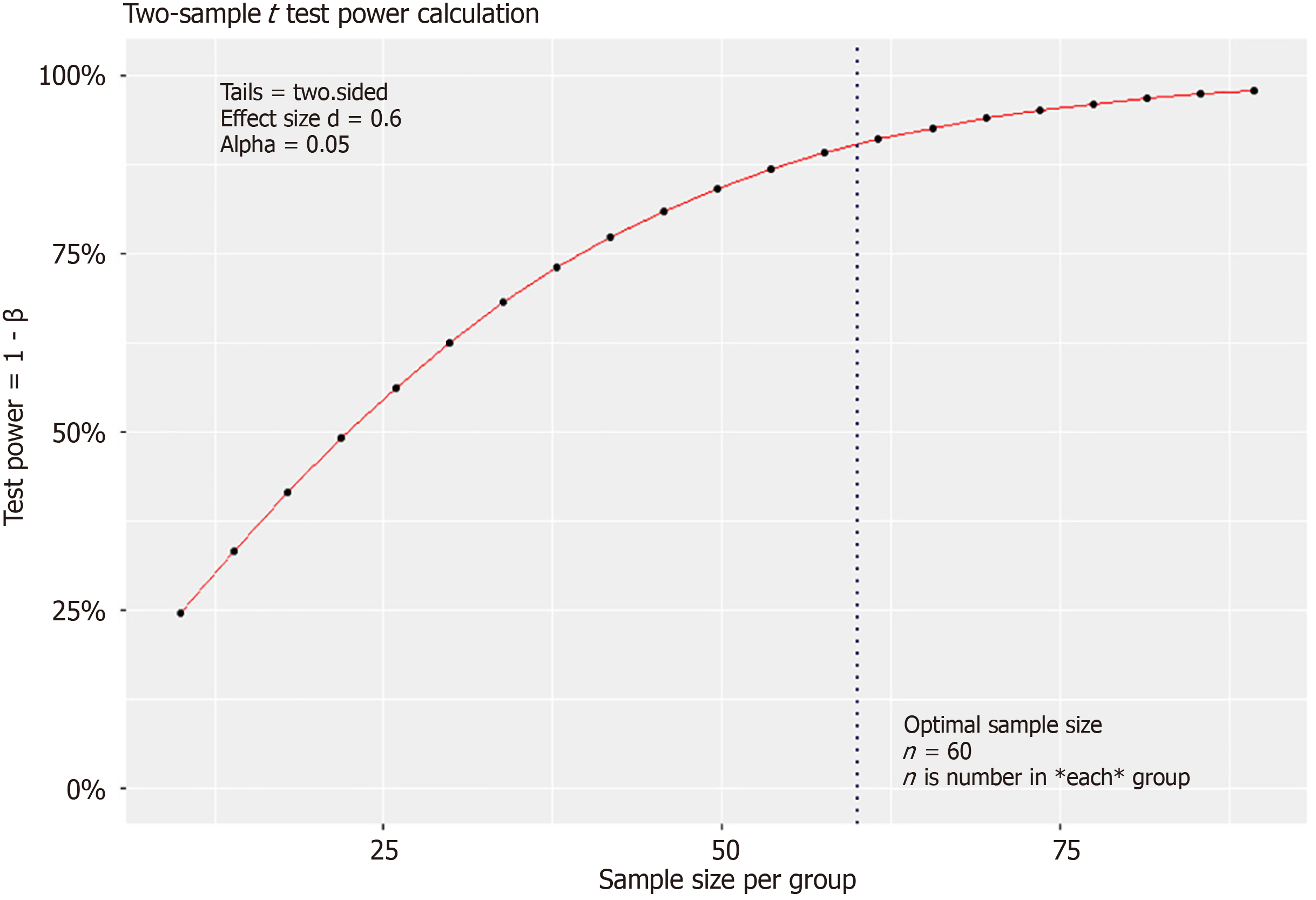
Statistical tests and cluster analyses were performed using R (version 4.4.1). In this correlation study, we used the effect size index d according to Cohen’s criterion, with d = ∆/σ where σ is the standard deviation (the square root of the variance) and ∆ is the influence index of the risk factor (e.g. treatment or genotype) on the population phenotype. With ∆ and the patient’s group μ1 and the control group μ2, we constructed the index ∆ = μ1 - μ2. The smaller the value of ∆, the smaller the difference, and the larger the sample size must be according to the formular: Where the two-sided confidence level is Zα/2. α is the probability of making a type I error and β is the probability of making a type II error. The power of the study is 1 – β. Cohen’s effect size d was calculated as follows: D = ∆/σ[14]. We set criterion d at a moderate level (d = 0.6), with a power of 90%, using a two-sided T test (two-sample) and a statistical significance level (sign level) of 0.05.
The distribution of genotypes between patients and controls was compared using the Chi-square test. Categorical and continuous variables were compared using the Chi-square test and student’s t-test, respectively. Data were expressed as the median (range) or number of cases. Statistical significance was defined as P < 0.05. Hardy-Weinberg equilibrium (HWE) for genotype distributions and the linkage disequilibrium (LD) of SNPs were examined. Chi-squared statistics were computed using Yates’ correction for continuity, generating Pyates. The Pearson or product-moment correlation coefficient is frequently used as the outcome measure for meta-analyses. Pearson’s method is advantageous when all or most of the nonzero parameters share the same sign. Pearson’s test proves useful in a genomic setting for screening age-related genes, which is also our objective. Two alternative criteria were a bias-corrected version of the correlation coefficient (Puncor) and Fisher’s r-to-z transformed correlation coefficient (Pfisher).
The main advantage of the HCA clustering concept is that it shows possible correlations between several factors to provide reference markers useful for diagnostic control and to improve outcome prevention. In particular, the association between genetic characteristics and clinical outcomes requires several in vitro studies; however, these have some limitations. It is critical to clean and prepare the dataset because HCA and K-means cannot operate with missing or noisy data. We combined and validated the data using K-means clustering, which provides several options for the optimal cluster number to produce a PCA cluster plot and define the principal component position. Because our data contained various types of statements, calculating the distance matrix in HCA and K-means proved challenging. We calculated the distance between each observation and the estimated cluster distances from the remaining statements. The distance between the elements can be complete, single, average, ward, squitty, or centroidal. A cluster tree was generated by computing the correlation between cophenetic distances and initial distance data. The number of clusters was determined using K-means, which calculates clustering indices and reallocates observations to the closest cluster. The K-means computation was optimized using 20 indices to construct a PCA cluster plot, which visualized the best cluster number. PCA is a dimensionality reduction method that can reduce the dimensionality of large datasets and transform them into smaller datasets that still contain most of the information in a large set[13].
Cluster analysis was used to evaluate the factors that may interact with gene variants to influence disease progression. Based on the clustering and decision tree results, we identified the first essential factors in each patient group. We maximized multiple metrics simultaneously to obtain the optimal cut-off point and present specific sensitivity and specificity values in the receiver operating characteristic (ROC) plot.
We set the criterion d at a moderate level (d = 0.6) with a power of 90% in a two-sided T test (two-sample), and a statistical significance level (sig. level) of 0.05. This indicates that the considered risk factor (HLA genotype) had a moderate or high level of influence on the phenotype (HCC or Cirrhosis progression). According to the formula in the Methods section and using the pwr.t.test function, the minimum expected sample size for each group was 86 (Figure 1)[14].
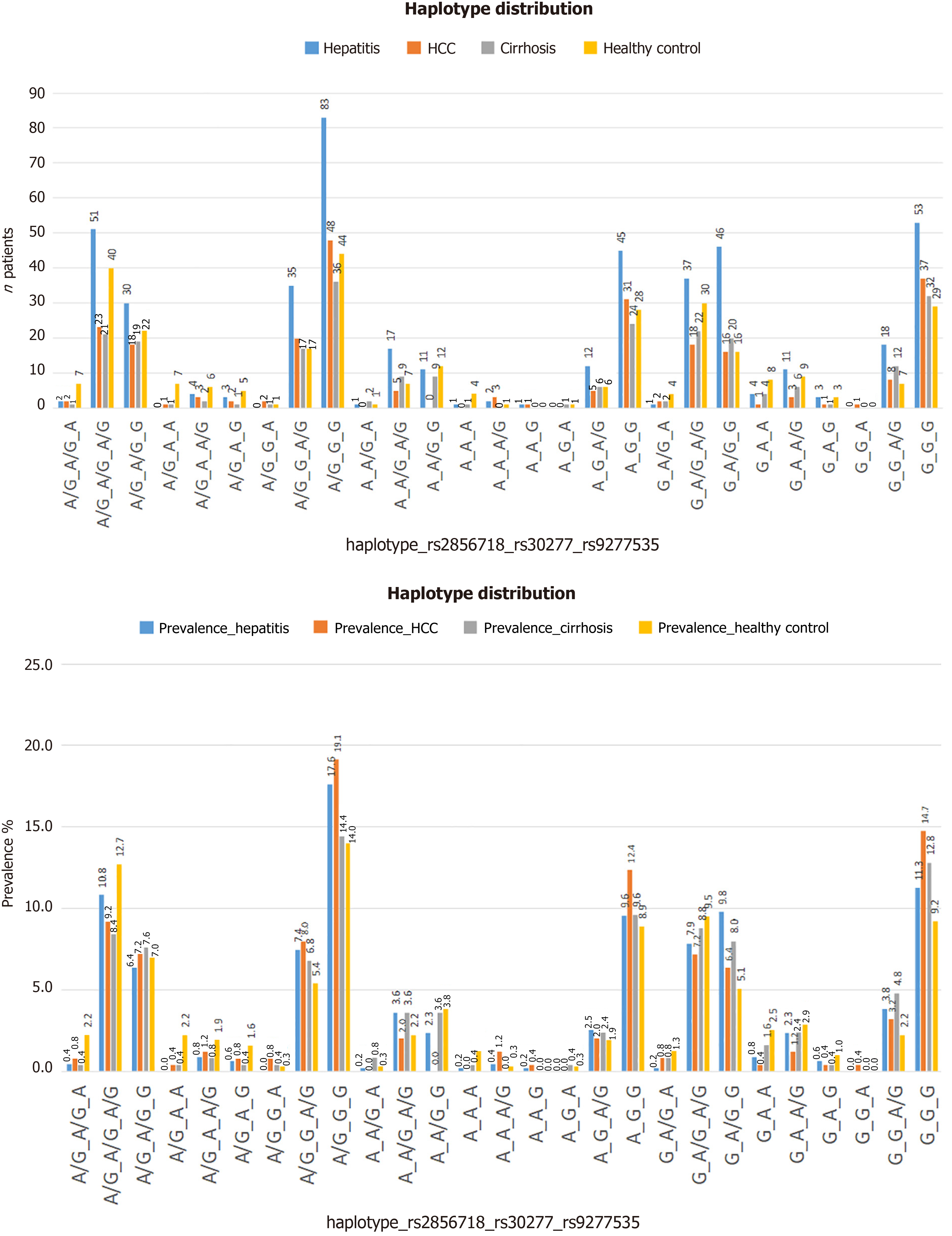
The average age of the patients and study participants in each group was similar. The liver enzyme indices ALT and aspartate aminotransferase (AST) tested in all dot groups showed significant differences between the pathological and healthy groups, as well as between the cirrhosis and remaining disease groups (Table 1). We determined the distribution of genotype or haplotype combinations in each patient/participant group (Figure 2), and found that haplotype A/G-G-G (in order of variation: Rs2856718, rs3077, and rs9277535) had the highest frequency, especially in the HCC group (19.1%, 48/250 patients) (Figure 2).
| Variable | Healthy control | CHB (n = 471) | HBV-related LC | HBV-related HCC | HBV carrier | |
| Age, mean ± SD | 52.78 ± 11.63 | 50.43 ± 14.67 | 54.75 ± 13.36 | 55.99 ± 9.91 | 52.98 ± 13.48 | |
| Age | < 60 | 237 | 353 | 165 | 165 | |
| ≥ 60 | 78 | 118 | 85 | 86 | ||
| Gender | Male | 244 | 367 | 190 | 212 | 769 |
| Female | 71 (22.5) | 104 (22.1) | 60 (24.0) | 39 (15.5) | 203 (20.9) | |
| ALT | 24.92 ± 11.93 | 87.79 ± 168.66 | 388.67 ± 442.21 | 73.19 ± 73.53 | ||
| AST | 24.91 ± 7.68 | 70.89 ± 112.36 | 362.19 ± 401.41 | 87.89 ± 177.25 | ||
Looking at each variation, we identified some critical markers for each specific group. For example, when comparing the risk of chronic hepatitis due to hepatitis B, we found that the AA genotype, the A allele of the rs3077 variant, and the AA genotype or the A allele of the rs9277535 variant could be protective factors with an odds ratio (OR) < 1 and P < 0.05, compared with the healthy control and hepatitis B groups in particular, or the group with liver disease in general (Table 2). In addition, we also observed that the AG of the rs3077 variant could be a protective factor against the risk of progression of hepatitis B or cirrhosis to liver cancer with OR < 1 (P < 0.05) (Table 3). The AA genotype of rs9277535 could also be a protective factor against the risk of developing cirrhosis based on chronic hepatitis B, with OR > 1 (P < 0.05) (Table 3).
| SNPs | Healthy control | HBV carrier | Healthy control vs CHB | Healthy control vs HBV carrier | ||||
| CHB | HBV-related LC | HBV-related HCC | OR (95%CI) | P value | OR (95%CI) | P value | ||
| rs2856718 | ||||||||
| GG | 106 (33.7) | 173 (36.7) | 100 (40.0) | 87 (34.7) | 1 | 1 | ||
| AG | 149 (47.3) | 208 (44.2) | 97 (38.8) | 119 (47.4) | 0.855 (0.621-1.179) | 0.3392 | 0.838 (0.630-1.115) | 0.2253 |
| AA | 60 (19.0) | 90 (19.1) | 53 (21.2) | 45 (17.9) | 0.919 (0.612-1.380) | 0.6840 | 0.923 (0.642-1.326) | 0.6631 |
| AA + AG | 209 (66.3) | 298 (63.3) | 150 (60.0) | 164 (65.3) | 0.874 (0.648-1.179) | 0.3767 | 0.862 (0.660-1.127) | 0.2774 |
| G allele | 361 (57.3) | 554 (58.8) | 297 (59.4) | 293 (58.4) | 1 | 1 | ||
| A allele | 269 (42.7) | 384 (41.2) | 203 (40.6) | 209 (41.6) | 0.930 (0.758-1.141) | 0.4882 | 0.939 (0.782-1.126) | 0.4937 |
| rs3077 | ||||||||
| GG | 133 (42.2) | 246 (52.2) | 128 (51.2) | 152 (60.5) | 1 | 1 | ||
| AG | 139 (44.1) | 196 (41.6) | 107 (42.8) | 84 (33.5 | 0.762 (0.563-1.032) | 0.0702 | 0.704 (0.537-0.924) | 0.0113 |
| AA | 43 (13.7) | 29 (6.2) | 15 (6.0) | 15 (6.0) | 0.365 (0.218-0.611) | 0.0001 | 0.347 (0.224-0.537) | < 0.0001 |
| AA + AG | 182 (57.8) | 225 (47.8) | 122 (48.8) | 99 (39.5) | 0.668 (0.501-0.891) | 0.0060 | 0.620 (0.479-0.801) | 0.0003 |
| G allele | 405 (64.3) | 688 (73.0) | 364 (72.5) | 388 (77.3) | 1 | 1 | ||
| A allele | 225 (35.7) | 254 (27.0) | 138 (27.5) | 114 (22.7) | 0.665 (0.535-0.826) | 0.0002 | 0.632 (0.521-0.765) | < 0.0001 |
| rs9277535 | ||||||||
| GG | 159 (50.5) | 275 (58.4) | 142 (56.8) | 154 (61.4) | 1 | 1 | ||
| AG | 123 (39.0) | 187 (39.7) | 95 (38.0) | 88 (35.0) | 0.879 (0.651-1.186) | 0.3993 | 0.838 (0.640-1.097) | 0.1972 |
| AA | 33 (10.5) | 9 (1.9) | 13 (5.2) | 9 (3.6) | 0.158 (0.074-0.338) | < 0.0001 | 0.262 (0.155-0.440) | < 0.0001 |
| AA + AG | 156 (49.5) | 196 (41.6) | 108 (43.2) | 97 (38.6) | 0.726 (0.545-0.968) | 0.0290 | 0.716 (0.555-0.924) | 0.0102 |
| G allele | 441 (70.0) | 737 (78.1) | 379 (75.8) | 396 (78.9) | 1 | 1 | ||
| A allele | 189 (30.0) | 205 (21.9) | 121 (24.2) | 106 (21.1) | 0.649 (0.516-0.817) | 0.0002 | 0.667 (0.545-0.815) | 0.0001 |
| SNPs | CHB vs HBV-related LC | CHB vs HBV-related HCC | HBV-related LC vs HBV-related HCC | |||
| OR (95%CI) | P value | OR (95%CI) | P value | OR (95%CI) | P value | |
| rs2856718 | ||||||
| GG | 1 | 1 | 1 | |||
| AG | 0.807 (0.572-1.139) | 0.2219 | 1.138 (0.808-1.602) | 0.4601 | 1.410 (0.952-2.089) | 0.0865 |
| AA | 0.019 (0.670-1.549) | 0.9307 | 1.006 (0.647-1.564) | 0.9795 | 0.976 (0.598-1.594) | 0.9224 |
| AA + AG | 0.871 (0.637-1.193) | 0.3891 | 1.094 (0.794-1.508) | 0.5813 | 1.257 (0.874-1.806) | 0.2170 |
| G allele | 1 | 1 | 1 | |||
| A allele | 0.986 (0.791-1.230) | 0.9011 | 1.029 (0.826-1.282) | 0.7984 | 1.044 (0.811-1.342) | 0.7396 |
| rs3077 | ||||||
| GG | 1 | 1 | 1 | |||
| AG | 1.049 (0.763-1.442) | 0.7673 | 0.694 (0.501-0.961) | 0.0278 | 0.661 (0.457-0.957) | 0.0284 |
| AA | 0.994 (0.514-1.921) | 0.9859 | 0.837 (0.435-1.612) | 0.5949 | 0.842 (0.397-1.789) | 0.6548 |
| AA + AG | 1.042 (0.767-1.416) | 0.7924 | 0.712 (0.522-0.972) | 0.0324 | 0.683 (0.479-0.974) | 0.0352 |
| G allele | 1 | 1 | 1 | |||
| A allele | 1.027 (0.805-1.310) | 0.8305 | 0.791 (0.613-1.02) | 0.0705 | 0.775 (0.582-1.032) | 0.0810 |
| rs9277535 | ||||||
| GG | 1 | 1 | 1 | |||
| AG | 0.984 (0.715-1.354) | 0.9204 | 0.840 (0.610-1.159) | 0.2883 | 0.854 (0.591-1.235) | 0.4022 |
| AA | 2.797 (1.168-6.702) | 0.0210 | 1.786 (0.694-4.593) | 0.2290 | 0.638 (0.265-1.539) | 0.3174 |
| AA + AG | 1.067 (0.783-1.455) | 0.6814 | 0.884 (0.646-1.209) | 0.4393 | 0.828 (0.580-1.183) | 0.3001 |
| G allele | 1 | 1 | 1 | |||
| A allele | 1.148 (0.888-1.484) | 0.2924 | 0.962 (0.739-1.254) | 0.7759 | 0.838 (0.623-1.128) | 0.2438 |
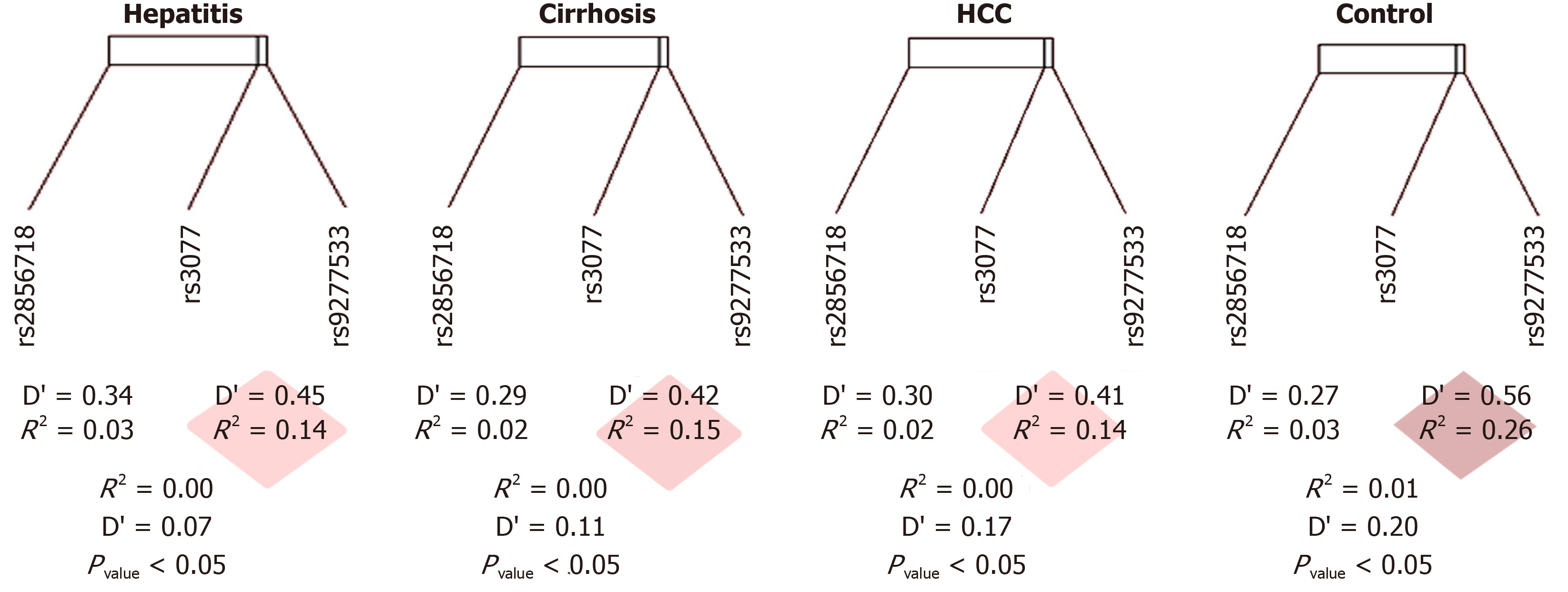
It is necessary to understand whether this OR result has an objective effect of the genotype on the pathological condition or is simply due to the distribution of allele frequencies in the general population. To this end, we conducted tests on link imbalance and distribution in the collected sample group according to the Hardy–Weinberg law. The genotype distributions of three SNPs (rs2856718, rs3077, and rs9277535) met the HWE criterion between the observed and expected frequencies of each genotype in the HCC, cirrhosis, chronic hepatitis B, and control groups (P > 0.05). The physical distance separating these SNPs was 29816 kB and the D’ value ranged from 0.07 to 0.34. The disequilibrium was strong when D’ ≥ 0.80, moderate when D’ was around 0.50, and weak when D’ was 0. Significantly, the close linkage between rs3077 and rs9277535 was due to the distance of 21 kB. We found a decrease in D’ from that of the healthy controls (D’ = 0.50, P < 0.05) in the hepatic disease group (D’ < 0.3, P < 0.05), indicating moderate to weak disequilibrium (Figure 3 and Table 4). The variant rs2856718 showed a moderate LD with rs3077 (D’ from 0.27 to 0.34) but very weak with rs9277535 (D’ from 0.07 to 0.20). This may predict the independence of this SNP in disease status, except for the specific genotype of rs3077.
| Hepatitis | rs3077 | rs9277535 | Cirrhosis | rs3077 | rs9277535 | HCC | rs3077 | rs9277535 | Healthy Control | rs3077 | rs9277535 | ||||
| rs2856718 | D | -0.038 | -0.006 | rs2856718 | D | -0.032 | -0.012 | rs2856718 | D | -0.028 | -0.015 | rs2856718 | D | -0.041 | -0.025 |
| D' | 0.340 | 0.069 | D' | 0.288 | 0.117 | D' | 0.298 | 0.166 | D' | 0.272 | 0.198 | ||||
| Corr.(R) | -0.173 | -0.030 | Corr.(R) | -0.146 | -0.055 | Corr.(R) | -0.136 | -0.073 | Corr.(R) | -0.175 | -0.112 | ||||
| R2 | 0.030 | 0.001 | R2 | 0.021 | 0.003 | R2 | 0.019 | 0.005 | R2 | 0.031 | 0.013 | ||||
| χ2 | 28.168 | 0.874 | χ2 | 10.691 | 1.498 | χ2 | 9.352 | 2.651 | χ2 | 19.260 | 7.903 | ||||
| P value | < 0.0001 | 0.350 | P value | 0.001 | 0.221 | P value | 0.002 | 0.103 | P value | < 0.0001 | 0.005 | ||||
| n | 471 | 471 | n | 250 | 250 | n | 251 | 251 | n | 315 | 315 | ||||
| rs3077 | D | 0.072 | rs3077 | D | 0.074 | rs3077 | D | 0.066 | rs3077 | D | 0.109 | ||||
| D' | 0.450 | D' | 0.419 | D' | 0.405 | D' | 0.565 | ||||||||
| Corr.(R) | 0.391 | Corr.(R) | 0.385 | Corr.(R) | 0.387 | Corr.(R) | 0.496 | ||||||||
| R2 | 0.153 | R2 | 0.148 | R2 | 0.150 | R2 | 0.246 | ||||||||
| χ2 | 143.729 | χ2 | 74.223 | χ2 | 75.122 | χ2 | 155.016 | ||||||||
| P value | < 2.2 × 10-16 | P value | < 2.2 × 10-16 | P value | < 2.2 × 10-16 | P value | < 2.2 × 10-16 | ||||||||
| n | 471 | n | 250 | n | 251 | n | 315 | ||||||||
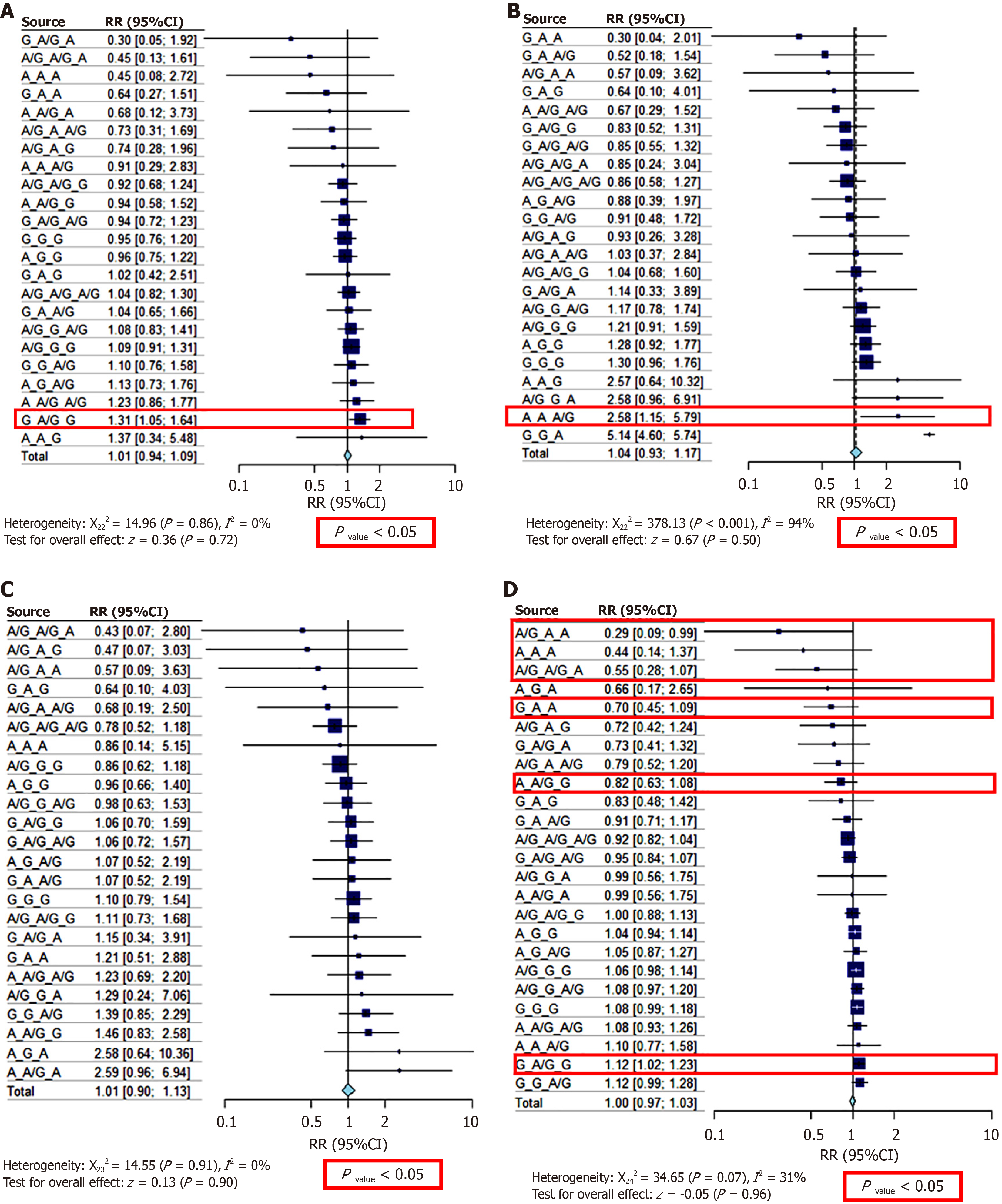
We conducted a risk ratio (RR) test to better understand the disease risk of the genotype or haplotype combinations of the three variants. Detailed results are presented in Table 4 and Figure 4. The G-A/G-G haplotype [RR = 1.31 (1.05; 1.64), P < 0.01] increased the risk of chronic hepatitis B by 31% when the group of patients with chronic hepatitis B was compared with the other groups and the healthy control group (Figure 4A). This haplotype also increased the risk of overall liver disease by 12% compared to healthy individuals, with RR = 1.12 [1.02; 1.23] (P < 0.05) (Figure 4D). In cases of liver cancer, haplotype A-A-A/G increased the risk of liver cancer by 1.58 times RR = 2.58 (1.15; 5.79), P < 0.05 (Figure 4B). The A/G-A-A, A-A-A, A/G-A/G-A, G-A-A, and A-A/G-G haplotypes significantly reduced the risk of liver disease from 18% [RR = 0.82 (0.63; 1.08)] to 71% [RR = 0.29 (0.09; 0.99)] (P < 0.01 and P < 0.05) (Figure 4D) (Table 5).
| Haplotype: Rs2856718, rs30277, rs9277535 | A/G_A/G_A | A/G_A/G_A/ | A/G_A/G_G | A/G_A_A | A/G_A_A/G | A/G_A_G | A/G_G_A | A/G_G_A/G | A/G_G_G | A_A/G_A | A_A/G_A/G | A_A/G_G | A_A_A | A_A_A/G | A_A_G | A_G_A | A_G_A/G | A_G_G | G_A/G_A | G_A/G_A/G | G_A/G_G | G_A_A | G_A_A/G | G_A_G | G_G_A | G_G_A/G | G_G_G | |
| Hepa | N | 2 | 51 | 30 | NA | 4 | 3 | NA | 35 | 83 | 1 | 17 | 11 | 1 | 2 | 1 | NA | 12 | 45 | 1 | 37 | 46 | 4 | 11 | 3 | NA | 18 | 53 |
| % | 0.4 | 10.8 | 6.4 | NA | 0.8 | 0.6 | NA | 7.4 | 17.6 | 0.2 | 3.6 | 2.3 | 0.2 | 0.4 | 0.2 | NA | 2.5 | 9.6 | 0.2 | 7.9 | 9.8 | 0.8 | 2.3 | 0.6 | NA | 3.8 | 11.3 | |
| Pfisher | 1.15 × 10-01 | 3.89 × 10-01 | 3.24 ×10-01 | 1.53 × 10-02 | 2.96 × 10-01 | 3.78 × 10-01 | 1.52 × 10-01 | 2.85 × 10-01 | 1.95 × 10-01 | 5.00 × 10-01 | 1.54 × 10-01 | 4.27 × 10-01 | 2.12 × 10-01 | 5.00 × 10-01 | 5.00 × 10-01 | 2.68 × 10-01 | 2.83 × 10-01 | 3.86 × 10-01 | 8.36 × 10-02 | 3.38 × 10-01 | 1.47 × 10-02 | 1.59 × 10-01 | 4.24 × 10-01 | 5.00 × 10-01 | 5.00 × 10-01 | 3.19 × 10-01 | 3.60 × 10-01 | |
| Pyates | 1.27 × 10-01 | 4.18 ×10-01 | 3.18 × 10-01 | 2.62 × 10-02 | 2.97 × 10-01 | 3.71 × 10-01 | 1.58 × 10-01 | 3.30 × 10-01 | 2.05 × 10-01 | 5.00 × 10-01 | 1.88 × 10-01 | 4.69 × 10-01 | 2.77 × 10-01 | 5.00 × 10-01 | 5.00 × 10-01 | 3.67 × 10-01 | 3.65 × 10-01 | 3.97 × 10-01 | 1.06 × 10-01 | 3.64 × 10-01 | 1.78 × 10-02 | 1.91 × 10-01 | 5.00 × 10-01 | 5.00 × 10-01 | 5.00 × 10-01 | 3.73 × 10-01 | 3.76 × 10-01 | |
| Puncor | 7.49 × 10-02 | 3.82 × 10-01 | 2.79 × 10-01 | 1.11 × 10-02 | 2.11 × 10-01 | 2.60 × 10-01 | 6.40 × 10-02 | 2.90 × 10-01 | 1.83 × 10-01 | 3.15 × 10-01 | 1.45 × 10-01 | 3.96 ×10-01 | 1.55 × 10-01 | 4.34 × 10-01 | 3.47 × 10-01 | 1.41 × 10-01 | 2.94 × 10-01 | 3.61 × 10-01 | 5.56 × 10-02 | 3.25 × 10-01 | 1.35 × 10-02 | 1.30 × 10-01 | 4.40 × 10-01 | 4.79 × 10-01 | 2.24 × 10-01 | 3.15 × 10-01 | 3.42 × 10-01 | |
| RR | 0.45 | 1.04 | 0.92 | NA | 0.73 | 0.74 | NA | 1.08 | 1.09 | 0.68 | 1.23 | 0.94 | 0.45 | 0.91 | 1.37 | NA | 1.13 | 0.96 | 0.3 | 0.94 | 1.31 | 0.64 | 1.04 | 1.02 | NA | 1.1 | 0.95 | |
| Low | 0.13 | 0.82 | 0.68 | NA | 0.31 | 0.28 | NA | 0.83 | 0.91 | 0.12 | 0.86 | 0.58 | 0.08 | 0.29 | 0.34 | NA | 0.73 | 0.75 | 0.05 | 0.72 | 1.05 | 0.27 | 0.65 | 0.42 | NA | 0.76 | 0.76 | |
| Upp | 1.61 | 1.3 | 1.24 | NA | 1.69 | 1.96 | NA | 1.41 | 1.31 | 3.73 | 1.77 | 1.52 | 2.72 | 2.83 | 5.48 | NA | 1.76 | 1.22 | 1.92 | 1.23 | 1.64 | 1.51 | 1.66 | 2.51 | NA | 1.58 | 1.2 | |
| HCC | N | 2 | 23 | 18 | 1 | 3 | 2 | 2 | 20 | 48 | NA | 5 | NA | NA | 3 | 1 | NA | 5 | 31 | 2 | 18 | 16 | 1 | 3 | 1 | 1 | 8 | 37 |
| % | 0.8 | 9.2 | 7.2 | 0.4 | 1.2 | 0.8 | 0.8 | 8 | 19.1 | NA | 2 | NA | NA | 1.2 | 0.4 | NA | 2 | 12.4 | 0.8 | 7.2 | 6.4 | 0.4 | 1.2 | 0.4 | 0.4 | 3.2 | 14.7 | |
| Pfisher | 5.00 × 10-01 | 2.46 × 10-01 | 4.45 × 10-01 | 5.00 × 10-01 | 5.00 × 10-01 | 5.00 × 10-01 | 8.65 × 10-02 | 2.44 × 10-01 | 1.08 × 10-01 | 5.00 × 10-01 | 2.04 × 10-01 | 6.15 × 10-04 | 3.02 × 10-01 | 4.62 × 10-02 | 1.76 × 10-01 | 5.00 × 10-01 | 5.00 × 10-01 | 7.96 × 10-02 | 3.45 × 10-01 | 2.63 × 10-01 | 2.53 × 10-01 | 1.10 × 10-01 | 1.70 × 10-01 | 5.00 × 10-01 | 9.75 × 10-02 | 5.00 × 10-01 | 5.09 × 10-02 | |
| Pyates | 5.00 × 10-01 | 2.58 × 10-01 | 4.84 × 10-01 | 4.15 × 10-01 | 5.00 × 10-01 | 5.00 × 10-01 | 1.81 × 10-01 | 2.76 × 10-01 | 1.14 × 10-01 | 3.62 × 10-01 | 2.14 × 10-01 | 4.75 × 10-03 | 2.44 × 10-01 | 8.48 × 10-02 | 4.22 × 10-01 | 5.00 × 10-01 | 4.71 × 10-01 | 9.65 × 10-02 | 5.00 × 10-01 | 2.73 × 10-01 | 2.44 × 10-01 | 1.32 × 10-01 | 1.53 × 10-01 | 4.79 × 10-01 | 2.21 × 10-01 | 4.58 × 10-01 | 6.16 × 10-02 | |
| Puncor | 4.02 × 10-01 | 2.22 × 10-01 | 4.29 × 10-01 | 2.62 × 10-01 | 4.81 × 10-01 | 4.56 × 10-01 | 6.16 × 10-02 | 2.32 × 10-01 | 9.65 × 10-02 | 1.62 × 10-01 | 1.58 × 10-01 | 2.40 × 10-03 | 1.13 × 10-01 | 2.94 × 10-02 | 1.38 × 10-01 | 2.43 × 10-01 | 3.78 × 10-01 | 7.79 × 10-02 | 4.18 × 10-01 | 2.32 × 10-01 | 2.05 × 10-01 | 7.68 × 10-02 | 1.04 × 10-01 | 3.08 × 10-01 | 2.11 × 10-02 | 3.83 × 10-01 | 4.94 × 10-02 | |
| RR | 0.85 | 0.86 | 1.04 | 0.57 | 1.03 | 0.93 | 2.58 | 1.17 | 1.21 | NA | 0.67 | NA | NA | 2.58 | 2.57 | NA | 0.88 | 1.28 | 1.14 | 0.85 | 0.83 | 0.3 | 0.52 | 0.64 | 5.14 | 0.91 | 1.3 | |
| Low | 0.24 | 0.58 | 0.68 | 0.09 | 0.37 | 0.26 | 0.96 | 0.78 | 0.91 | NA | 0.29 | NA | NA | 1.15 | 0.64 | NA | 0.39 | 0.92 | 0.33 | 0.55 | 0.52 | 0.04 | 0.18 | 0.1 | 4.6 | 0.48 | 0.96 | |
| Upp | 3.04 | 1.27 | 1.6 | 3.62 | 2.84 | 3.28 | 6.91 | 1.74 | 1.59 | NA | 1.52 | NA | NA | 5.79 | 10.32 | NA | 1.97 | 1.77 | 3.89 | 1.32 | 1.31 | 2.01 | 1.54 | 4.01 | 5.75 | 1.72 | 1.76 | |
| Cirr | N | 1 | 21 | 19 | 1 | 2 | 1 | 1 | 17 | 36 | 2 | 9 | 9 | 1 | NA | NA | 1 | 6 | 24 | 2 | 22 | 20 | 4 | 6 | 1 | NA | 12 | 32 |
| % | 0.4 | 8.4 | 7.6 | 0.4 | 0.8 | 0.4 | 0.4 | 6.8 | 14.4 | 0.8 | 3.6 | 3.6 | 0.4 | NA | NA | 0.4 | 2.4 | 9.6 | 0.8 | 8.8 | 8 | 1.6 | 2.4 | 0.4 | NA | 4.8 | 12.8 | |
| Pfisher | 2.40 × 10-01 | 1.26 × 10-01 | 3.38 × 10-01 | 5.00 × 10-01 | 3.74 × 10-01 | 3.51 × 10-01 | 2.89 × 10-01 | 5.00 × 10-01 | 1.96 × 10-01 | 8.59 × 10-02 | 2.66 × 10-01 | 1.27 × 10-01 | 5.00 × 10-01 | 3.02 × 10-01 | 5.00 × 10-01 | 1.75 × 10-01 | 4.07 × 10-01 | 4.53 × 10-01 | 3.45 × 10-01 | 3.99 × 10-01 | 3.95 × 10-01 | 3.78 × 10-01 | 4.07 × 10-01 | 5.00 × 10-01 | 5.00 × 10-01 | 1.24 × 10-01 | 2.92 × 10-01 | |
| Pyates | 2.71 × 10-01 | 1.39 × 10-01 | 3.68 ×10-01 | 4.17 ×10-01 | 3.93 × 10-01 | 3.13 × 10-01 | 5.00 ×10-01 | 5.00 × 10-01 | 1.97 × 10-01 | 1.80 × 10-01 | 3.21 × 10-01 | 1.51 × 10-01 | 5.00 × 10-01 | 2.46 × 10-01 | 5.00 × 10-01 | 4.21 × 10-01 | 5.00 × 10-01 | 4.66 × 10-01 | 5.00 × 10-01 | 4.28 × 10-01 | 4.51 × 10-01 | 4.51 × 10-01 | 5.00 × 10-01 | 4.81 × 10-01 | 5.00 × 10-01 | 1.45 × 10-01 | 3.17 × 10-01 | |
| Puncor | 1.65 × 10-01 | 1.15 × 10-01 | 3.17 × 10-01 | 2.63 × 10-01 | 2.74 × 10-01 | 1.92 × 10-01 | 3.89 × 10-01 | 4.68 × 10-01 | 1.71 × 10-01 | 6.08 × 10-02 | 2.50 × 10-01 | 1.04 × 10-01 | 4.32 ×10-01 | 1.14 × 10-01 | 2.44 × 10-01 | 1.37 × 10-01 | 4.31 × 10-01 | 4.19 × 10-01 | 4.16 × 10-01 | 3.78 × 10-01 | 3.99 × 10-01 | 3.33 × 10-01 | 4.31 × 10-01 | 3.10 × 10-01 | 3.12 × 10-01 | 1.06 × 10-01 | 2.80 × 10-01 | |
| RR | 0.43 | 0.78 | 1.11 | 0.57 | 0.68 | 0.47 | 1.29 | 0.98 | 0.86 | 2.59 | 1.23 | 1.46 | 0.86 | NA | NA | 2.58 | 1.07 | 0.96 | 1.15 | 1.06 | 1.06 | 1.21 | 1.07 | 0.64 | NA | 1.39 | 1.1 | |
| Low | 0.07 | 0.52 | 0.73 | 0.09 | 0.19 | 0.07 | 0.24 | 0.63 | 0.62 | 0.96 | 0.69 | 0.83 | 0.14 | NA | NA | 0.64 | 0.52 | 0.66 | 0.34 | 0.72 | 0.7 | 0.51 | 0.52 | 0.1 | NA | 0.85 | 0.79 | |
| Upp | 2.8 | 1.18 | 1.68 | 3.63 | 2.5 | 3.03 | 7.06 | 1.53 | 1.18 | 6.94 | 2.2 | 2.58 | 5.15 | NA | NA | 10.36 | 2.19 | 1.4 | 3.91 | 1.57 | 1.59 | 2.88 | 2.19 | 4.03 | NA | 2.29 | 1.54 | |
| Heal | N | 7 | 40 | 22 | 7 | 6 | 5 | 1 | 17 | 44 | 1 | 7 | 12 | 4 | 1 | NA | 1 | 6 | 28 | 4 | 30 | 16 | 8 | 9 | 3 | NA | 7 | 29 |
| % | 2.2 | 12.7 | 7 | 2.2 | 1.9 | 1.6 | 0.3 | 5.4 | 14 | 0.3 | 2.2 | 3.8 | 1.3 | 0.3 | NA | 0.3 | 1.9 | 8.9 | 1.3 | 9.5 | 5.1 | 2.5 | 2.9 | 1 | NA | 2.2 | 9.2 | |
| Pfisher | 6.13 × 10-03 | 7.00 × 10-02 | 5.00 × 10-01 | 5.62 × 10-04 | 1.11 × 10-01 | 7.52 × 10-02 | 5.00 × 10-01 | 1.26 × 10-01 | 9.50 × 10-02 | 5.00 × 10-01 | 2.24 × 10-01 | 4.77 × 10-02 | 1.73 × 10-02 | 5.00 ×10-01 | 5.00 × 10-01 | 2.15 × 10-01 | 4.14 × 10-01 | 2.58 × 10-01 | 1.17 × 10-01 | 2.05 × 10-01 | 2.55 × 10-02 | 2.15 × 10-02 | 1.94 × 10-01 | 2.07 × 10-01 | 5.00 × 10-01 | 1.08 × 10-01 | 6.52 × 10-02 | |
| Pyates | 8.12 × 10-03 | 8.59 × 10-02 | 5.00 ×10-01 | 4.14 × 10-04 | 1.35 × 10-01 | 1.01 × 10-01 | 5.00 × 10-01 | 1.37 × 10-01 | 1.05 × 10-01 | 5.00 × 10-01 | 2.45 × 10-01 | 6.34 × 10-02 | 2.66 × 10-02 | 5.00 × 10-01 | 5.00 × 10-01 | 4.93 × 10-01 | 3.97 × 10-01 | 2.70 × 10-01 | 1.56 × 10-01 | 2.18 × 10-01 | 3.36 × 10-02 | 2.90 × 10-02 | 2.70 × 10-01 | 3.27 × 10-01 | 5.00 × 10-01 | 1.07 × 10-01 | 6.65 × 10-02 | |
| Puncor | 3.06 × 10-03 | 7.05 × 10-02 | 4.78 × 10-01 | 9.49 × 10-05 | 7.98 × 10-02 | 5.20 × 10-02 | 4.90 × 10-01 | 1.11 × 10-01 | 9.04 × 10-02 | 4.90 × 10-01 | 1.89 × 10-01 | 4.13 × 10-02 | 7.99 × 10-03 | 3.28 × 10-01 | 2.10 × 10-01 | 2.00 × 10-01 | 3.16 × 10-01 | 2.35 × 10-01 | 8.10 × 10-02 | 1.85 × 10-01 | 2.55 × 10-02 | 1.46 × 10-02 | 2.03 × 10-01 | 1.95 × 10-01 | 2.85 × 10-01 | 7.83 × 10-02 | 5.44 × 10-02 | |
| RR | 0.55 | 0.92 | 1.00 | 0.29 | 0.79 | 0.72 | 0.99 | 1.08 | 1.06 | 0.99 | 1.08 | 0.82 | 0.44 | 1.10 | 1.32 | 0.66 | 1.05 | 1.04 | 0.73 | 0.95 | 1.12 | 0.70 | 0.91 | 0.83 | 1.32 | 1.12 | 1.08 | |
| Low | 0.28 | 0.82 | 0.88 | 0.09 | 0.52 | 0.42 | 0.56 | 0.97 | 0.98 | 0.56 | 0.93 | 0.63 | 0.14 | 0.77 | 1.28 | 0.17 | 0.87 | 0.94 | 0.41 | 0.84 | 1.02 | 0.45 | 0.71 | 0.48 | 1.28 | 0.99 | 0.99 | |
| Upp | 1.07 | 1.04 | 1.13 | 0.99 | 1.20 | 1.24 | 1.75 | 1.20 | 1.14 | 1.75 | 1.26 | 1.08 | 1.37 | 1.58 | 1.37 | 2.65 | 1.27 | 1.14 | 1.32 | 1.07 | 1.23 | 1.09 | 1.17 | 1.42 | 1.37 | 1.28 | 1.18 | |
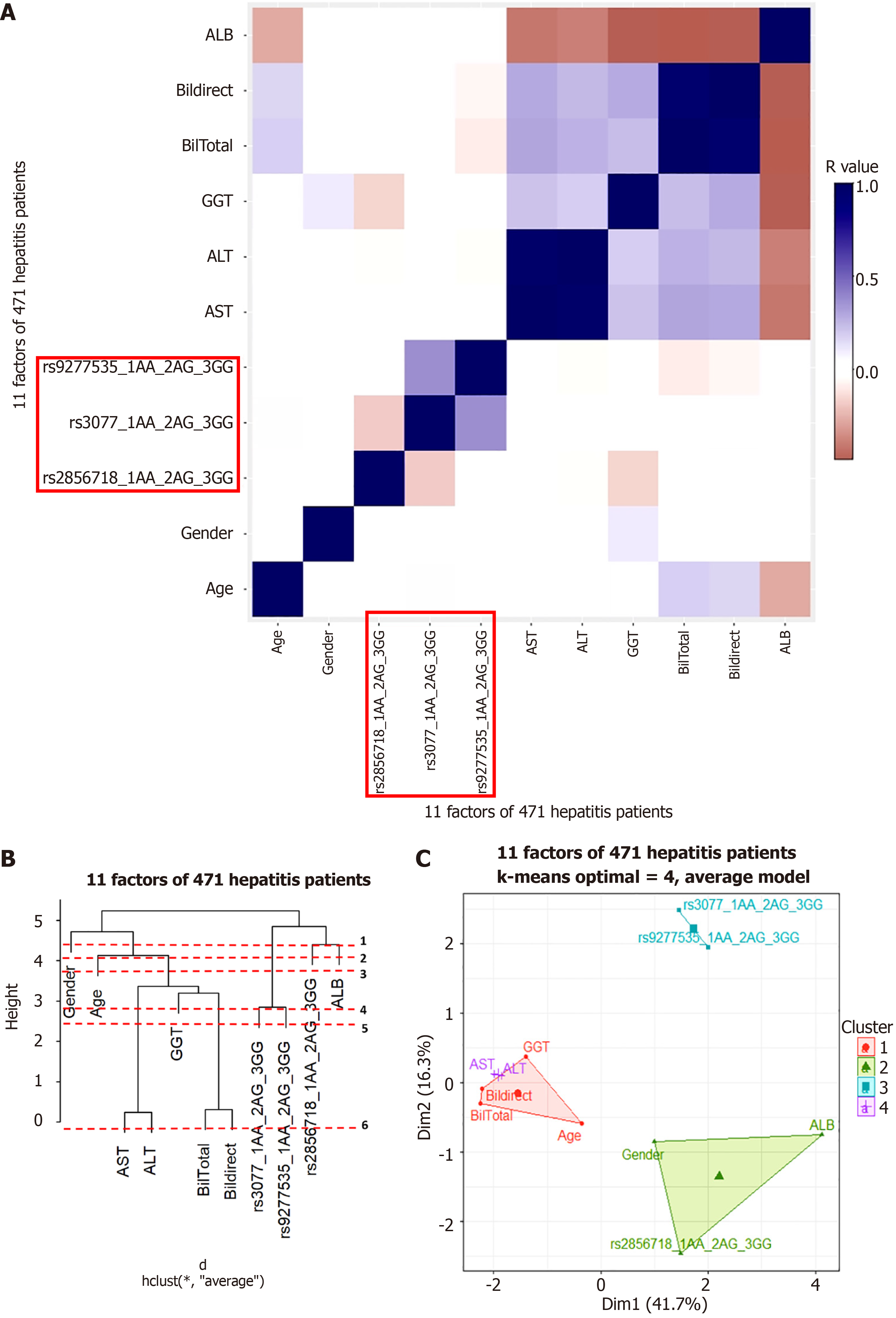
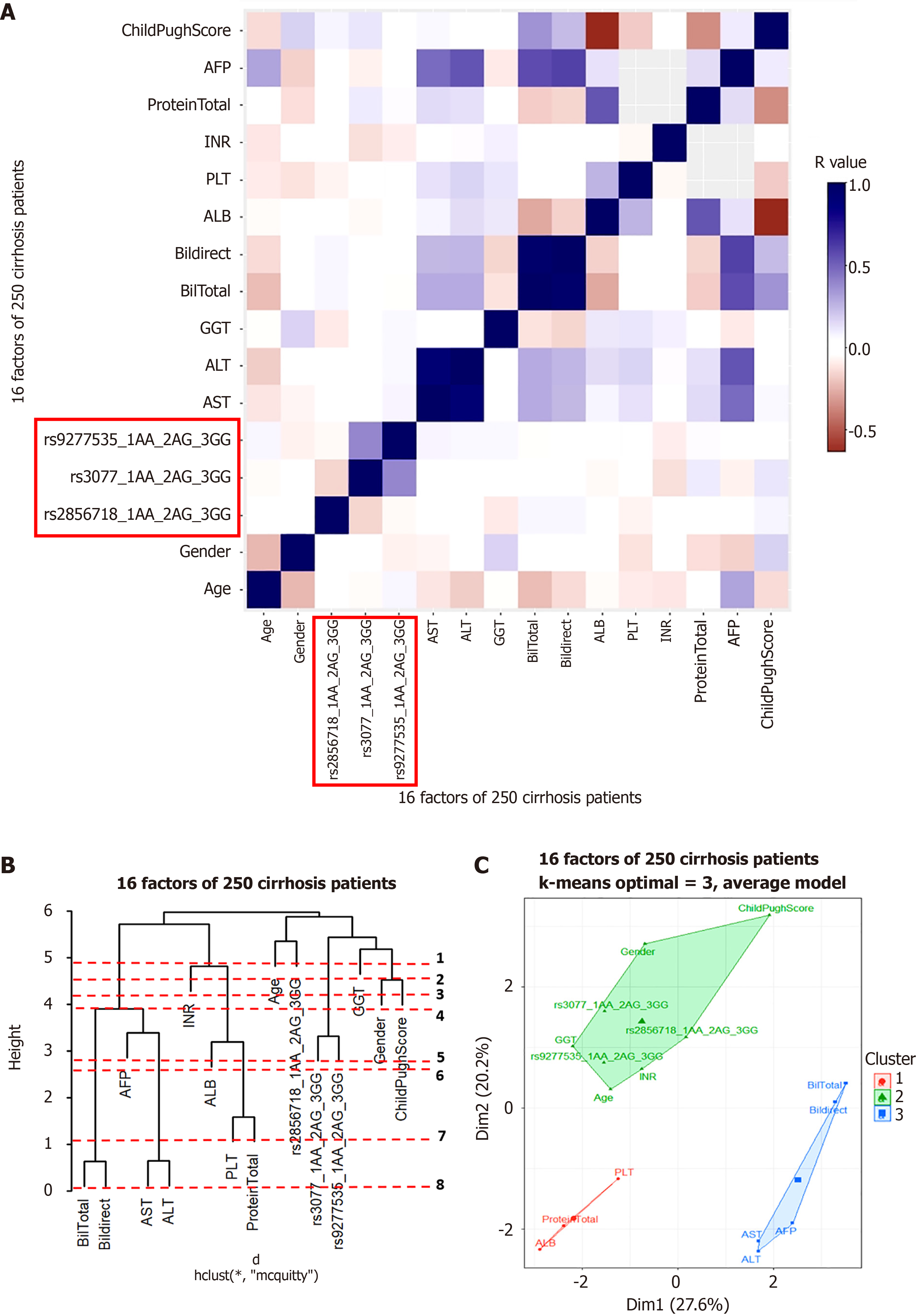
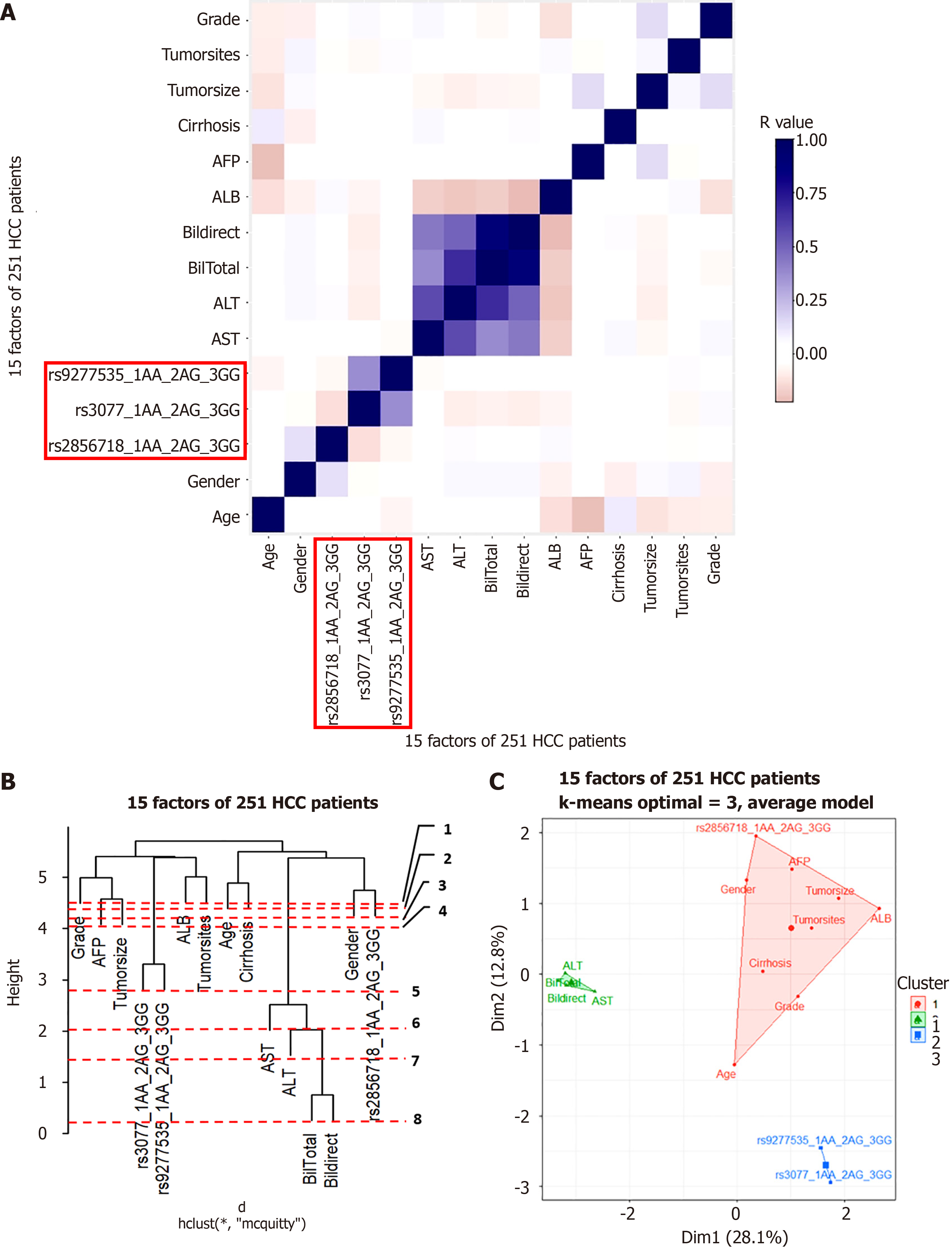
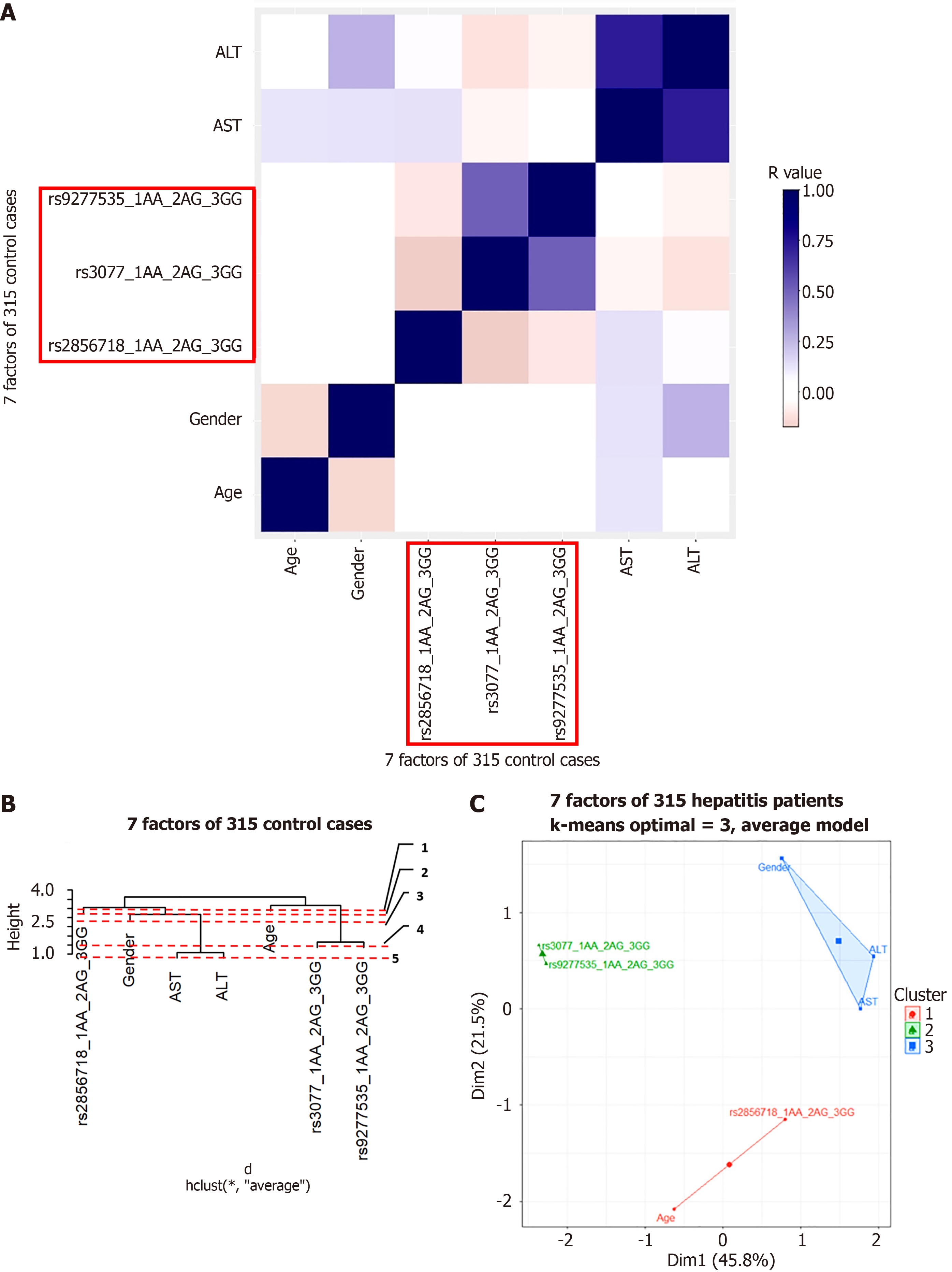
Cluster analysis was used to understand the factors that may interact with gene variants to influence disease progression. Pearson’s correlation was tested for each group. The results are shown in Figures 5A, 6A, 7A, and 8A for the hepatitis B, cirrhosis, liver cancer, and healthy control groups, respectively. In all groups, we saw a significant positive correlation of the two variants rs3077-rs9277535 with R from 0.5 (in the healthy control group) to 0.3 in the disease groups (P < 0.01). A weak, stable negative correlation was found in two pairs of rs2856718-rs3077 mutations with R from –0.17 to –0.12 (P < 0.001) (Figures 5A, 6A, 7A, and 8A).
To identify the most important factors in each patient group, we constructed a decision tree based on the results of the Pearson correlation test. Hierarchical clustering grouped similar objects in a dendrogram, iteratively merging similar clusters, starting with each data point as a separate cluster. This creates a tree-like structure that shows the relationships between clusters and their hierarchy. The height represents the distance between clusters, and the optimal number of clusters is where the height changes the most. The positions of the clades and leaves provide information regarding which element is closest to the root, which is an important factor in determining the following threshold values. This was ordered from 1 (the leaf closest to the root) in the dendrogram (Figures 5B, 6B, 7B, and 8B).
PCA plots help to better model the relationships between biomarkers. PCA allowed us to summarize and visualize information in a dataset containing individuals/observations described by multiple intercorrelated quantitative variables. Each variable can be considered as a different dimension. We selected the results from 20 different index values, where the cluster number could distinguish the clusters and appropriately avoid overlaps. We applied K-means cluster testing with ML algorithms to obtain the optimal number of clusters with the Euclidean distance as a measurement. This chart clearly shows that closely related markers were in the same cluster (Figures 5C, 6C, 7C, and 8C). The dendrogram and PCA map completed all our databases, in which the studied variants could combine and be necessary for the outcomes.
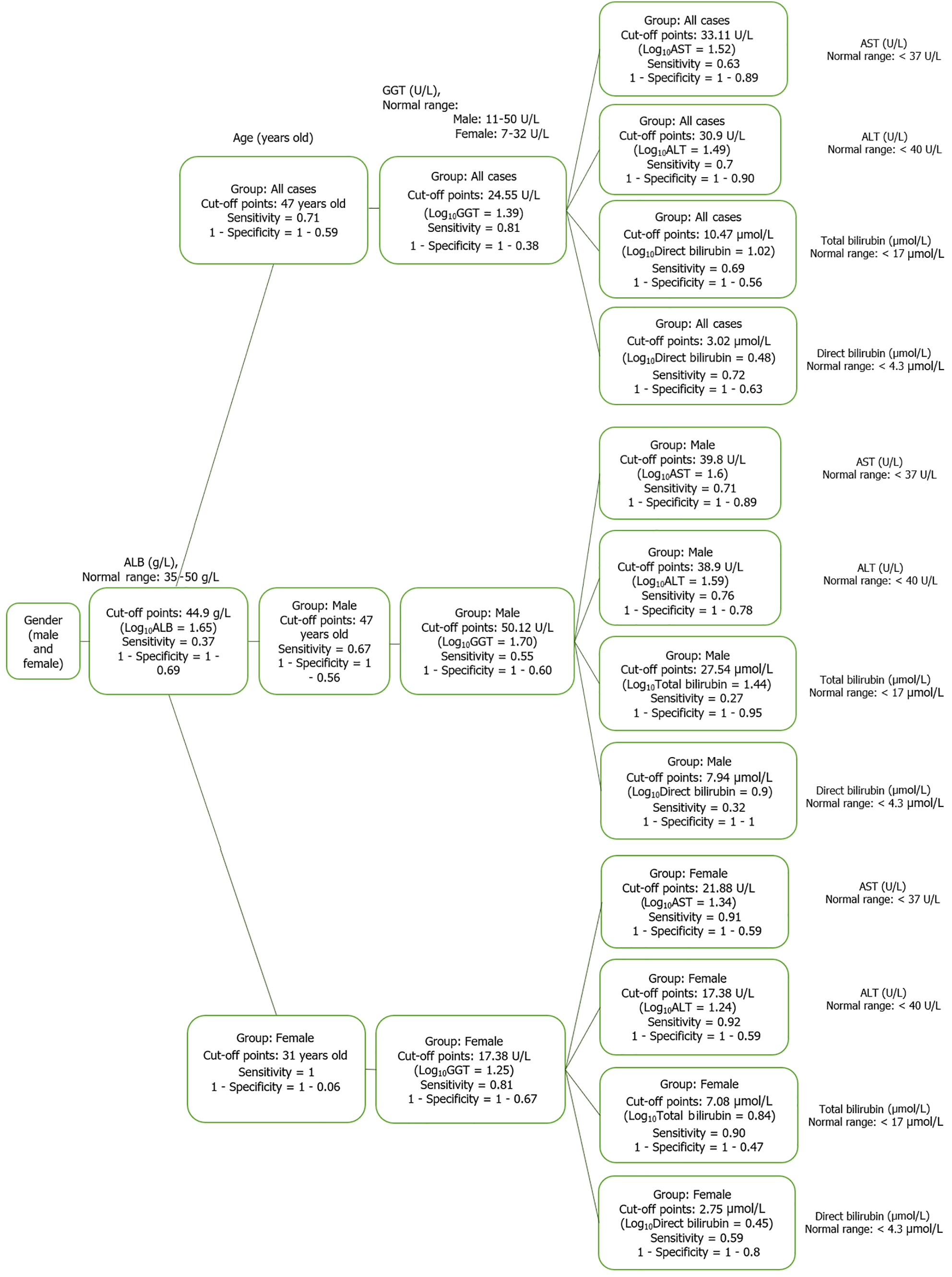
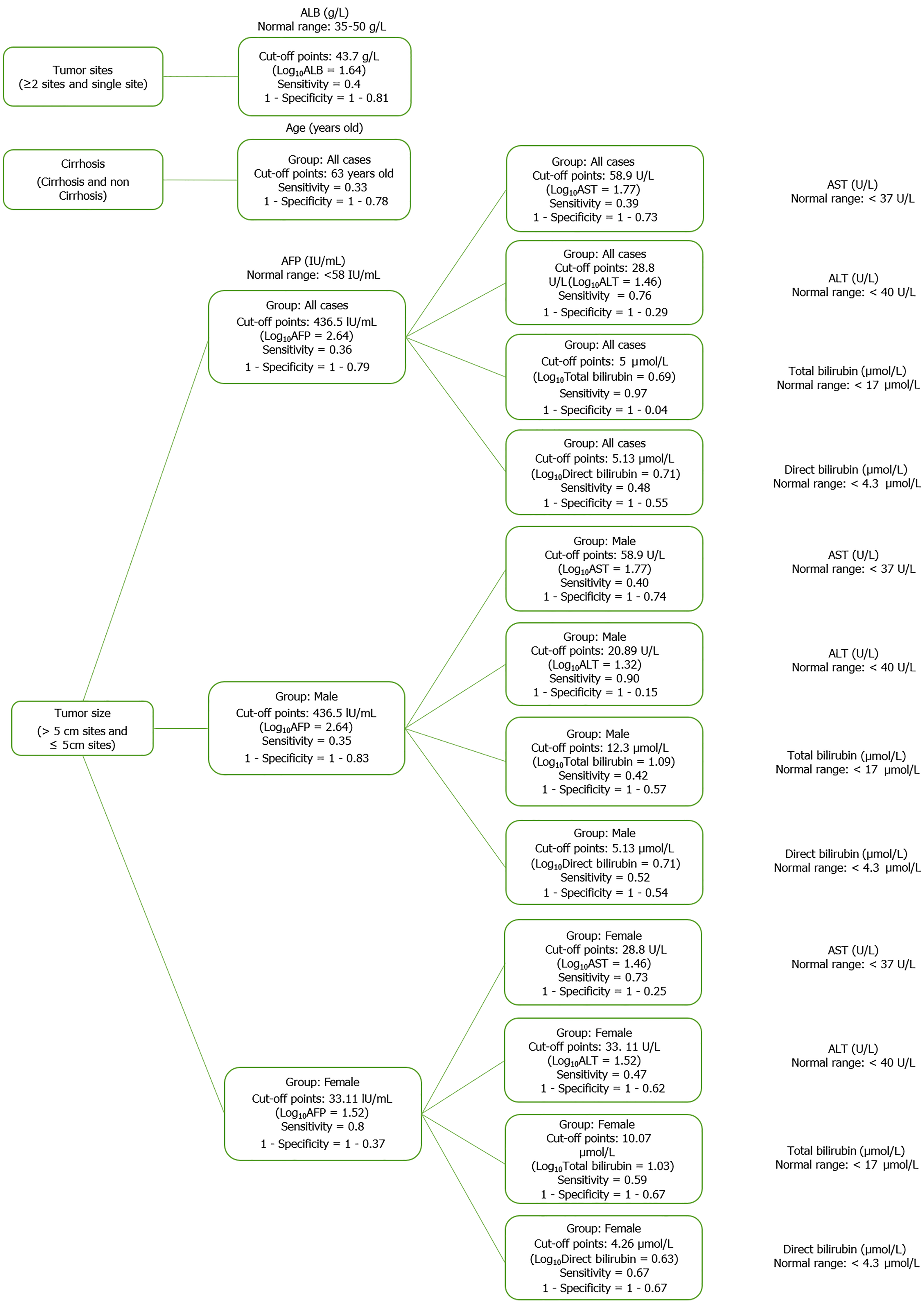
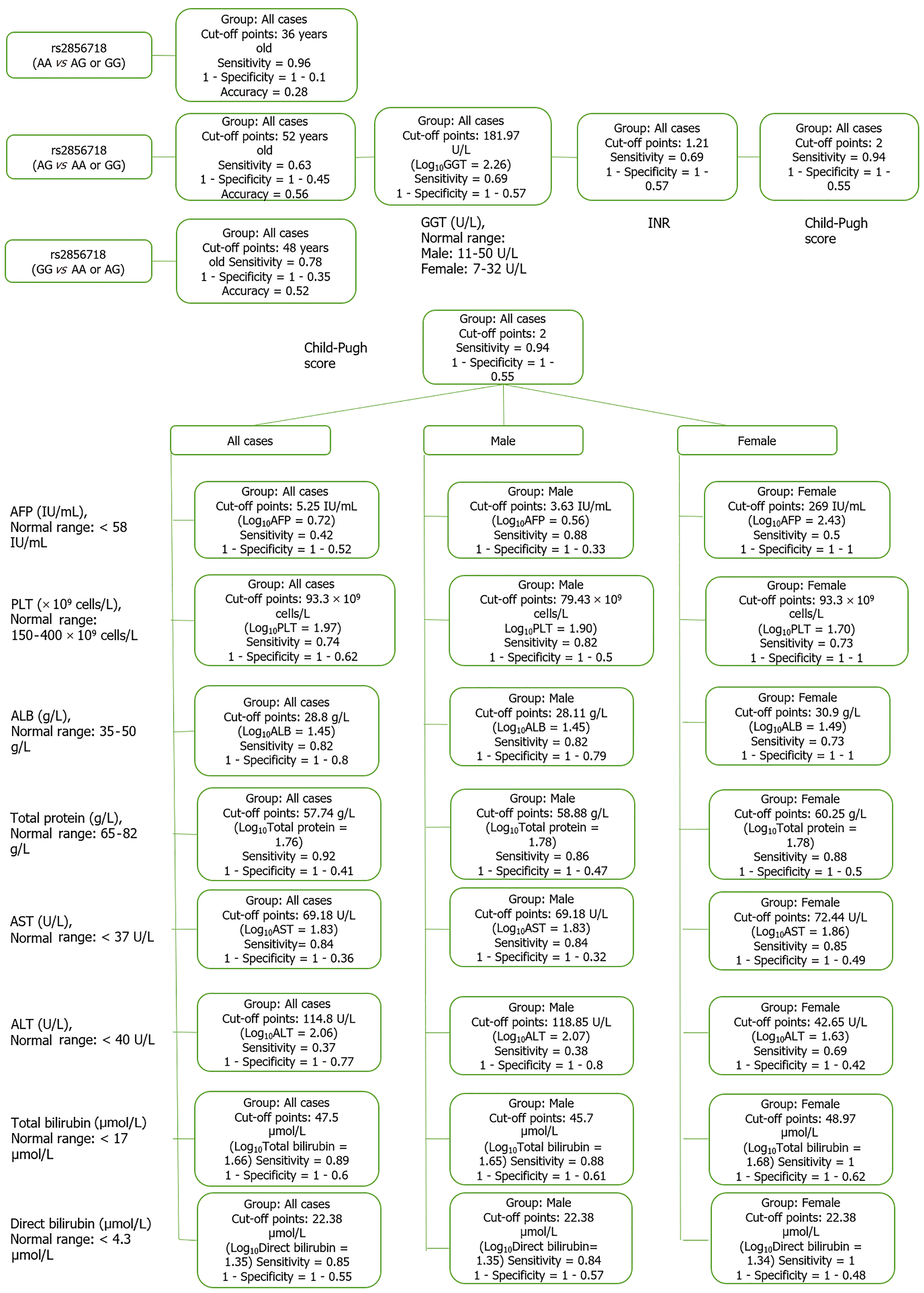
Based on the results of the cluster analysis and decision trees, we identified the most important first factors in each patient group. For example, sex and albumin (ALB) concentration were the most important factors in a hepatitis B patient group (Figure 9). In the liver cancer group, cirrhosis status, tumor status (size and differentiation), ALB concentration, sex, and age were the primary factors monitored (Figure 10). In the cirrhosis group, the genotype of the rs2856718 variant, age, international normalized ratio, gamma glutamyl transferase (GGT) concentration, and the Child-Pugh score were considered the most important factors. Running bootstrapping on two genotype groups (AG vs GG/AA) provided the best accuracy (acc = 0.56) for an optimal age cut-off of 52 years (Figure 11). We maximized multiple metrics simultaneously to obtain the optimal cut-off point and presented specific sensitivity and specificity values in the ROC plot, their RR in the dot and forest plot (ROC plot Supplementary Figures 1-7 with their related dot and forest plots Supplementary Figures 8-10, ROC plot Supplementary Figures 11-18 with their related dot and forest Supplementary Figures 19-28, and ROC plot Supplementary Figures 29-40 with their related dot and forest plots Supplementary Figures 41 and 42).
To better understand the diagnostic significance of genetic variants, we examined the RR of the haplotype with each directly related factor (based on the results of cluster analysis and PCA map) and cut-off points. Specifically, in the female hepatitis patient group, the AG/GG/GG haplotype reduced the risk of total bilirubin greater than 7.08 μmol/L by about 30% [RR = 0.72 (0.49; 1.06), P < 0.05]. In this group, the AA/GG/GG haplotype reduced the risk of alpha-fetoprotein (AFP) > 33.11 U/mL by 56% with RR = 0.44 (0.20; 0.99), P < 0.01; the GG/AG/AG haplotype reduced the risk of chronic hepatitis from the age of 31 years by 40% with RR = 0.63 (0.31; 1.28), P < 0.05. The AG/AG/GG haplotype was 40% more frequent in women with hepatitis B than that in male patients [RR = 0.76 (0.56; 1.02), P < 0.05] (Table 6).
| Preclinical factors | Groups | Female | Male | All | ||
| Total bilirubin (µmol/L); Normal range: < 17 µmol/L | Hepatitis | AG/GG/GG | ||||
| RR = 0.72 (0.49; 1.06) | ||||||
| Cutpoint = 7.08 µmol/L | ||||||
| Nupper/Nlower = 10/6 | ||||||
| Pfisher = 0.017, Pyates = 0.027, Puncor = 0.011 | ||||||
| HCC | AG/GG/GG | |||||
| RR = 1.45 (1.05; 1.99) | ||||||
| Cutpoint = 43.6 µmol/L | ||||||
| Nupper/Nlower = 25/19 | ||||||
| Pfisher = 0.02, Pyates= 0.03, Puncor = 0.02 | ||||||
| AFP (IU/mL); Normal range: < 5.8 IU/mL | Hepatitis | AA/GG/GG | ||||
| RR = 0.44 (0.20; 0.99) | ||||||
| Cutpoint = 33.11 U/mL | ||||||
| Nupper/Nlower = 4/7 | ||||||
| Pfisher = 0.008, Pyates = 0.008, Puncor = 0.002 | ||||||
| ALB (g/L); Normal range: 35-50 g/L | HCC | GG/AG/AG | ||||
| RR = 2.20 (1.24; 3.92) | ||||||
| Cutpoint = 43.6 g/L | ||||||
| Nupper/Nlower = 8/10 | ||||||
| Pfisher = 0.016, Pyates = 0.018, Puncor = 0.008 | ||||||
| Direct bilirubin (µmol/L); Normal range: < 4.3 µmol/L | HCC | GG/AG/GG | ||||
| RR = 1.57 (1.09; 2.25) | ||||||
| Cutpoint = 5.13 µmol/L | ||||||
| Nupper/Nlower = 11/5 | ||||||
| Pfisher = 0.034, Pyates = 0.046, Puncor = 0.026 | ||||||
| Tumor size (cm) | HCC | GG/AA/AG | AA/GG/AG | |||
| RR = 2.2 (1.94; 2.56) | RR = 2.25 (1.96; 2.59) | |||||
| Cutpoint = 5 cm | Cutpoint = 5 cm | |||||
| Nupper/NloweRR = 5/0 | Nupper/NloweRR = 3/0 | |||||
| Pfisher = 0.009, Pyates = 0.02, Puncor = 0.006 | Pfisher = 0.046, Pyates = 0.092, Puncor = 0.028 | |||||
| ALT (U/L); Normal range: < 40 U/L at 37 oC | HCC | GG/AG/AG | ||||
| RR = 0.74 (0.49; 1.13) | ||||||
| Cutpoint = 28.8 U/L | ||||||
| Nupper/Nlower = 10/8 | ||||||
| Pfisher = 0.048, Pyates = 0.068, Puncor = 0.038 | ||||||
| AST (U/L); Normal range: < 37 U/L at 37 oC | HCC | GG/GG/GG | GG/GG/GG | |||
| RR = 0.52 (0.25; 1.10) | RR = 0.53 (0.27; 1.06) | |||||
| Cutpoint = 58.9 U/L | Cutpoint = 58.9 U/L | |||||
| Nupper/Nlower = 6/26 | Nupper/Nlower = 7/30 | |||||
| Pfisher = 0.034, Pyates = 0.043, Puncor = 0.028 | Pfisher = 0.029, Pyates = 0.036, Puncor = 0.024 | |||||
| Age (years old) | Hepatitis | GG/AG/AG | ||||
| RR = 0.63 (0.31; 1.28) | ||||||
| Cutpoint = 31 years old | ||||||
| Nupper/Nlower = 3/2 | ||||||
| Pfisher = 0.013, Pyates = 0.0086, Puncor = 0.0004 | ||||||
| Cirrhosis | AA/AG/AG | |||||
| RR = 0.38 (0.11; 1.31) | ||||||
| Cutpoint = 52 years old | ||||||
| Nupper/Nlower = 2/7 | ||||||
| Pfisher = 0.023, Pyates = 0.038, Puncor = 0.018 | ||||||
| HCC | GG/GG/GG | |||||
| RR = 1.63 (0.98; 2.71) | ||||||
| Cutpoint = 63 years old | ||||||
| Nupper/Nlower = 13/24 | ||||||
| Pfisher = 0.045, Pyates = 0.055, Puncor = 0.035 | ||||||
| Gender (Female, male) | Hepatitis | AG/AG/GG | ||||
| RR = 0.76 (0.56; 1.02) | ||||||
| Cutpoint = Male | ||||||
| Nmale/Nfemale = 18/12 | ||||||
| Pfisher = 0.03, Pyates = 0.01, Puncor = 0.007 | ||||||
| HCC | AA/GG/GG | AG/AG/GG | ||||
| RR = 0.74 (0.567; 0.96) | RR = 1.20 (1.13; 1.27) | |||||
| Cutpoint = Male | Cutpoint = Male | |||||
| Nmale/Nfemale = 12/11 | Nmale/Nfemale = 18/0 | |||||
| Pfisher = 0.001, Pyates = 0.001, Puncor = 0.0005 | Pfisher = 0.04, Pyates = 0.06, Puncor = 0.02 | |||||
| GGT (U/L); Male: 11-50 U/L at | Cirrhosis | AG/GG/GG | ||||
| RR = 1.71 (1.09; 2.70) | ||||||
| Cutpoint = 181.97 U/L | ||||||
| Nupper/Nlower = 15/21 | ||||||
| Pfisher = 0.02, Pyates = 0.024, Puncor = 0.014 | ||||||
In the group of male liver cancer patients, the AG/GG/GG haplotype increased the risk of total bilirubin > 43.6 μmol/L by 45% [RR = 1.45 (1.05; 1.99), P < 0.05]. The GG/AG/AG haplotype increased the risk of ALB > 43.6 g/L by 1.2 times [RR = 2.20 (1.24; 3.92), P < 0.05] and reduced the risk of ALT > 28.8 U/L by 30% [RR = 0.74 (0.49; 1.13), P < 0.05]. The GG/GG/GG haplotype reduced the risk of AST > 58.9 U/L by 50% [RR = 0.52 (0.25; 1.10), P < 0.05]. In the HCC group, the GG/AA/AG and AA/GG/AG haplotypes increased the risk of tumors > 5 cm by 1.2 times [RR = 2.23 (1.94; 2.56) and 2.25 (1.96; 2.59), P < 0.05]. Haplotype GG/AG/GG increased the risk of direct bilirubin > 5.13 μmol/L by 43% [RR = 1.57 (1.09; 2.25), P < 0.05]. The GG/GG/GG haplotype reduced the risk of AST > 58.9 U/L by 50% [RR = 0.53 (0.27; 1.06), P < 0.05]. This haplotype was associated with an increased risk of liver cancer from the age of 63 years by 63% [RR = 1.63 (0.98; 2.71), P < 0.05]. The AA/GG/GG haplotype was 26% less frequent in male patients with liver cancer than that in female patients, whereas the AG/AG/GG haplotype was 20% more frequent in male patients (Table 6).
In patients with cirrhosis, the AA/AG/AG haplotype reduced the risk of HBV-infected patients developing cirrhosis from the age of 52 years by 60% [RR = 0.38 (0.11; 1.31), P < 0.05] (Table 6, Supplementary Figure 41). The AG/GG/GG haplotype increased the risk of GGT > 181.97 U/L by 64% [RR = 1.71 (1.09; 2.70), P < 0.05] (Table 6).
In clinical and biological studies, effect size, which is the difference between two or more groups, is important. Statistical significance is necessary but not sufficient to conclude whether the relationship or effect is real. The standard deviation determines the stability of statistical data around the mean value. The lower the standard deviation, the greater the stability of the data, and the smaller the fluctuation around the mean value. The higher the standard deviation, the smaller the stability of the data, and the greater the fluctuation around the mean value. The number of patient samples obtained from each group ensured that our initial hypothesis was met, with a Cohen's d value of 0.6, indicating that the genotypes of HLA variants may moderate the progression of liver disease (HCC and cirrhosis). Subsequent analyses confirmed this hypothesis.
HLA class I expression is higher in HCC cells than in normal hepatic cells[15]. We also observed a clear influence of HLA variants in our study, which belong to HLA class II, on the development of liver diseases, particularly liver cancer and cirrhosis. Multivariate analysis revealed that the three alleles had suppressive or additive effects on the risk of developing liver cancer or cirrhosis due to hepatitis B. We began with a LD test to determine whether there was an influence at the physical level, that is, the location of the variants on the chromosome, on the risk of developing the disease. Combinations of two or three SNPs revealed significant associations between HLA variants and susceptibility to HBV-related diseases. Maximal LD occurs when haplotypes have a null frequency, and intermediate levels of LD imply recombination between markers or recurrent mutations. For each marker pair, the strength of association (R2) was measured using a standard contingency table χ2 test. R2 > 0.5 indicates a high association, 0.3-0.5 a moderate level, 0.1-0.3 a low association, and 0-0.1 no association. In this study, all R2 values were very low, indicating that these haplotypes were rare, their distribution followed the Hardy-Weinberg equilibrium, and their LD moderately depended on the disease or phenotype status. (D’ was from 0.27 in disease cases to 0.56 in healthy control) (Figure 3 and Table 4).
Variants of the HLA-DP locus have been studied and have shown a link with HBV infection in Asian populations. The most significant alleles were the A alleles of HLA-DPA1 rs3077 and HLA-DPB1 rs9277535, which confer a decreased risk of HBV infection in Caucasian[16]. In the GWAS study in 2011 from the Japanese group, a significant association of two SNPs (rs2856718 and rs7453920) within the HLA-DQ locus was found (overall P value of 5.98 × 10-28 and 3.99 × 10-37). The association of CHB with rs2856718 and rs7453920 remained significant even after stratification with rs3077 and rs9277535, indicating independent effect of HLA-DQ variants on CHB susceptibility (P value of 1.52 × 10-21 – 2.38 × 10-30). In the Japanese cohort, the SNP rs2856718 was located in a LD block, including HLA-DQB1 and HLA-DQA1 genes (R2 = 0.1, D’ = 0.73). Similar to HLA-DPs, HLA-DQs are highly polymorphic, especially in exon two, which encodes an antigen-binding site[5]. The SNP rs9277535 in 3’-UTR of the HLA-DPB1 gene (approximately 22 kB from rs3077) augmented the risk of chronic hepatitis B and reduced the production of HLA-DPB1 (P = 10-15). Only weak LD was observed between rs9277535 and rs3077 in Europeans (R2 = 0.09, D’ = 0.50; HapMap CEU) and Asians (R2 = 0.24, D’ = 0.54), suggesting that the effects of these SNPs are likely independent[17]. The LD results indicated that the relationships between SNPs in this genomic region were comparable between Asian and European ancestry groups, even though the allele frequencies differed[17]. These findings indicate that variants in the HLA-DP-DQ antigen-binding regions contribute to the risk of persistent HBV infection.
After separating each variant, we determined how each genotype affected the risk of disease development (Tables 2 and 3). When combining the variants, we observed that the genotypes of two SNPs, rs3077 and rs9277535, were AA, and the risk of liver disease was reduced by 70% [RR = 0.29 (0.09; 0.99), P < 0.001] (Table 5). The rs3077 and rs9277535 variants influence the immune response by affecting the binding of HLA class II to CD4+ T cells. A significant positive correlation was observed between the SNP rs3077 and rs9277535 in HLA-DP and Systemic Lupus Erythematosus susceptibility. The rs3077 polymorphism was significantly correlated to IL-17, INF-γ, and cutaneous vasculitis. Additionally, carriers of the rs3077-AA genotype showed lower concentrations of inflammatory cytokines than those of the other two genotypes[18]. The rs3077-G allele was associated with a higher risk of chronic hepatitis B infection and decreased HLA-DPA1 expression, consistent with an additive genetic model. The SNPs rs3077 and rs9277535 situated in 3’-UTR regions of HLA-DPA1 and HLA-DPB1 are good candidates for allelic expression imbalance testing, suggesting that the mRNA expression associations found in the human liver cohort may change between the non-risk A allele and others. For both SNPs, the proportion of the non-risk A allele was significantly higher in the cDNA than in the DNA of heterozygous samples, indicating reduced expression of the risk alleles. These SNPs were highly associated with chronic HBV infection. Together, these independent results strongly implicate lower levels of HLA-DPA1 and HLA-DPB1 expression with an increased risk of chronic HBV infection[17].
HLA-DQ rs2856718, HLA-DP rs3077, and HLA-DP rs9277535 are significantly associated with a decreased risk of HCC and an increased risk of LC[7]. Some studies have shown that SNP rs2856718 differentially affects LC and HCC risk in chronic HBV-infected patients with HBV mutations[7]. In our study, we showed the effect of this phenotypic variant on the genotypes of rs3077 and rs9277535. The more A allele in the haplotype, the lower the risk of disease (hepatitis, cirrhosis, or HCC-related HBV) [A-A-A haplotype, RR = 0.44 (0.14; 1.37), P < 0.05]. In contrast, the more G allele in the haplotype, the greater the risk of hepatic disease [G-A/G-G haplotype, RR = 1.12 (1.02; 1.23), P < 0.05] (Table 5).
To better understand when genetic factors are essential for diagnosis, all relevant risk factors must be considered and arranged in the specific context of each disease group. By applying the K-means cluster analysis strategy and generating the Euclidean distance, we can better understand the above relationships by repeating this process until there is no further movement between clusters[19]. The ROC curve shows the clinical sensitivity and specificity of every possible cut-off value. An ROC curve plots the actual positive rate (sensitivity) and false-positive positive (100-Specificity) for the cut-off points. The points on the ROC curve represent the sensitivity/specificity pair for a particular decision threshold. This imputation also provides the accuracy, determines the percentage of correct predictions made by the model, and measures how well a single model performs. This is one of the most commonly used metrics.
Significant differences were observed in the optimal cut-off value for each patient group and the average value in healthy people. In this study, we discuss this trait when combined with different genotypes of the three variant combinations. In the hepatitis patient group, the AG/GG/GG haplotype reduced the risk of total bilirubin greater than 7.08 μmol/L by about 30% [RR = 0.72 (0.49; 1.06), P < 0.05]. In the group of liver cancer patients, the AG/GG/GG haplotype increased the risk of total bilirubin > 43.6 μmol/L by 45% [RR = 1.45 (1.05; 1.99), P < 0.05]. The normal range of total bilirubin was < 17 μmol/L. The GG/AG/GG haplotype increased the risk of direct bilirubin > 5.13 μmol/L by 43% [RR = 1.57 (1.09; 2.25), P < 0.05], while the normal level was inferior (4.3 μmol/L). These may be important markers that indicate liver damage status based on bilirubin levels. High bilirubin levels indicate an issue in the liver or gallbladder. For example, high levels of direct bilirubin may indicate that the liver does not properly clear bilirubin. Therefore, these results are indicative of liver damage or disease.
In the female hepatitis patient group, the AA/GG/GG haplotype reduced the risk of AFP > 33.11 IU/mL by 40% [RR = 0.44 (0.20; 0.99), P < 0.01]. An AFP level between 0 and 40 ng/mL or < 58 IU/mL is within the normal range for adults. A very high level of AFP in the blood (> 400 ng/mL or 332 IU/mL) is a sign of a liver tumor. We found that the optimal cut-off point of AFP in our patients was 436.5 IU/mL (log10AFP = 2.64) (Supplementary Figure 14). In the liver cancer group, the AA/GG/GG haplotype was much higher in female patients (28%, 11/39 patients) than in male patients (9.4%, 20/212 patients) [RR = 0.74 (0.56; 0.96), P < 0.01]. However, the AG/AG/GG haplotype was 20% more frequent in male patients (Supplementary Figure 25). The AG/AG/GG haplotype was 40% more frequent in women with hepatitis B than in male patients [RR = 0.76 (0.56; 1.02), P < 0.05] (Supplementary Figure 10).
The GG/AG/AG haplotype reduced the risk of chronic hepatitis from the age of 31 years to 40% [RR = 0.63 (0.31; 1.28) (P < 0.05) in female patients (Table 6). The GG/GG/GG haplotype was associated with an increased risk of liver cancer from the age of 63 years by 63% [RR = 1.63 (0.98; 2.71), P < 0.05]. In patients with cirrhosis, the AA/AG/AG haplotype reduced the risk of HBV-infected patients developing cirrhosis from the age of 52 years by 60% [RR = 0.39 (0.11; 1.31), P < 0.05] (Supplementary Figure 29, Supplementary Figure 41). The role of HLA in longevity may be challenging owing to significant methodological problems, such as serological and molecular typing of different loci, sample sizes, inclusion criteria, and age cut-off. However, within this complex scenario, some data emerge; on the whole, the (sex-specific) link of longevity with alleles or haplotypes of several genes may be the risk factors for a variety of diseases (cardiovascular diseases, cancer), including HLA alleles and haplotypes, were not unexpected based on previous studies on the genetics of longevity in centenarians[20].
In the liver cancer group, the GG/GG/GG haplotype reduced the risk of AST > 58.9 U/L by 50% [RR = 0.53 (0.27; 1.06); P < 0.05]. The GG/AA/AG and AA/GG/AG haplotypes increased the risk of tumor size > 5 cm by 1.2 times [RR = 2.23 (1.94; 2.56) and 2.25 (1.96; 2.59), P < 0.05]. In the cirrhotic group, The AG/GG/GG haplotype increased the risk of GGT > 181.97 U/L by 71% [RR = 1.71 (1.09; 2.70), P < 0.05]. The variant rs2856718 showed its effect on the age of patients with the best accuracy (0.58) when we ran the resampling in two groups having AG and AA/GG genotypes, as shown by the haplotype of three variants; the AA/AG/AG haplotype reduced the risk of HBV/infected patients developing cirrhosis from the age of 52 years by 60% [RR = 0.38 (0.98; 1.31), P < 0.05] (Table 6). HLA linkages with clinical expression and outcomes in the King’s College cohort showed that the baseline AST levels were most elevated in those expressing HLA- DRB1*03 and DRB1*07 (type 1 and 2 autoimmune hepatitis susceptibilities, respectively) compared with those expressing HLA-DRB1*13 (susceptibility to juvenile autoimmune sclerosing cholangitis). HLA-DRB1*13 cases showed increased baseline levels of alkaline phosphatase and GGT. This is indirect evidence that HLA genotypes contribute to disease outcomes. Further evidence of the role of HLA was the fact that more significant histological inflammation and fibrosis appeared in children with either the HLA-DRB1*03 or DRB1*13 genotypes compared with those with different HLA-DR genotypes[21].
This research method, combining paraclinical and bioinformatics, or more specifically, developing from biochemical results to LD analysis and then multi-cluster/multivariate, can be widely developed for pre- ML research in medicine, especially to move closer to personalized treatment strategies.
Combining data on clinical parameters, immune phenotyping data, genetic information, and ML models shows promise for effectively predicting the likelihood of a functional cure in patients with HBV. However, the limited size of multidimensional datasets and the characteristics of HBV cases pose challenges for machine-learning systems and meta-clustering, which typically require substantial data for pattern recognition[11]. Clustering helps to analyze an unlabeled dataset to group data points based on similarity. However, complex algorithmic adjustments are required to replicate these strategies in pathological studies. Accordingly, branching and iteration with complex algorithms must ensure logic and consistency with the statistical tests.
Rs9277535 affects liver fibrosis due to HBV infection, while rs3077 is associated with the risk of HBV-related HCC. In this study, the link between rs2856718, rs3077, and rs9277535 and disease risk was determined using a multi-clustering analysis. Using a key ML method, namely the cluster analysis method, decision trees were produced for each disease group. Ultimately, a complex relationship was observed between HLA variations and disease risk that occurred indirectly through other key markers, including bilirubin, GGT, AST, ALT, and AFP.
We thank the patients and participants for their consent and permission to publish our data. We thank our colleagues at the National Cancer Hospital in Hanoi, the National Hospital of Tropical Diseases, Thanh Nhan Hospital, and Thai Nguyen Hospital who supported us in patient recruitment and clinical data supply.
| 1. | World Health Organization. Hepatitis B. Available from: https://www.who.int/news-room/fact-sheets/detail/hepatitis-b. |
| 2. | Cooke GS, Andrieux-Meyer I, Applegate TL, Atun R, Burry JR, Cheinquer H, Dusheiko G, Feld JJ, Gore C, Griswold MG, Hamid S, Hellard ME, Hou J, Howell J, Jia J, Kravchenko N, Lazarus JV, Lemoine M, Lesi OA, Maistat L, McMahon BJ, Razavi H, Roberts T, Simmons B, Sonderup MW, Spearman CW, Taylor BE, Thomas DL, Waked I, Ward JW, Wiktor SZ; Lancet Gastroenterology & Hepatology Commissioners. Accelerating the elimination of viral hepatitis: a Lancet Gastroenterology & Hepatology Commission. Lancet Gastroenterol Hepatol. 2019;4:135-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 395] [Article Influence: 65.8] [Reference Citation Analysis (0)] |
| 3. | Flower B, Du Hong D, Vu Thi Kim H, Pham Minh K, Geskus RB, Day J, Cooke GS. Seroprevalence of Hepatitis B, C and D in Vietnam: A systematic review and meta-analysis. Lancet Reg Health West Pac. 2022;24:100468. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 4. | Kamatani Y, Wattanapokayakit S, Ochi H, Kawaguchi T, Takahashi A, Hosono N, Kubo M, Tsunoda T, Kamatani N, Kumada H, Puseenam A, Sura T, Daigo Y, Chayama K, Chantratita W, Nakamura Y, Matsuda K. A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nat Genet. 2009;41:591-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 396] [Cited by in RCA: 430] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 5. | Mbarek H, Ochi H, Urabe Y, Kumar V, Kubo M, Hosono N, Takahashi A, Kamatani Y, Miki D, Abe H, Tsunoda T, Kamatani N, Chayama K, Nakamura Y, Matsuda K. A genome-wide association study of chronic hepatitis B identified novel risk locus in a Japanese population. Hum Mol Genet. 2011;20:3884-3892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 189] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 6. | Hu L, Zhai X, Liu J, Chu M, Pan S, Jiang J, Zhang Y, Wang H, Chen J, Shen H, Hu Z. Genetic variants in human leukocyte antigen/DP-DQ influence both hepatitis B virus clearance and hepatocellular carcinoma development. Hepatology. 2012;55:1426-1431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 143] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 7. | Ji X, Zhang Q, Li B, Du Y, Yin J, Liu W, Zhang H, Cao G. Impacts of human leukocyte antigen DQ genetic polymorphisms and their interactions with hepatitis B virus mutations on the risks of viral persistence, liver cirrhosis, and hepatocellular carcinoma. Infect Genet Evol. 2014;28:201-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 8. | Al-Qahtani AA, Al-Anazi MR, Abdo AA, Sanai FM, Al-Hamoudi W, Alswat KA, Al-Ashgar HI, Khalaf NZ, Eldali AM, Viswan NA, Al-Ahdal MN. Association between HLA variations and chronic hepatitis B virus infection in Saudi Arabian patients. PLoS One. 2014;9:e80445. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 48] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 9. | Wasityastuti W, Yano Y, Ratnasari N, Triyono T, Triwikatmani C, Indrarti F, Heriyanto DS, Yamani LN, Liang Y, Utsumi T, Hayashi Y. Protective effects of HLA-DPA1/DPB1 variants against Hepatitis B virus infection in an Indonesian population. Infect Genet Evol. 2016;41:177-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 10. | Zhang X, Zheng C, Zhou ZH, Li M, Gao YT, Jin SG, Sun XH, Gao YQ. Relationship between HLA-DP gene polymorphisms and the risk of hepatocellular carcinoma: a meta-analysis. Genet Mol Res. 2015;14:15553-15563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 11. | Dost S, Rivas A, Begali H, Ziegler A, Aliabadi E, Cornberg M, Kraft AR, Vidal M. Unraveling the Hepatitis B Cure: A Hybrid AI Approach for Capturing Knowledge about the Immune System's Impact. Proceedings of the 12th Knowledge Capture Conference 2023. NY, United States: Association for Computing Machinery. 2023. [DOI] [Full Text] |
| 12. | Chu YJ, Yang HI, Hu HH, Liu J, Lin YL, Chang CL, Luo WS, Jen CL, Chen CJ. HBV genotype-dependent association of HLA variants with the serodecline of HBsAg in chronic hepatitis B patients. Sci Rep. 2023;13:359. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 13. | Nguyen TT, Ho CT, Bui HTT, Ho LK, Ta VT. Multidimensional Machine Learning for Assessing Parameters Associated With COVID-19 in Vietnam: Validation Study. JMIR Form Res. 2023;7:e42895. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 14. | R-bloggers. Calculating required sample size in R and SAS. 2024. Available from: https://www.r-bloggers.com/2017/02/calculating-required-sample-size-in-r-and-sas/. |
| 15. | Akazawa Y, Nobuoka D, Takahashi M, Yoshikawa T, Shimomura M, Mizuno S, Fujiwara T, Nakamoto Y, Nakatsura T. Higher human lymphocyte antigen class I expression in early-stage cancer cells leads to high sensitivity for cytotoxic T lymphocytes. Cancer Sci. 2019;110:1842-1852. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 16. | Vermehren J, Lötsch J, Susser S, Wicker S, Berger A, Zeuzem S, Sarrazin C, Doehring A. A common HLA-DPA1 variant is associated with hepatitis B virus infection but fails to distinguish active from inactive Caucasian carriers. PLoS One. 2012;7:e32605. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 39] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 17. | O'Brien TR, Kohaar I, Pfeiffer RM, Maeder D, Yeager M, Schadt EE, Prokunina-Olsson L. Risk alleles for chronic hepatitis B are associated with decreased mRNA expression of HLA-DPA1 and HLA-DPB1 in normal human liver. Genes Immun. 2011;12:428-433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 84] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 18. | Zhang J, Zhan W, Yang B, Tian A, Chen L, Liao Y, Wu Y, Cai B, Wang L. Genetic Polymorphisms of rs3077 and rs9277535 in HLA-DP associated with Systemic lupus erythematosus in a Chinese population. Sci Rep. 2017;7:39757. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 19. | Nichols L, Taverner T, Crowe F, Richardson S, Yau C, Kiddle S, Kirk P, Barrett J, Nirantharakumar K, Griffin S, Edwards D, Marshall T. In simulated data and health records, latent class analysis was the optimum multimorbidity clustering algorithm. J Clin Epidemiol. 2022;152:164-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 18] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 20. | Caruso C, Candore G, Colonna Romano G, Lio D, Bonafè M, Valensin S, Franceschi C. HLA, aging, and longevity: a critical reappraisal. Hum Immunol. 2000;61:942-949. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 50] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 21. | Mack CL. HLA Associations in pediatric autoimmune liver diseases: Current state and future research initiatives. Front Immunol. 2022;13:1019339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |