Published online Dec 7, 2024. doi: 10.3748/wjg.v30.i45.4817
Revised: September 20, 2024
Accepted: October 15, 2024
Published online: December 7, 2024
Processing time: 245 Days and 18.9 Hours
Obstructed defecation syndrome (ODS) represents the most prevalent form of chronic constipation, affecting a diverse patient population, leading to numerous complications, and imposing a significant burden on healthcare resources. Most ODS patients have insufficient rectal propulsion, but the exact mechanism underlying the pathogenesis of ODS remains unclear.
To explore the molecular mechanism underlying the pathogenesis of ODS.
A total of 30 pairs of rectal samples were collected from patients with ODS (ODS group) or grade IV prolapsed hemorrhoids without constipation (control group) for quantitative proteomic and bioinformatic analysis. Subsequently, 50 pairs of paraffin-embedded rectal specimens were selected for immunohistochemistry and immunofluorescence studies to validate the analysis results. Human intestinal smooth cell contractile function experiments and electrophysiological experiments were conducted to verify the physiological functions of target proteins. Cellular ultrastructure was detected using transmission electron microscopy.
In comparison to the control group, the expression level of dystrophin (DMD) in rectal specimens from ODS patients was markedly reduced. This finding was corroborated using immunohistochemistry and immunofluorescence techniques. The diminished expression of DMD compromised the contractile function of intestinal smooth muscle cells. At the molecular level, nucleoporin protein 153 and L-type voltage-gated calcium channel were found to be overexpressed in intestinal smooth muscle cells exhibiting downregulated DMD expression. Electrophysiological experiments confirmed an excessive influx of calcium ions into these cells. Moreover, vacuolar-like structures which may be associated with excessive calcium influx were observed in the cells by transmission electron microscopy.
Decreased DMD expression in intestinal smooth muscle may upregulate L-type voltage-gated calcium channel expression, leading to excessive calcium influx which may cause a decrease in rectal propulsion, thereby contributing to the pathogenesis of ODS.
Core Tip: The long-term outcomes of numerous treatments for obstructed defecation syndrome (ODS) are now unsatisfactory. The reason is a lack of knowledge about ODS’s molecular abnormalities. Our study indicated a connection between the pathophysiology of ODS and reduced dystrophin expression. The overexpression of nucleoporin protein 153 and L-type voltage-gated calcium channel was triggered by the downregulation of dystrophin, which caused a significant flow of calcium ions into the intestinal smooth muscle cells. Overabundance of calcium ion inflow may reduce muscle contractility, which manifested as weakened rectal propulsion in ODS.
- Citation: Li WZ, Xiong Y, Wang TK, Chen YY, Wan SL, Li LY, Xu M, Tong JJ, Qian Q, Jiang CQ, Liu WC. Quantitative proteomics analysis reveals the pathogenesis of obstructed defecation syndrome caused by abnormal expression of dystrophin. World J Gastroenterol 2024; 30(45): 4817-4835
- URL: https://www.wjgnet.com/1007-9327/full/v30/i45/4817.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i45.4817
Obstructed defecation syndrome (ODS) is responsible for between 30% and 60% of cases of chronic constipation globally[1]. According to two United States surveys, the financial impact of persistent constipation is $12.7 billion in direct medical bills and $3 million in doctor visits. The indirect costs of persistent constipation, such as lost productivity and impairment at work, are difficult to quantify in addition to these costs[2,3]. Moreover, mental health conditions including anxiety and depression frequently occur with persistent constipation[4]. Additionally, studies have shown that individuals who experience persistent constipation are more likely to develop rectal cancer and cardiovascular events[5,6]. ODS consequently has a major negative impact on the health and quality of life of its sufferers.
Current ODS therapies, whether conservative or surgical (Table 1), have not proven to be long-term satisfactory, according to recent studies[7]. This therapeutic difficulty is a result of the pathophysiology of ODS being complex. According to the ‘iceberg theory’, which has been put out by certain researchers, concealed lesions are known as the ‘underwater rocks’, and the morphological alterations seen in ODS are only the ‘tip of the iceberg’[8]. Anatomical anomalies, pelvic floor dysfunction, inadequate rectal propulsion, and decreased rectal sensation are a few potential pathogenic causes of ODS[9]. The pathophysiology of persistent constipation has also been linked to colonic propulsion caused by smooth muscle contractions[10]. Anorectal manometry in many ODS patients reveals insufficient rectal propulsion[11,12], and a study has demonstrated that intestinal smooth muscle contractile dysfunction is the primary cause contributing to insufficient intestinal propulsion, resulting in slow transit constipation[13]. Therefore, although the precise molecular pathways underlying the development of ODS are yet unknown, smooth muscle contractile failure in the rectal tract may be strongly linked to it.
| Conservative management | |
| Lifestyle management | Appropriate water intake, diet rich in fibers |
| Medical treatment | Colonic lavage or rectal irrigation |
| Pelvic floor exercises with biofeedback | |
| Surgical management | |
| Ventral rectopexy | |
| Stapled transanal rectal resection | |
| Tissue selecting therapy stapled transanal rectal resection | |
| Rectocele repair | |
Recent advances in proteomics have allowed for the identification of certain biomarkers that have shed light on the pathophysiology of a number of complex diseases[14,15]. Furthermore, proteomics based on isobaric tag for relative and absolute quantification (iTRAQ), which offers benefits like high-throughput capability, high sensitivity, and high accuracy, has been used to identify important pathophysiology of cardiovascular illnesses and malignant tumors[16,17]. Few studies have been done thus far using proteomics to investigate the pathophysiology of ODS. In order to explore the possible pathogenesis of ODS, the goal of this work was to identify important proteins in the pathogenesis of ODS using iTRAQ proteomics. This study screened for differentially expressed proteins using iTRAQ-based proteomic analysis, and then used bioinformatics to further examine the roles of these proteins. Personalized bioinformatics research identified dystrophin (DMD) as a putatively important pathogenic protein in ODS.
The DMD gene, which spans roughly 2.3 Mb (Xp21.2-p21.1), is the biggest gene related to humans on the X chromosome[18]. Producing DMD, a cytoskeletal protein that stabilizes and shields muscle fibers, is its main function. Owing to its size, the DMD gene can have duplications, point mutations, and deletions[19]. The majority of these mutations cause the open reading frame to be disrupted, which causes a coding frame shift that results in a shortened, non-functional DMD protein. This in turn affects the function of muscle contraction[20]. However, there has been limited research on the DMD gene’s impact on intestinal smooth muscle and more attention was paid to how it affects cardiomyocytes and skeletal muscle cells.
To further confirm DMD’s function in ODS, we performed immunohistochemistry (IHC) and immunofluorescence (IF) tests on clinical samples. Furthermore, we looked into DMD’s biological role in human primary intestinal smooth muscle cells. Dysregulated expression of nucleoporin protein 153 (NUP153) and an L-type voltage-gated calcium channel (Cav1.2) on the cell membrane may arise from aberrant expression of DMD. This might then lead to an excessive influx of calcium ions into the cells, ultimately causing contractile failure. In conclusion, our research uncovered new molecular indicators for ODS and disclosed signaling pathways linked to aberrant DMD expression in ODS, offering novel targets for ODS treatment.
From January to May 2016, 30 patients with ODS (ODS group) and 30 patients with IV-degree prolapsed hemorrhoids without constipation symptoms (control group) were identified who received transanal surgical procedure at Department of Colorectal and Anal Surgery, Zhongnan Hospital of Wuhan University (Table 1). Where available, rectal specimens for quantitative proteomics analysis were obtained from these patients [ODS group (n = 30) and control group (n = 30)]. In addition, from June to December 2016, another 20 patients with ODS (ODS group) and 20 patients with IV-degree prolapsed hemorrhoids without constipation symptoms (control group) were identified who received transanal surgical procedure at Department of Colorectal and Anal Surgery, Zhongnan Hospital of Wuhan University (Table 1). IHC and IF staining was performed for all these 50 pairs of formalin-fixed paraffin-embedded rectal specimens [ODS group (n = 50) and control group (n = 50)]. The diagnosis of ODS was based on the Rome IV criteria for functional constipation. The pathological diagnosis was confirmed by pathologists. Data collection was conducted by using Patient Information Form and Wexner Constipation Score. The clinical details of ODS patients are summarized in Table 2. Ethical permission for the study was authorized by the Research Ethics Committee of Zhongnan Hospital of Wuhan University (No. 2015048 and No. 2022156K). The present study was performed in accordance with the principles of the Declaration of Helsinki. All patients agreed to participate in the study. And an informed consent form was signed by all the participants before being included in the study.
| Characteristics | Categories | n (%) |
| Number of patients | 50 | |
| Age (years) | > 60 | 32 (64) |
| < 60 | 18 (36) | |
| Gender | Male | 11 (22) |
| Female | 39 (78) | |
| Surgical procedure | STARR | 44 (88) |
| TST-STARR | 6 (12) | |
| Oxford grade for rectal prolapse | RRI (Grade I) | 2 (4) |
| RRI (Grade II) | 7 (14) | |
| RAI (Grade III) | 14 (28) | |
| RAI (Grade IV) | 16 (32) | |
| ERP (Grade V) | 11 (22) | |
| Wexner constipation score | mean ± SD | 11.34 ± 2.43 |
All collected rectal specimens were washed with cold phosphate buffered saline (PBS) and thoroughly ground in liquid nitrogen. The ground powder was then transferred to centrifuge tubes. After adding an appropriate amount of lysis buffer (1% phenylmethylsulfonyl fluoride in PBS) to the samples, the tubes were placed in an ice bath and sonicated for 5 min (a period of 2 s with an interval of 3 s). After centrifugation at 12000 × g for 10 min at 4 °C, the supernatant was collected and an equal volume of 100% acetonitrile (ACN) solution was added. The mixture was then left on ice for 1 h with vortexing at 20-min intervals. Following precipitation, the tubes were centrifuged at 10000 × g for 10 min, and the entire supernatant was collected. After reducing the volume by half through lyophilization, we centrifuged pre-wetted 10 kD ultrafiltration tubes (Merck Millipore, UFC501096, Germany) at 10000 g at 4 °C for 20 min, and then added 200 μL of ultrapure water to repeat the above step. The filtrate was collected, and 1% formic acid was added to adjust the pH to 2-3. The prepared specimens were stored at -80 °C and divided into two groups for experiments: The ODS group and the control group, with all specimens mixed in each group.
One hundred microgram of protein was dissolved in U2 Lysis buffer (8 M urea/100 mmol/L TEAB) to approximately 1 μg/μL and diluted in 5 volumes of 100 mmol/L tetraethylammonium bromide. Then 1.2 μL of 0.5 M CaCl2 was added to the tube and mixed well by shaking and centrifuging. The protein was trypsinised (pancreatic enzyme:protein = 1:100) and incubated at 37 °C for over 8 h, with 10 mg of C18 column per 100 μg of peptide sample. One milliliter of methanol was centrifuged with shaking to activate the column material, and the supernatant was discarded. Then 1 mL of 0.1% formic acid was added into the samples and centrifuged, and the supernatant was discarded. The peptide samples were acidified with an equal volume of 0.1% formic acid, mixed thoroughly by vortexing, spun down into a centrifuge tube, allowed to stand for 30 min to mix, and centrifuged. After the supernatant was discarded, the samples were washed twice with 0.1% formic acid + 3% ACN for desalting and eluted with 1 mL of 0.1% formic acid + 80% ACN. The eluted peptides were dried using a vacuum concentrator.
The peptide samples were dissolved in 20 μL of dissolution buffer (0.5 M TEAB) and mixed thoroughly with 70 μL of isopropanol by shaking and centrifuging. The samples were labeled and mixed according to the specifications of the iTRAQ-8 Kit (SCIEX), then the mixed peptides were fractionated using the Ultimate 3000 HPLC System (Thermo DINOEX, United States). A Welch C18 column (250 mm × 4.6 mm, 5 μm) was used as the chromatographic column. Peptide isolation was achieved by progressively increasing the ACN concentrations under alkaline conditions. The eluent was combined into 12 components, which were desalted and vacuum dried on the Strata-X column.
The peptide samples were diluted in the machine to a concentration of 1 μg/μL and the scanning mode was set to 8 μL for 90 min. Peptides with a mass-to-charge ratio (m/z) ranging from 350-1200 in the sample were scanned. Mass spectrometry data acquisition was performed using the Triple TOF 5600+ liquid chromatography-tandem mass spectrometry system (AB SCIEX, United States). The peptide samples were dissolved in 2% CAN/0.1% formic acid and analyzed using the Triple TOF 5600+ mass spectrometer coupled with the Eksigent nano-olc system (AB SCIEX, United States). The peptide solution was added to a C18 capture column (3 μm, 350 μm × 0.5 mm, AB Sciex, United States), and then gradient eluted for 90 m at a flow rate of 300 nL/min on a C18 analytical column (3 μm, 75 μm × 150 mm, Welch Materials, Inc). The two mobile phases were buffer A (2% CAN/0.1% formic acid/98% H2O) and buffer B (98% CAN/0.1% formic acid/2% H2O). For information dependent acquisition, primary mass spectrometry scans were performed with an ion accumulation time of 250 ms, and secondary mass spectrometry scans were obtained from 30 precursor ions with an ion accumulation time of 50 ms. MS1 spectra were acquired in the m/z range of 350 to 1200, and MS2 spectra were acquired in the m/z range of 100 to 1500. The dynamic exclusion time for precursor ions was set to 15 s.
Bioinformatics analysis was performed on significantly up-regulated or down-regulated proteins, indicating the fold change in protein (ratio > 1.5 or < 0.6667) with a P value < 0.05. These identified proteins were categorized as differentially expressed proteins. Subsequently, Gene Ontology analysis was conducted on these differential proteins, including cellular component, molecular function, and biological process. In addition, studies on the Kyoto Encyclopedia of Genes and Genomes (KEGG) and protein-protein interaction (PPI) were carried out using R (Version 4.0.2). The raw mass spectrometry data (*.wiff) was analyzed using ProteinPilot software (Version 4.5), with protein identification and iTRAQ quantification based on the Uniprot homo database (Version 2019.6). The identification parameters were as follows: The search type was set to iTRAQ 8plex (Peptide Labeled), the instrument was TripleTOF 5600, enzyme = trypsin, bias correction was set to True, and background correction was set to “True”.
Human colon smooth muscle cells (humm-iCell-d011, iCell Bioscience Company, Shanghai, China) were cultured in a complete primary smooth muscle cell culture medium (primedi-iCell-004, iCell Bioscience Company, Shanghai, China), supplemented with 10% fetal bovine serum (141215, Tianhang Biotechnology Company, Hangzhou, China). The cells were maintained in a humidified incubator with 5% CO2 at 37 °C. The cells were transfected with negative control small interfering RNA (siR-NC) or siR-DMD. For cell transfection, 5 μL of Lipofectamine 2000 (Invitrogen) was used to transfect 5 μL of small interfering RNA (20 μM, ELK Biotechnology, Wuhan, China) in a 6-well plate. After 6 h, the cell culture medium was replaced with complete culture medium. The sequence of the small interfering RNA for DMD is: TGGA
Total RNA was extracted using TRIzol reagent (Invitrogen, USA), and cDNA was synthesized using the M-MLV reverse transcriptase kit (ELK Biotechnology, EQ002). Real-time polymerase chain reaction (PCR) reactions were performed using the SYBR® Premix Ex Taq™ kit (TaKaRa, Shanghai, China). The StepOne™ Real-Time PCR System (Life Technologies) was used for amplification and detection. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) served as the internal reference housekeeping gene. All samples were run in triplicate, with each replicate repeated three times. The 2−ΔΔCT method was used for data analysis. The primer sequences used for real-time quantitative PCR (RT-qPCR) are listed in Table 3.
| Gene | Forward 5’-3’ | Reverse 5’-3’ |
| GAPDH | CATCATCCCTGCCTCTACTGG | GTGGGTGTCGCTGTTGAAGTC |
| DMD | AATGGAAACAGTAACTACGGTGAC | TAATCCAGCTGTGAAGTTCAGTTAT |
| NUP153 | GGACCATCTGGTATATGCCGA | GGCTCCGATGAAGAGAAGGC |
| Cav1.2 | ACGTTCCGATGTGTGAGGATC | GGTTCATCTTCACTTCATAGGTCTC |
The cell contraction assay was performed using the CytoSelect™ 24-well Cell Contractility Assay Kit (Floating Matrix Model, Cell Biolabs) according to the manufacturer’s protocol. Human intestinal smooth muscle cells were resuspended in a 2 mL EP tube. The cell suspension was then mixed with the collagen gel working solution at a ratio of 2:8 to prepare the cell contraction matrix, which was allowed to polymerize for 1 h at 37 °C. After the gel solidified, 1 mL of complete culture medium was added and incubated for 24 h. Half of the culture medium was replaced every 24 h. After 48 h, the gels were released from the sides of the wells, and the images were captured using the MicroPublisher (Q-IMAGING). The size of the collagen gel was analyzed using ImageJ (NIH).
Total protein was extracted from all intestinal smooth muscle cells using the RIPA Protein Extraction Kit (AS1004, Aspen, China). The total protein amount used per well was 40 mg. After performing sodium-dodecyl sulfate gel electrophoresis on the proteins, they were transferred to a polyvinylidene difluoride membrane via electroblotting. The membrane was blocked with 1% casein in 1 × TBS blocking buffer at room temperature for 1 h, then incubated overnight at 4 °C with gentle shaking using primary antibodies against the following proteins: DMD (1:1000, 68120-1-Ig, Proteintech), NUP153 (1:1000, ab171074, Abcam), Cav1.2 (1:500, ab81980, Abcam), and GAPDH (1:10000, ab181602, Abcam). The membrane was washed five times, for 5 min each, with TBS containing 5% Tween 20 (TBST), followed by incubation with secondary antibodies at room temperature for 1 h. After washing with TBST for 5 min, the membrane was incubated with ECL chemiluminescent solution (AS1059, Aspen, China). The protein bands were visualized using a chemiluminescence detection system (Canon, #LiDE110).
Paraffin-embedded samples were prepared as 5 μm sections. Following deparaffinization, rehydration, and antigen retrieval, endogenous peroxidase activity was quenched. IHC staining was performed using primary antibodies against DMD (68120-1-Ig, Proteintech Group), NUP153 (CSB-RA986790A0HU, Cusabio), and Cav1.2 (21774-1-AP, Proteintech Group) at 4 °C overnight. Then, the sections were incubated with biotinylated secondary antibodies and stained with the DAB kit (GTVision, China) to complete the IHC staining. The percentage of positive staining area was assessed with Image J based on three randomly captured fields by technicians blinded to the sample groups. Brown staining was considered as specific antigen-antibody binding, with normal stromal cells excluded from the analysis.
Paraffin sections were dewaxed to water and then placed into 0.01 M citrate buffer (pH = 6.0). The tissue sections were microwaved for antigen retrieval, and repaired over medium-high heat for 3 min. When cooled to room temperature, circles were drawn around the tissue with a histochemistry pen to prevent the incubation liquid from flowing away later in the process. The tissue was then washed with PBS. Antibodies were diluted with 5% BSA at certain ratios. An appropriate amount of primary antibody working solution was added, including those against DMD (68120-1-Ig, Proteintech Group), NUP153 (CSB-RA986790A0HU, Cusabio), and (21774-1-AP, Proteintech Group), and incubated overnight at 4 °C. After being rewarmed, the sections were washed 3 times with PBS, 5 min each time. An appropriate amount of secondary antibody working solution was added to the sections. The sections were then incubated in a 37 °C water bath in the dark for 40 min and washed with PBS three times, each for 5 min. Next, the nuclei was stained with DAPI (Sigma, Fluoroshield with DAPI). The sections were incubated in the dark at room temperature for 20-30 min and washed with PBS. Finally, the sections were sealed with an anti-fluorescence quenching mounting agent. The expression of DMD was analyzed using a fluorescence microscopy system (Olympus).
The human intestinal smooth muscle cells on a coverslip were placed in a recording chamber filled with extracellular solution, and the cells were imaged on a monitor screen with real-time observation through an inverted microscope. The recording electrodes were made from glass capillaries with an outer diameter of 1.5 mm, an inner diameter of 0.89 mm, and a length of 10 mm, drawn on a horizontal plotter. For whole-cell patch clamp experiments, the tip diameter of the electrode tip was usually about 2 μm, with an impedance of approximately 3-5 MΩ.
Under high magnification, a cell with a smooth surface and good refractive index was selected, and the recording electrode filled with internal electrode solution was moved close to the cell. As the electrode approached the cell, the impedance of the electrode increased, and a negative pressure trend was observed. The holding potential was adjusted to -70 mV to easily form a high-resistance seal. After the seal was stably formed, the negative pressure was removed, the electrode capacitance was compensated, and then mechanical negative pressure was applied to rupture the cell membrane at the tip of the electrode, thus forming a whole cell recording configuration. All electrophysiological experiments were conducted at room temperature (20-25 °C). All voltage clamp recordings were made with an Axon 700B patch clamp amplifier (Axon Instruments, Foster City, CA, United States). Experimental parameter settings, data acquisition, and stimulation protocols were controlled with Clampex 10.7 software (Molecular Devices). The extracellular solution contained: NaCl (110 mM), TEA (25 mM), KCl (5 mM), MgCl2 (1 mM), CaCl2 (1.2 mM), HEPES (10 mM), glucose (10 mM), TTX (0.005 mM), with pH adjusted to 7.4 with NaOH. The intracellular solution contained: CsCl (135 mM), MgCl2 (3 mM), HEPES (10 mM), EGTA (10 mM), Na (2 mM), ATP (4 mM), with pH adjusted to 7.2-7.4 with CsOH.
Cell samples were placed in an embedding plate containing 812 embedding medium (90529-77-4, SPI), and polymerized at 65 °C for 48 h. Then, the samples were stained in the dark with 2% (w/v) uranyl acetate for 10 min, followed by staining with lead acetate for another 10 min. The samples were then analyzed using a transmission electron microscope (TECNAI G2 SPIRIT, FEI, United States) at an accelerating voltage of 100 kV.
Statistical analyses and graphing were performed using GraphPad Prism v9.0. Unpaired, two-sided Student’s t-test was performed for comparisons between two groups, with data presented as the mean ± SEM. Spearman’s correlation analysis was used for correlation analyses. All quantitative experimental data were derived from at least three repetitions. A P value < 0.05 was considered statistically significant.
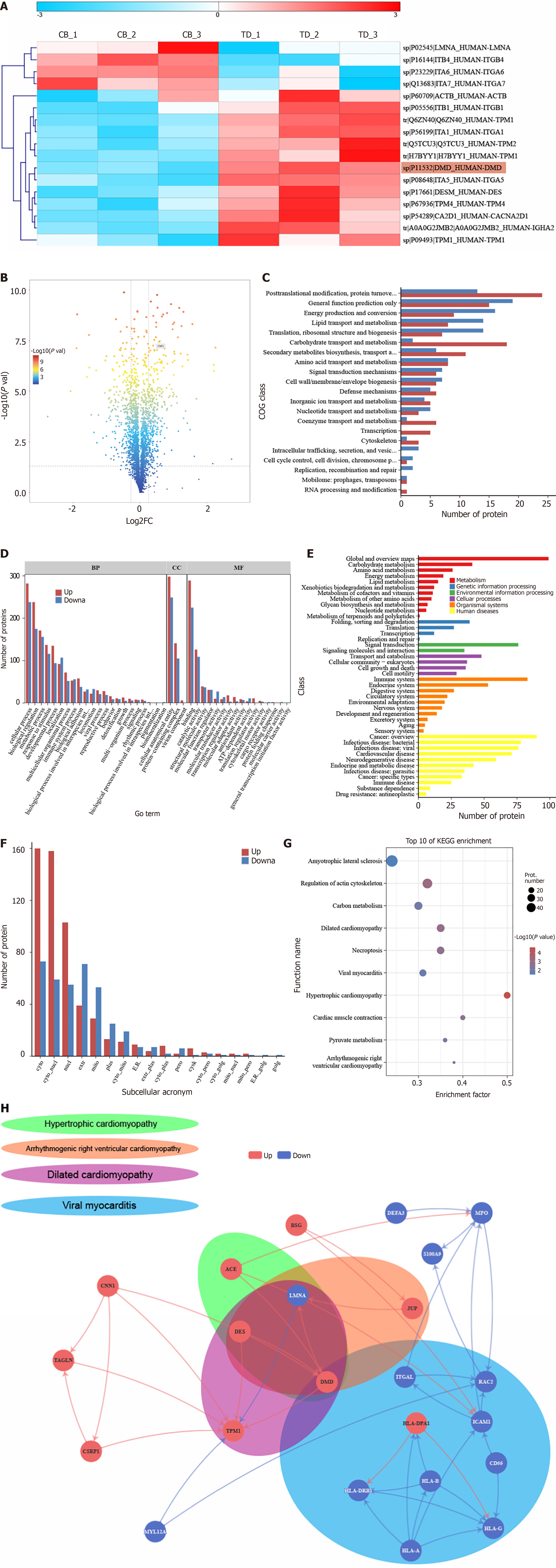
The collected rectal specimens were divided into experimental and control groups, with each group containing 30 pairs, randomly distributed into three replicates. The three experimental groups and three control groups were labeled as TD-1, TD-2, TD-3, CB-1, CB-2, and CB-3, respectively. Based on the quantitative results from proteomics, a standard deviation coefficient of 0.05 was used for comparison between TD and CB, and normalized variation coefficient [fold change (FC)] along with corrected hypothesis probability (adjusted P value) thresholds were applied for differential filtering. The screening thresholds used in this study were FC ≥ 1.5 or FC ≤ 0.667 and adjusted P value ≤ 0.05, based on which a total of 164 significantly differentially expressed proteins (88 upregulated and 76 downregulated) were identified. A heatmap was constructed based on the differentially expressed proteins that may be related to muscle contraction dysfunction (Figure 1A). For the differentially expressed proteins, the volcano plot allowed for direct visualization of all differentially expressed proteins marked with DMD (Figure 1B). Functional classifications of upregulated and downregulated proteins were compared and analyzed, with Figure 1C and D listing the results of Cluster of Orthologous Groups of proteins functional classification annotations and Gene Ontology classification annotations, respectively. KEGG pathway enrichment analysis showed that most differentially expressed proteins are involved in pathways related to global and overview maps, cancer overview, immune system, and signal transduction (Figure 1E). Additionally, subcellular localization prediction of the differentially expressed proteins suggested that they are primarily located in the cytoplasm and nucleus (Figure 1F). KEGG pathway enrichment analysis (Figure 1G) indicated that the differentially expressed carbonylated proteins in both the TD and CB groups belong to pathways associated with amyotrophic lateral sclerosis, actin cytoskeleton regulation, carbon metabolism, dilated cardiomyopathy, necrotic bowel disease, viral myocarditis, hypertrophic cardiomyopathy, cardiac muscle contraction, pyruvate metabolism, and arrhythmogenic right ventricular cardiomyopathy. Among the significantly enriched pathways, multiple pathways related to muscle dysfunction such as hypertrophic cardiomyopathy, amyotrophic lateral sclerosis, dilated cardiomyopathy, and viral myocarditis were identified. Considering that ODS manifests as rectal propulsion defects, we conducted PPI analysis on these pathways highly compatible with muscle contraction disorders and annotated all four pathways to DMD (Figure 1H). In summary, we used an iTRAQ-based quantitative proteomics approach to screen for the key differential protein DMD from patient samples.
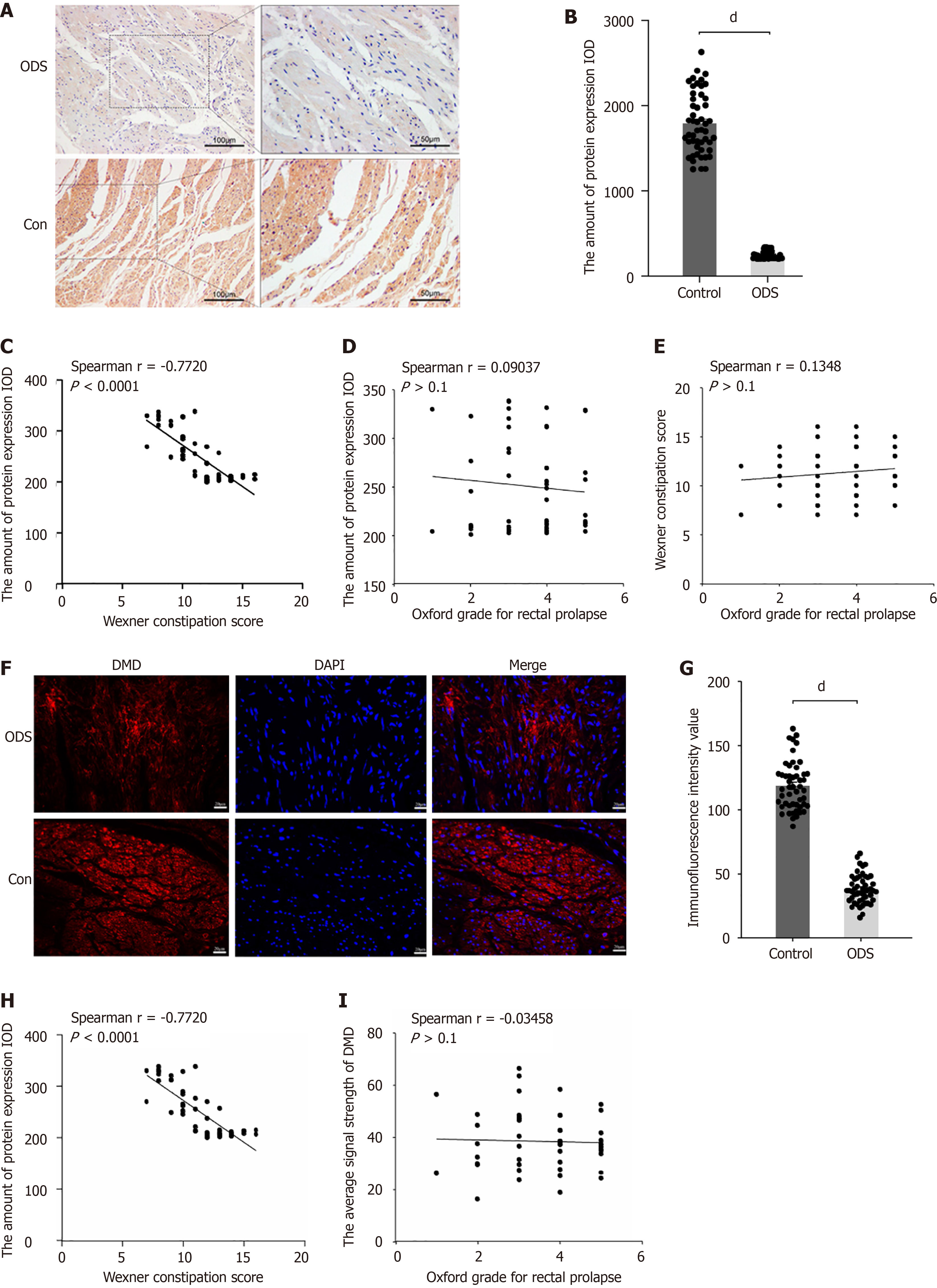
To verify the difference in DMD expression between the ODS and control groups, we conducted immunohistochemical studies on clinical samples collected strictly according to the inclusion criteria. The immunohistochemical results showed that DMD expression was significantly downregulated in the intestinal smooth muscle cells of ODS patients (Figure 2A and B). To investigate whether DMD expression could indicate the severity of ODS, we analyzed the correlation between the integrated optical density values and Wexner constipation severity scores of the corresponding patients. The results showed that there was a negative correlation between DMD expression and the severity of ODS (Figure 2C). However, in the corresponding patients, there was no significant correlation between the integrated optical density (IOD) values and the severity of anatomical lesions (Oxford grade rectal prolapse) (Figure 2D). Similarly, the severity of constipation symptoms in ODS patients did not necessarily reflect the degree of anatomical severity (Figure 2E). Additionally, IF studies further confirmed that DMD expression was significantly reduced in ODS patients (Figure 2F and G). Similar to IOD values, the mean fluorescence intensity of each sample also showed a negative correlation with the severity of constipation symptoms in the corresponding patients, and there was no significant correlation between the mean fluorescence intensity and the severity of anatomical conditions (Figure 2H and I).
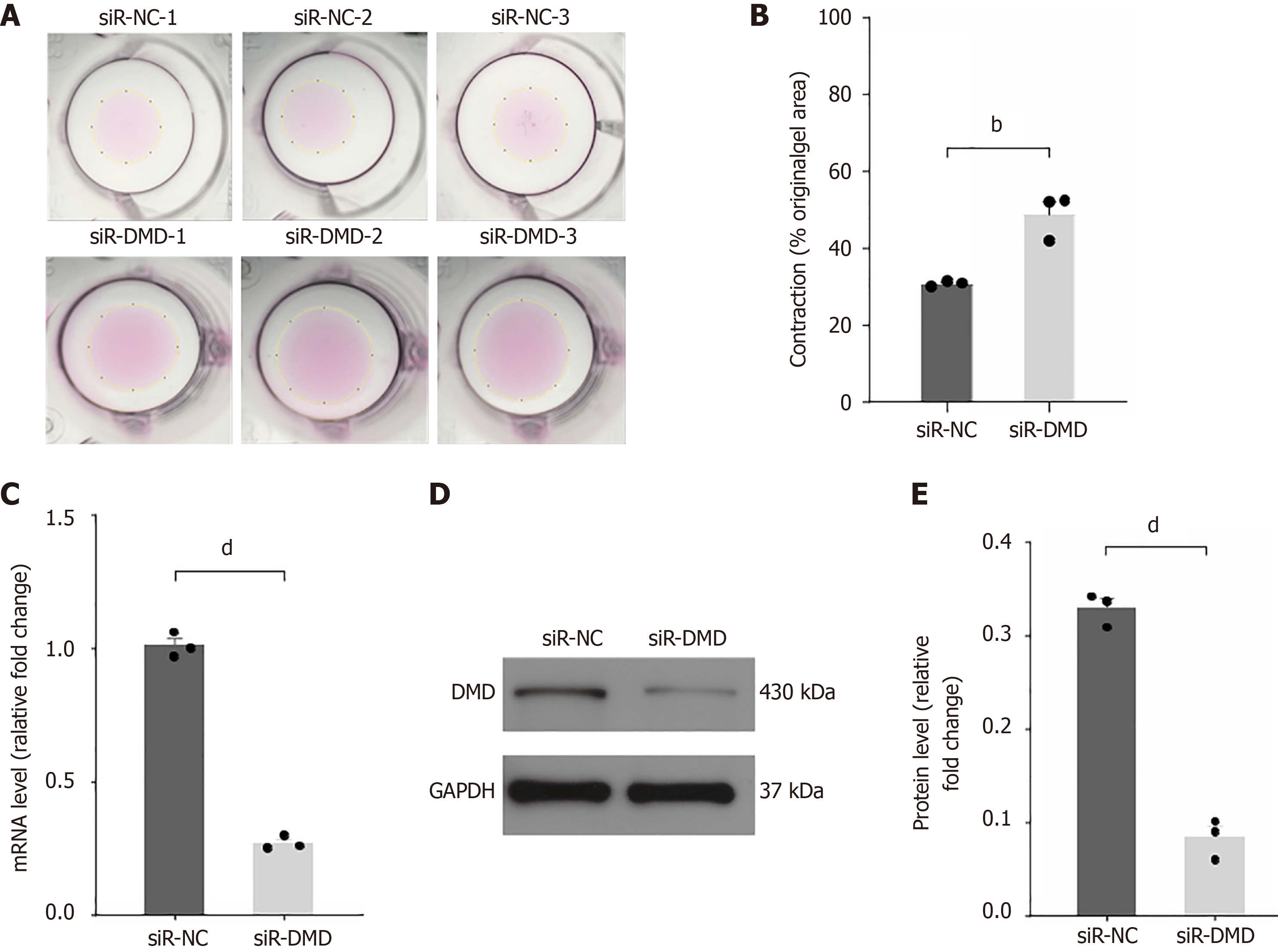
To further investigate the regulatory role of DMD expression on rectal contractile function, we performed collagen gel contraction experiments to assess the impact of reduced DMD expression on the contractile function of human intestinal smooth muscle cells. Due to the lack of a primary human intestinal smooth muscle cell line, we chose primary human colonic smooth muscle cells with similar functions as experimental materials. The results showed that the surface area of the gel after contraction by cells transfected with siR-DMD was larger than that of the control group (Figure 3A and B), indicating that the contractile function of primary intestinal smooth muscle cells with reduced DMD expression was weakened. The downregulation of DMD expression in the cells transfected with siR-DMD was verified by RT-qPCR (Figure 3C) and Western blot (Figure 3D and E).
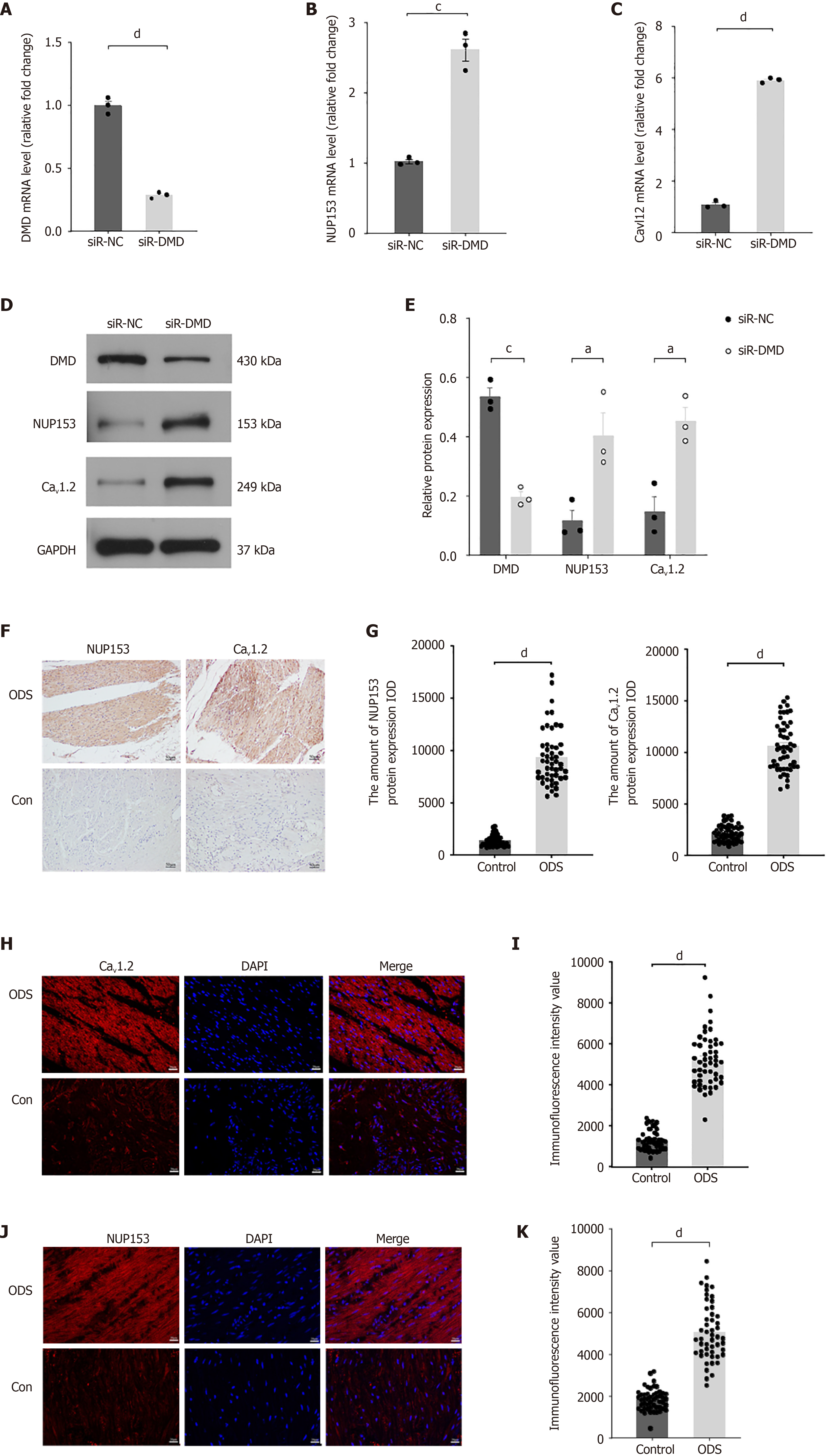
We confirmed that reducing DMD levels can enhance the mRNA and protein levels of NUP153 and Cav1.2 in intestinal smooth muscle cells (Figure 4A-E). In addition, immunohistochemical and IF studies further confirmed the elevated levels of NUP153 and Cav1.2 in ODS patient samples (Figure 4F-K).
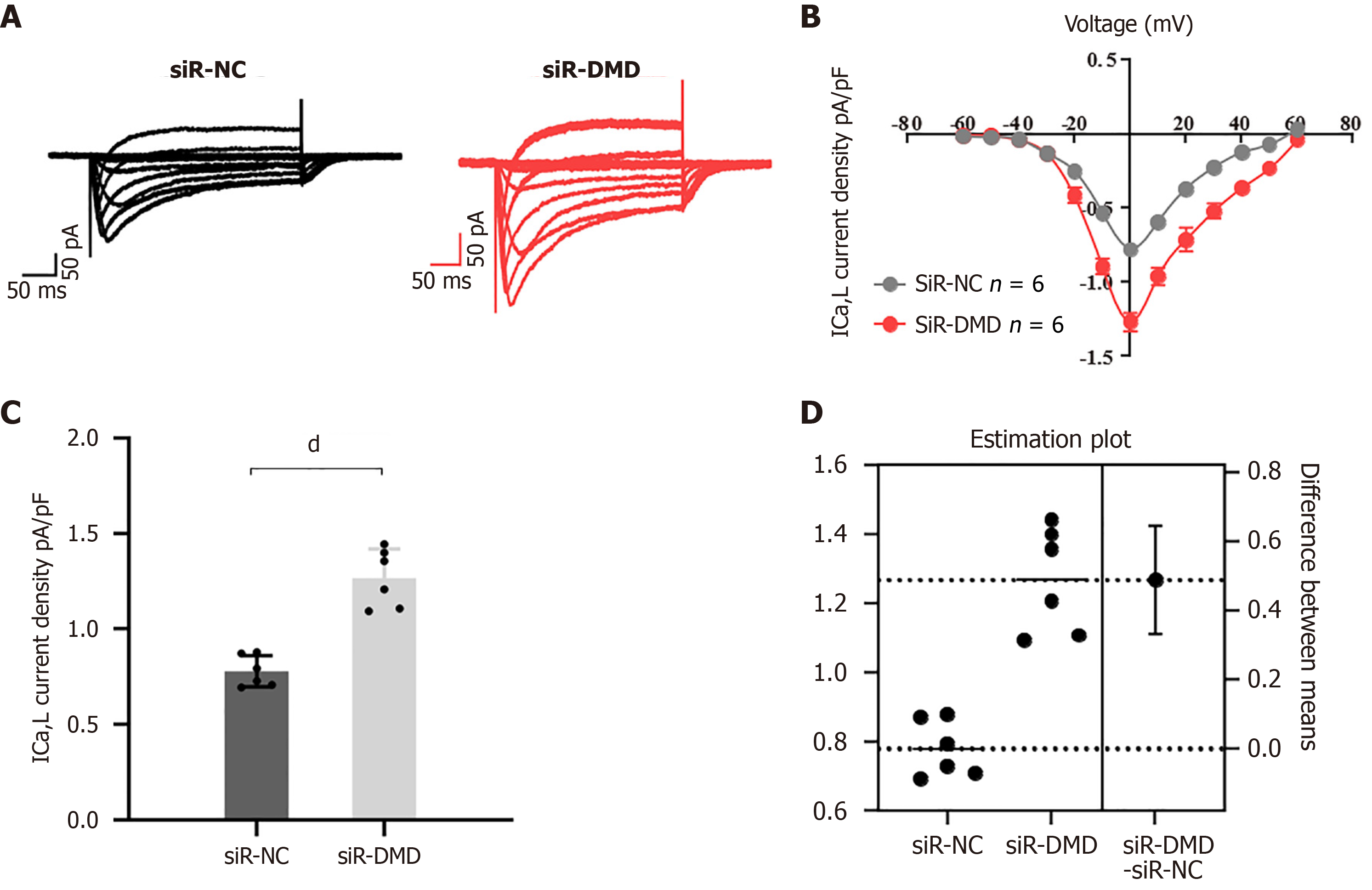
To investigate whether the downregulation of DMD expression affects calcium influx in human intestinal smooth muscle cells, whole-cell currents were recorded using patch-clamp techniques in cells transfected with siR-DMD or siR-NC. After siR-DMD transfection, the amplitude of the calcium tail currents at a repolarization voltage of -30 mV was increased compared to the control (Figure 5A and B; in Figure 5A, the X-axis represents the time in milliseconds and the Y-axis represents the current magnitude in pico-amps). The peak intracellular calcium concentration in cells transfected with siR-DMD was significantly higher than that of the control group (Figure 5C and D).
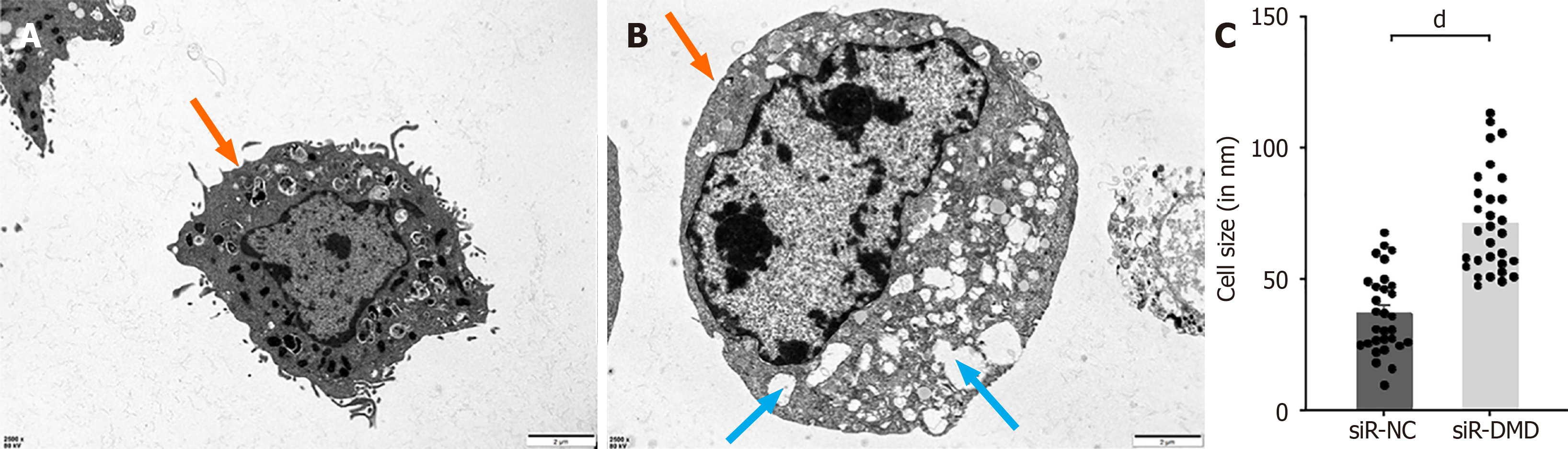
To investigate whether there is any change in the cell membrane structure of intestinal smooth muscle cells after the reduction of DMD expression, we observed the cell membranes of normal and DMD-deficient cells using a transmission electron microscope. We found that the difference between normal and DMD-deficient human intestinal smooth muscle cells lies not in the cell membrane structure but in the cytoplasm. The cell membranes of the experimental group were as intact as those of the control group. Interestingly, vacuolar-like structures were observed in the most cells of the experimental group, but this phenomenon was rarely observed in the control group (Figure 6A and B). Furthermore, a recurring finding was the difference in the area size between the two groups of cells (Figure 6C).
Currently, the long-term outcomes of several ODS therapies remain inadequate. The main reason is a paucity of knowledge about the molecular abnormalities involved in ODS pathogenesis[9]. However, there has been few studies reported on the mechanisms of the molecular abnormalities in the pathogenesis of ODS. The clinical samples used in the present study were derived from patients with ODS patients (ODS group, n = 50), as well as from patients with grade IV hemorrhoids without constipation symptoms (control group, n = 50) who underwent transanal surgery at Department of Colorectal and Anal Surgery, Zhongnan Hospital of Wuhan University. These specimens were available for capture after clinical testing was completed and just prior to when the specimen was normally discarded. To control potential confounding factors, all patients underwent transanal surgery that was performed by the same team of surgeons. All rectal specimens in the present study involved the complete rectal wall with consistent size, volume, and scope. Besides, the rectal specimens obtained from both groups of patients were similar in terms of anatomical abnormalities. However, all patients with grade IV hemorrhoids (control group) were free of constipation symptoms. During the study, all relevant pathological diagnoses were independently completed by pathology experts, and proteomics data analysis was independently conducted by experts in molecular biology and bioinformatics.
In this study, we examined the differences in protein expression between rectal tissue samples from ODS patients and control people using iTRAQ-based proteomic approaches. The findings indicated that 88 proteins were upregulated and 76 proteins were downregulated in the ODS group in comparison to the control group; 216 pathways were associated with these differentially expressed proteins, according to functional annotation clustering. PPI research revealed that DMD may be a key player in the pathways linked to muscular dysfunction in the pathophysiology of ODS. Among all the proteins with differential expression, DMD expression was notably lower in the ODS group. We then verified that DMD expression was considerably reduced in the intestinal smooth muscle tissues of ODS patients using IHC and IF analysis. Research has indicated that there is only a modest relationship between the severity of ODS and anatomical anomalies[21,22]. Similarly, there was no discernible relationship between the severity of ODS and the degree of rectal prolapse in this investigation. It is interesting to note that the Wexner constipation score, which indicates the severity of ODS, was strongly adversely linked with both IOD and mean fluorescence intensity values. As a result, DMD and the intensity of ODS symptoms may be correlated as a diagnostic marker.
We are aware that a reduction in colonic propulsion may occur from abnormalities in the contractile action of intestinal smooth muscle cells. Functional constipation is the final result of inadequate defecation dynamics[9,10]. We speculated that there may be some pathological alterations in the contractile function of intestinal smooth muscle in ODS, since insufficient propulsion of the colon is the most frequent aberrant functional presentation in the disease. We thus carried out additional cellular and molecular biology tests to investigate the impact of aberrant DMD expression on the functionality of intestinal smooth muscle cells, based on the findings of the previously mentioned study. Collagen gel was utilized to quantify the cellular contractility. After cell contraction, the gel’s surface area visually represents the cells’ ability to contract[23]. Our study’s cell contraction testing results demonstrated that decreased DMD expression significantly impairs intestinal smooth muscle cell contraction. Our experimental data suggest that DMD may be a major biological element in the etiology of ODS.
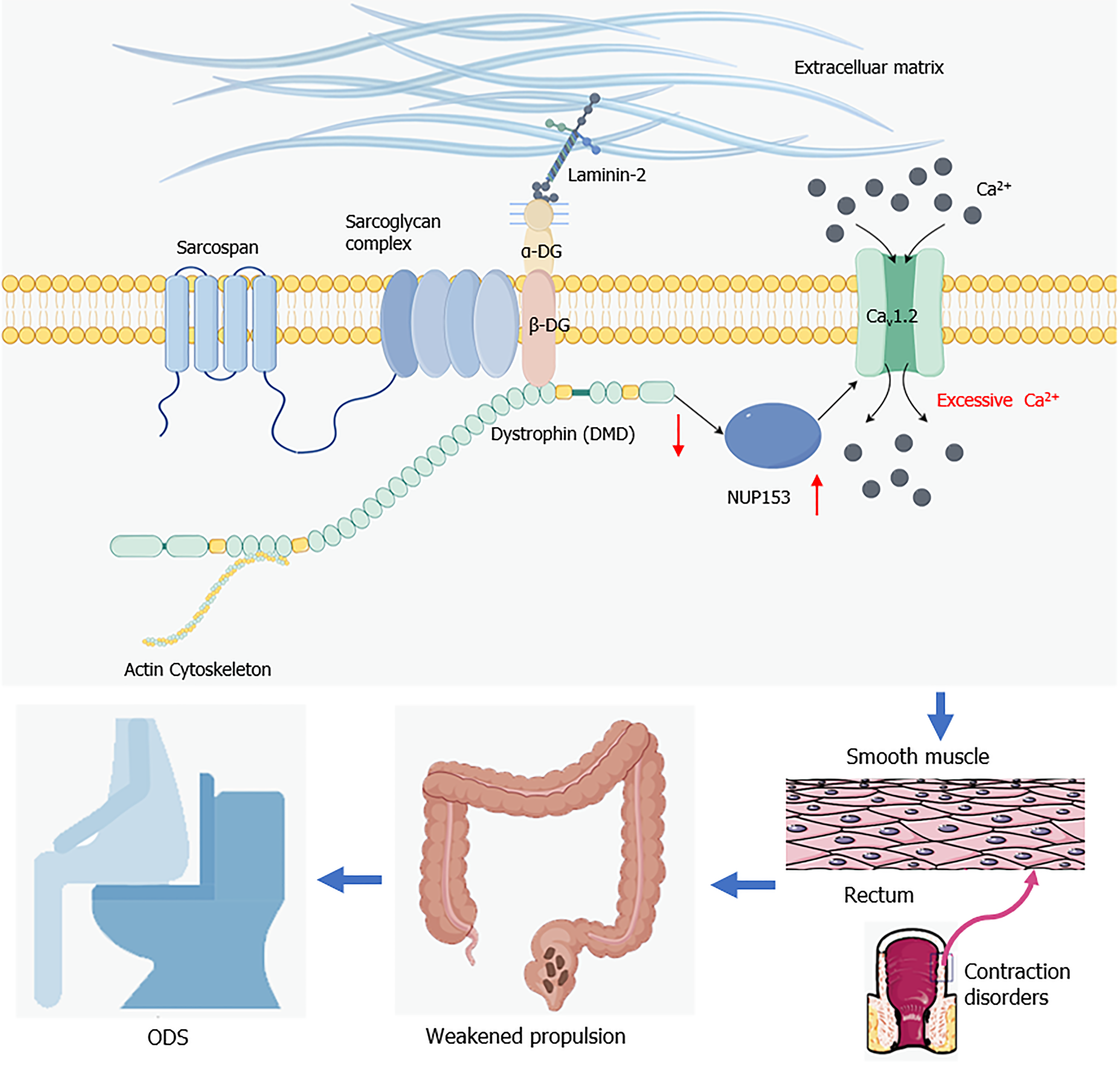
It has been reported that several protein molecules play an important role in regulating muscle contraction function. For example, mutations in the structural proteins that make up the muscle filaments themselves, such as actin, tropomyosin, and troponin, can directly cause muscle contraction dysfunction[24-26]. In addition, nuclear misalignment is one of the characteristics of muscle dysfunction, and the absence of some proteins that regulate nuclear localization, such as Sunday Driver, can lead to this condition[27]. Furthermore, maintaining muscle cell stability is a necessary prerequisite for normal muscle function. Loss of expression or structural changes in DMD-associated protein complex (DPC) members such as DMD and β-dystroglycan can disrupt muscle cell stability. It has been proven that DMD mutations can cause progressive skeletal and myocardial degeneration[28]. Another study has proven that abnormal hydrolysis of β-dystroglycan under the processing of certain matrix metalloproteinases can lead to the loss of DMD, and the combined loss of both exacerbates muscle function degradation[29]. Froehner et al[30] found that DMD deficiency inhibits nitric oxide (NO) signal transduction by blocking the normal expression of neuronal nitric oxide synthase mu, which can cause muscle damage and movement disorders. At present, there have been limited research reports on the interactions between DMD and other proteins related to muscle function. And there has been few studies describing the roles played by these biomolecules in ODS (Figure 7).
The DPC, which consists of dystroglycan, DMD, and syntrophin subcomplexes, is also found in skeletal and cardiac muscles. The precise mechanism of DMD’s stable association with the DPC and its relation to cell membrane stability may have close ties to biomechanics[31,32]. The entry of calcium ions is one of the main elements causing biomechanical alterations. It has been established that calcium-dependent proteases and cellular contractile dysfunction are related[33]. In addition, there are other factors that may be related to muscle contractile dysfunction. Previous studies have demon
Previous research indicates that DMD silencing increases NUP153 expression. Cav1.2 is activated by overexpressed NUP153, which increases calcium ion inflow into cardiomyocytes[38]. This signaling pathway may be a DMD downstream signaling pathway. It is not yet known, though, whether the variations in DMD expression in intestinal smooth muscle cells are similarly connected to related signaling networks. In this investigation, we discovered that NUP153 and Cav1.2 were significantly upregulated in response to a decrease in DMD expression. Furthermore, we recorded L-type calcium currents (ICa-L) from human intestinal smooth muscle cells transfected with siR-NC or siR-DM using the patch-clamp techniques, and the biological consequences of reduced DMD expression were further confirmed by electrophysiological tests, which led to a notable rise in intracellular Ca2+ in rectal smooth muscle cells. An excessive amount of Cav1.2, which is found on the cell membrane surface, may be activated by overexpression of NUP153. This might lead to a significant calcium ion inflow into the cells, which would eventually impair cell contraction. A study has demonstrated that the total calcium content in the muscles of mice lacking DMD expression was significantly higher than that observed in the control group[39]. Furthermore, another study on myocardial disease indicated overexpression of L-type calcium channels, which resulted in a significant increase in calcium influx in cardiomyocytes deficient in DMD expression[38]. The potential causal relationship between the deficiency of DMD expression and an imbalance in cellular calcium influx necessitates validation through more rigorous future experiments.
By using transmission electron microscopy, we were able to validate the impairment of cell contractile function by seeing that, on average, the experimental group’s cell area was bigger than that of the control group. However, we did not notice any visible alterations, like vacuoles, cracks, or deletions, in the cell membrane of the experimental group. Notably, the cytoplasm of the cells in the experimental group frequently had a large number of vacuole-like formations. The cytoplasm and muscular membrane appear to have large concentrations of calcium ions, according to electrophysiological testing, and the development of these vacuoles is most likely caused by an excessive inflow of calcium ions into the cells[40]. Squeezing the cellular contents due to an abundance of vacuoles in the cytoplasm results in skeletal muscle atrophy and increasing muscle weakness[41,42]. Another recent study confirmed, through a combination of molecular biology experiments and optical and electron microscopy examinations, that mutations in the supervillin protein, a large multi-domain protein that interacts with DMD, can lead to a structural myopathy. And characteristic autophagic vacuoles can be observed in the muscle fiber tissue of this muscle disease[43]. However, the specific formation process of vacuoles and the mechanisms underlying muscle contraction remain uncertain, which needs further investigation. In this investigation, transmission electron microscopy also revealed similar findings. More research is needed to determine whether the vacuolization of muscle cells and the increased input of calcium ions are connected to the decrease in intestinal smooth muscle contractility.
In conclusion, this work elucidates the pathophysiology of ODS resulting from abnormal DMD expression. NUP153 and Cav1.2 expression in intestinal smooth muscle cells is upregulated in response to downregulation of DMD expression. The overabundance of calcium ions that ensues from this ultimately impairs the colonic smooth muscle cells’ ability to contract. As a result, ODS and impaired rectal propulsion may result from this functional defect. The findings of this study may offer different approaches to treating ODS as well as specific diagnostic markers. Further research is necessary to determine if point mutations and duplications have a role in the pathophysiology of ODS, as we have only discovered the absence of the DMD gene in this condition. Besides, the sample size included in this study was limited, indicating a need for further expansion in future research. Additionally, this study did not encompass all subtypes of ODS. Future investigations should aim to include a broader range of ODS subtypes to further validate our research findings.
We would like to express our gratitude to Prof. Qiao Huang for his valuable guidance and advice on biostatistics, and to Prof. Yi-Ning Wan for his insightful guidance and suggestions regarding language.
| 1. | Picciariello A, Rinaldi M, Grossi U, Trompetto M, Graziano G, Altomare DF; SICCR ODS Study Group, Gallo G. Time trend in the surgical management of obstructed defecation syndrome: a multicenter experience on behalf of the Italian Society of Colorectal Surgery (SICCR). Tech Coloproctol. 2022;26:963-971. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 2. | Schmier JK, Miller PE, Levine JA, Perez V, Maki KC, Rains TM, Devareddy L, Sanders LM, Alexander DD. Cost savings of reduced constipation rates attributed to increased dietary fiber intakes: a decision-analytic model. BMC Public Health. 2014;14:374. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 3. | Singh G, Lingala V, Wang H, Vadhavkar S, Kahler KH, Mithal A, Triadafilopoulos G. Use of health care resources and cost of care for adults with constipation. Clin Gastroenterol Hepatol. 2007;5:1053-1058. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 71] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 4. | Liang J, Zhao Y, Xi Y, Xiang C, Yong C, Huo J, Zou H, Hou Y, Pan Y, Wu M, Xie Q, Lin Q. Association between Depression, Anxiety Symptoms and Gut Microbiota in Chinese Elderly with Functional Constipation. Nutrients. 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 23] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 5. | Shen L, Li C, Li N, Shen L, Li Z. Abnormal bowel movement frequency increases the risk of rectal cancer: evidence from cohort studies with one million people. Biosci Rep. 2020;40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 6. | Sumida K, Molnar MZ, Potukuchi PK, Thomas F, Lu JL, Yamagata K, Kalantar-Zadeh K, Kovesdy CP. Constipation and risk of death and cardiovascular events. Atherosclerosis. 2019;281:114-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 173] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 7. | Nojkov B, Baker JR, Chey WD, Saad R, Watts L, Armstrong M, Collins K, Ezell G, Phillips C, Menees S. Age- and Gender-Based Differences in Anorectal Function, Gastrointestinal Symptoms, and Constipation-Specific Quality of Life in Patients with Chronic Constipation. Dig Dis Sci. 2023;68:1403-1410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 8. | Pescatori M, Zbar AP, Ayabaca SM. Tailoring surgery for obstructed defecation syndrome to the 'iceberg diagram': Long-term results. Surgery. 2022;172:1636-1641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 9. | Bunni J, Laugharne MJ. Pathophysiological basis, clinical assessment, investigation and management of patients with obstruction defecation syndrome. Langenbecks Arch Surg. 2023;408:75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 10. | Lalwani N, El Sayed RF, Kamath A, Lewis S, Arif H, Chernyak V. Imaging and clinical assessment of functional defecatory disorders with emphasis on defecography. Abdom Radiol (NY). 2021;46:1323-1333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 11. | Brusciano L, Tolone S, Limongelli P, Del Genio G, Messina F, Martellucci J, Lanza Volpe M, Longo A, Docimo L. Anatomical and Functional Features of the Internal Rectal Prolapse With Outlet Obstruction Determined With 3D Endorectal Ultrasonography and High-Resolution Anorectal Manometry: An Observational Case-Control Study. Am J Gastroenterol. 2018;113:1247-1250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 4] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 12. | Prichard DO, Lee T, Parthasarathy G, Fletcher JG, Zinsmeister AR, Bharucha AE. High-resolution Anorectal Manometry for Identifying Defecatory Disorders and Rectal Structural Abnormalities in Women. Clin Gastroenterol Hepatol. 2017;15:412-420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 61] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 13. | Mazzone A, Strege PR, Gibbons SJ, Alcaino C, Joshi V, Haak AJ, Tschumperlin DJ, Bernard CE, Cima RR, Larson DW, Chua HK, Graham RP, El Refaey M, Mohler PJ, Hayashi Y, Ordog T, Calder S, Du P, Farrugia G, Beyder A. microRNA overexpression in slow transit constipation leads to reduced Na(V)1.5 current and altered smooth muscle contractility. Gut. 2020;69:868-876. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 14. | Chai YN, Qin J, Li YL, Tong YL, Liu GH, Wang XR, Liu CY, Peng MH, Qin CZ, Xing YR. TMT proteomics analysis of intestinal tissue from patients of irritable bowel syndrome with diarrhea: Implications for multiple nutrient ingestion abnormality. J Proteomics. 2021;231:103995. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 15. | Zhang Y, Bhosle A, Bae S, McIver LJ, Pishchany G, Accorsi EK, Thompson KN, Arze C, Wang Y, Subramanian A, Kearney SM, Pawluk A, Plichta DR, Rahnavard A, Shafquat A, Xavier RJ, Vlamakis H, Garrett WS, Krueger A, Huttenhower C, Franzosa EA. Discovery of bioactive microbial gene products in inflammatory bowel disease. Nature. 2022;606:754-760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 54] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 16. | Tan M, Ma J, Yang X, You Q, Guo X, Li Y, Wang R, Han G, Chen Y, Qiu X, Wang X, Zhang L. Quantitative proteomics reveals differential immunoglobulin-associated proteome (IgAP) in patients of acute myocardial infarction and chronic coronary syndromes. J Proteomics. 2022;252:104449. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 17. | Yao B, Li Y, Chen T, Niu Y, Wang Y, Yang Y, Wei X, Liu Q, Tu K. Hypoxia-induced cofilin 1 promotes hepatocellular carcinoma progression by regulating the PLD1/AKT pathway. Clin Transl Med. 2021;11:e366. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 38] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 18. | Min YL, Bassel-Duby R, Olson EN. CRISPR Correction of Duchenne Muscular Dystrophy. Annu Rev Med. 2019;70:239-255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 120] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 19. | De Palma FDE, Nunziato M, D'Argenio V, Savarese M, Esposito G, Salvatore F. Comprehensive Molecular Analysis of DMD Gene Increases the Diagnostic Value of Dystrophinopathies: A Pilot Study in a Southern Italy Cohort of Patients. Diagnostics (Basel). 2021;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 20. | Kornegay JN, Spurney CF, Nghiem PP, Brinkmeyer-Langford CL, Hoffman EP, Nagaraju K. Pharmacologic management of Duchenne muscular dystrophy: target identification and preclinical trials. ILAR J. 2014;55:119-149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 40] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 21. | Hawkins AT, Olariu AG, Savitt LR, Gingipally S, Wakamatsu MM, Pulliam S, Weinstein MM, Bordeianou L. Impact of Rising Grades of Internal Rectal Intussusception on Fecal Continence and Symptoms of Constipation. Dis Colon Rectum. 2016;59:54-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 41] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 22. | Hicks CW, Weinstein M, Wakamatsu M, Pulliam S, Savitt L, Bordeianou L. Are rectoceles the cause or the result of obstructed defaecation syndrome? A prospective anorectal physiology study. Colorectal Dis. 2013;15:993-999. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 23. | Kim J, Ushida T, Montagne K, Hirota Y, Yoshino O, Hiraoka T, Osuga Y, Furuakwa KS. Acquired contractile ability in human endometrial stromal cells by passive loading of cyclic tensile stretch. Sci Rep. 2020;10:9014. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 24. | Parker F, Baboolal TG, Peckham M. Actin Mutations and Their Role in Disease. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 44] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 25. | Lambert MR, Gussoni E. Tropomyosin 3 (TPM3) function in skeletal muscle and in myopathy. Skelet Muscle. 2023;13:18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 10] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 26. | Na I, Kong MJ, Straight S, Pinto JR, Uversky VN. Troponins, intrinsic disorder, and cardiomyopathy. Biol Chem. 2016;397:731-751. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 27. | Schulman VK, Folker ES, Rosen JN, Baylies MK. Syd/JIP3 and JNK signaling are required for myonuclear positioning and muscle function. PLoS Genet. 2014;10:e1004880. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 28. | Finsterer J, Cripe L. Treatment of dystrophin cardiomyopathies. Nat Rev Cardiol. 2014;11:168-179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 51] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 29. | Yamada H, Saito F, Fukuta-Ohi H, Zhong D, Hase A, Arai K, Okuyama A, Maekawa R, Shimizu T, Matsumura K. Processing of beta-dystroglycan by matrix metalloproteinase disrupts the link between the extracellular matrix and cell membrane via the dystroglycan complex. Hum Mol Genet. 2001;10:1563-1569. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 117] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 30. | Froehner SC, Reed SM, Anderson KN, Huang PL, Percival JM. Loss of nNOS inhibits compensatory muscle hypertrophy and exacerbates inflammation and eccentric contraction-induced damage in mdx mice. Hum Mol Genet. 2015;24:492-505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 31. | Farrokhi V, Walsh J, Palandra J, Brodfuehrer J, Caiazzo T, Owens J, Binks M, Neelakantan S, Yong F, Dua P, Le Guiner C, Neubert H. Dystrophin and mini-dystrophin quantification by mass spectrometry in skeletal muscle for gene therapy development in Duchenne muscular dystrophy. Gene Ther. 2022;29:608-615. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 32. | Kaplan KM, Morgan KG. The importance of dystrophin and the dystrophin associated proteins in vascular smooth muscle. Front Physiol. 2022;13:1059021. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 33. | Allen DG, Gervasio OL, Yeung EW, Whitehead NP. Calcium and the damage pathways in muscular dystrophy. Can J Physiol Pharmacol. 2010;88:83-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 132] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 34. | Colussi C, Gurtner A, Rosati J, Illi B, Ragone G, Piaggio G, Moggio M, Lamperti C, D'Angelo G, Clementi E, Minetti G, Mozzetta C, Antonini A, Capogrossi MC, Puri PL, Gaetano C. Nitric oxide deficiency determines global chromatin changes in Duchenne muscular dystrophy. FASEB J. 2009;23:2131-2141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 62] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 35. | Houang EM, Sham YY, Bates FS, Metzger JM. Muscle membrane integrity in Duchenne muscular dystrophy: recent advances in copolymer-based muscle membrane stabilizers. Skelet Muscle. 2018;8:31. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 43] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 36. | Kyrychenko V, Poláková E, Janíček R, Shirokova N. Mitochondrial dysfunctions during progression of dystrophic cardiomyopathy. Cell Calcium. 2015;58:186-195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 46] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 37. | Lovering RM, De Deyne PG. Contractile function, sarcolemma integrity, and the loss of dystrophin after skeletal muscle eccentric contraction-induced injury. Am J Physiol Cell Physiol. 2004;286:C230-C238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 116] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 38. | Nanni S, Re A, Ripoli C, Gowran A, Nigro P, D'Amario D, Amodeo A, Crea F, Grassi C, Pontecorvi A, Farsetti A, Colussi C. The nuclear pore protein Nup153 associates with chromatin and regulates cardiac gene expression in dystrophic mdx hearts. Cardiovasc Res. 2016;112:555-567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 39. | Matsumura CY, Taniguti AP, Pertille A, Santo Neto H, Marques MJ. Stretch-activated calcium channel protein TRPC1 is correlated with the different degrees of the dystrophic phenotype in mdx mice. Am J Physiol Cell Physiol. 2011;301:C1344-C1350. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 40. | Yan C, Tanaka M, Sugie K, Nobutoki T, Woo M, Murase N, Higuchi Y, Noguchi S, Nonaka I, Hayashi YK, Nishino I. A new congenital form of X-linked autophagic vacuolar myopathy. Neurology. 2005;65:1132-1134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 41. | Ban R, Zhang Y, Li K, Shi Q. A Case of Myotonic Dystrophy Type I With Rimmed Vacuoles in Skeletal Muscle Pathology. J Clin Rheumatol. 2021;27:S771-S772. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 42. | Ban R, Lu X, Pu C, Wang H, Liu H, Zhang Y, Shi Q. Novel GNE mutations in three Chinese patients with typical GNE myo-pathy. J Pak Med Assoc. 2020;70:913-916. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 43. | Hedberg-Oldfors C, Meyer R, Nolte K, Abdul Rahim Y, Lindberg C, Karason K, Thuestad IJ, Visuttijai K, Geijer M, Begemann M, Kraft F, Lausberg E, Hitpass L, Götzl R, Luna EJ, Lochmüller H, Koschmieder S, Gramlich M, Gess B, Elbracht M, Weis J, Kurth I, Oldfors A, Knopp C. Loss of supervillin causes myopathy with myofibrillar disorganization and autophagic vacuoles. Brain. 2020;143:2406-2420. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |