Published online Nov 21, 2024. doi: 10.3748/wjg.v30.i43.4609
Revised: September 6, 2024
Accepted: September 13, 2024
Published online: November 21, 2024
Processing time: 142 Days and 22.1 Hours
The early diagnosis rate of esophageal cancer (EC), one of the most prevalent digestive tract cancers worldwide, remains low.
To investigate the utility of plasma SHOX2, SEPTIN9, EPO, and RNF180 methy
Plasma samples were collected from 210 patients at Hubei Cancer Hospital, and TaqMan polymerase chain reaction was employed to detect plasma SHOX2, SEPTIN9, RNF180, and EPO methylation. The area under the curve was used to estimate their diagnostic value for EC. Cox and logistic regression analyses were used to esti
The sensitivity and specificity of combined assessment of plasma SHOX2, SEPTIN9, RNF180, and EPO methylation for adenocarcinoma, squamous cell carcinoma (SCC), and EC detection were 66.67% and 86.27%, 77.40% and 85.29%, and 76.19% and 86.27%, respectively; the area under the curve values for diagnosing adenocarcinoma, SCC, and EC were 0.737 [95% confidence interval (CI): 0.584–0.89], 0.824 (95%CI: 0.775–0.891), and 0.864 (95%CI: 0.809–0.92), respectively.
According to our findings, plasma SHOX2, SEPTIN9, RNF180, and EPO methylation exhibits appreciated sensi
Core Tip: This study for the first time analyzed the clinical value of detecting plasma SHOX2, SEPTIN9, EPO, and RNF180 methylation for diagnosis and monitoring of esophageal cancer (EC). We revealed that the sensitivity and specificity of combined assessment of methylation of the four genes in plasma for detecting EC were 76.19% and 86.27%, respectively; the area under the curve value of this combination was 0.864 (95% confidence interval: 0.809–0.92). The accurate measurement of methylation of these four genes in plasma might aid monitoring of therapy efficacy in both esophageal adenocarcinoma and squamous cell carcinoma.
- Citation: Liu XJ, Pi GL, Wang S, Kai JD, Yu HF, Shi HW, Yu J, Zeng H. Plasma DNA methylation detection for early screening, diagnosis, and monitoring of esophageal adenocarcinoma and squamous cell carcinoma. World J Gastroenterol 2024; 30(43): 4609-4619
- URL: https://www.wjgnet.com/1007-9327/full/v30/i43/4609.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i43.4609
Esophageal cancer (EC), a prevalent digestive tract tumor, claims the lives of approximately 604000 individuals worldwide annually. It exhibits varying morbidity and mortality rates across different countries. China reportedly has the highest incidence of EC globally, with an average annual death toll of approximately 576000 people[1]. In recent years, EC has surged rapidly in China, particularly affecting the younger population[1]. Progressive dysphagia is the hallmark symptom of EC; however, its diagnosis is frequently delayed due to nonspecific symptoms in the early stages of tumor development. Dysphagia onset often correlates with an advanced disease, carrying a 5-year survival rate of < 15%[1,2].
Clinical diagnosis of EC primarily relies on double-contrast barium X-ray and endoscopy[3]. However, endoscopic population screening lacks widespread acceptance in China[4]. The early diagnosis rate of EC remains low, contributing to poor prognoses for patients with EC diagnosed at advanced clinical stages[5,6]. Currently, there are no established tumor markers for EC. Traditional markers for esophageal squamous cell carcinoma (SCC) and adenocarcinoma, such as SCC antigen (SCCA), carbohydrate antigen 199 (CA199), and carcinoembryonic antigen (CEA), exhibit limited sensitivity and specificity. In recent years, the clinical significance of circulating tumor DNA (ctDNA) as tumor markers in cancer diagnosis has gained recognition. ctDNA is released into the bloodstream by tumor cells, encompassing cancer-related genetic mutations and epigenetic variations[7-9]. DNA methylation variations emerge early in cancer, representing dynamic epigenetic changes that evolve with tumor progression. ctDNA methylation in liquid biopsy offers advantages such as non-invasiveness, stable target molecules, cost-effectiveness, high diagnostic performance, and broad applicability[10,11]. Monitoring ctDNA methylation levels aids in early tumor diagnosis and prognosis assessment, with proven application and potential in the clinical diagnosis of various cancers. Noteworthy examples include the use of SHOX2 promoter methylation for lung cancer screening[12,13]. Further, an analysis of EPO promoter methylation revealed frequent hypermethylation of regulatory sequences in tumors, leading to epigenetic silencing of EPO in cancer cells[14]. SHOX2 exhibits high methylation levels in patients with lung cancer, with its dynamic increase markedly associated with elevated cancer risk in high-risk populations[15,16]. Moreover, SEPT9 methylation, employed for colorectal cancer (CRC) screening, stands out as the first ctDNA liquid biopsy indicator approved by the United States Food and Drug Administration. It demonstrates superior diagnostic performance than protein tumor markers and fecal immunochemical tests[17,18], with its sensitivity progressively increasing with cancer stage. According to previous studies, the sensitivity and specificity of circulating methylated Septin 9 (mSEPT9) are estimated to be 50%-70% and ≥ 90%, respectively, for detecting CRC. RNF180 is a novel preferentially methylated gene in gastric adenocarcinoma[19,20], and the area under the curve (AUC) values of methylated RNF180 is 0.723 [95% confidence interval (CI): 0.694-0.752] [sensitivity: 46.2% (95%CI: 42.3%-50.1%); specificity: 87.3% (95%CI: 84.1%-89.9%)]. The sensitivity of mSEPT9 for detecting EC was 43.1% (AUC = 0.69), with a 95.6% specificity[21,22]. Despite these promising findings, the application of methylation detection in early screening for EC remains limited in reported studies. Consequently, it is imperative to develop non-invasive detection methods that facilitate effective early screening of EC. The present study for the first time explored the clinical utility of combined detection of methylation of plasma SHOX2, SEPTIN9, EPO, and RNF180 in diagnosis and monitoring of esophageal adenocarcinoma and SCC.
Cancer genome atlas data were retrieved from the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) and The Cancer Genome Atlas (http://www.ualcan.path.uab.edu) databases.
We retrospectively enrolled patients (n = 210) from Hubei Cancer Hospital, China between June 2022 and July 2023 to evaluate the clinical performance of plasma SHOX2, SEPTIN9, EPO, and RNF180 gene methylation in diagnosing EC. The screening population had one of the following characteristics: Barrett’s esophagus history; family history of EC; dysphagia; chronic uncured gastroesophageal reflux disease; and corrosive esophageal burn. All patients underwent an upper endoscopy, and some received pathological examination if necessary. Thirty-five patients were initially diagnosed as having EC, and the other 175 EC patients were diagnosed based on pathological evidence according to World Health Organization criteria and TNM classification. Patients who had not undergone prior anticancer therapy were included. Healthy controls (n = 72) were obtained from the health examination center, and cases of benign esophageal diseases (n = 30) were collected from Wuhan Hospital of Integrated Traditional Chinese and Western Medicine, China. Ethical approval was obtained from the Hubei Cancer Hospital ethics committee.
To ensure sample integrity, whole blood samples (10 mL/case) were collected from each participant with Streck Cell-Free DNA BCT (Streck, United States) and centrifuged at 1600 g for 20 min at room temperature to obtain the plasma. All plasma samples were stored at -80 °C. To mitigate the impact of sample storage duration on DNA quality, all samples were analyzed within 1 mo after collection. The BioChain Circulating Nucleic Acid Kit (Qiagen, Valencia, CA, United States) was used to extract cfDNA from plasma. CpG island cytosine methylation in SHOX2, SEPTIN9, RNF180, and EPO genes in human plasma was detected by TaqMan polymerase chain reaction (PCR) after treating DNA with sulfite. Unmethylated cytosine was converted to uracil using sodium bisulfite to ensure that only methylated cytosine was replicated. Bisulfite-converted DNA was then captured on the magnetic particles, purified by washing steps and eluted in a 55-μL elution buffer for real-time PCR. DCt, calculated as Ct (gene of interest) − Ct (internal control), was used to assess the co-methylation levels of these genes. Samples were included in our analyses when 18 ≤ CtACTB ≤ 30. They were classified as methylation-positive when DNA methylation levels of at least one of the four genes met the following quantitative criteria: CtSHOX2 < 32 and DCtSHOX2 ≤ 9; CtEPO < 35 and DCtEPO ≤ 12; CtSEPTIN9 < 40 and DCtSEPTIN9 ≤ 9; and CtRNF180 < 40 and DCtRNF180 ≤ 9. All other samples were classified as methylation-negative. Methylation reagents for SHOX2 and EPO genes were sourced from LungMe Assay (Tellgen Corporation, Shanghai, China), and those for SEPTIN9 and RNF180 were from a methylation test kit (Biochain Science Technology Inc, Beijing, China).
Serum levels of SCCA, CEA, and CA199 were detected using a chemiluminescence immunoassay and associated reagents (Snibe Diagnostic Company Ltd., Shenzhen, China). Abnormal levels were determined based on the following predefined thresholds: SCCA > 2.7 mg/L, CEA > 5 μg/mL, and CA199 > 41 U/mL.
SPSS 25 (IBM, Armonk, NY, United States) was used for statistical analyses. The normality of the distribution was determined using the Shapiro–Wilk test. Differences between groups were analyzed using the Mann–Whitney U-test, Kruskal–Wallis H-test, one-way analysis of variance, independent t-test, or χ2 test, as appropriate. The association between individual patient variables was analyzed with Spearman’s rank correlation significance test. P < 0.05 indicated statistical significance. Specificity and sensitivity were calculated using receiver operating characteristic (ROC) curves. Differences between groups with different results were analyzed using the t-test or χ2 test. Univariate analysis and multivariate Cox regression were employed to identify the association of different variables with EC.
This study comprised 210 patients with EC and 102 controls from the screening population at Hubei Cancer Hospital between June 2022 and July 2023. The study cohort consisted of 122 men [average age: 65.6 years (51–87 years)] and 88 women [average age: 69.9 years (54–89 years)]. Disease characteristics of the cohort are presented in Table 1.
| Clinicopathological data | n | SHOX2 methylation | Septin9 methylation | EPO methylation | RNF180 methylation | Combined methylation detection |
| Tumor pathology | ||||||
| Adenocarcinoma | 33 | |||||
| Squamous cell carcinoma | 177 | |||||
| P value | 0.354 | 0.19 | 0.524 | 0.098 | 0.527 | |
| Gender | ||||||
| Male | 122 | |||||
| Female | 88 | |||||
| Male vs female | 0.416 | 0.379 | 0.439 | 0.279 | 0.464 | |
| Age at diagnosis | ||||||
| ≤ 60 years | 73 | |||||
| > 60 years | 137 | |||||
| Median age (years) | 65.6 | |||||
| Mean age (years) | 63.5 | |||||
| ≤ 60 vs > 60 years | 0.665 | 0.011 | 0.021 | 0.211 | 0.001 | |
| Tumor stage | ||||||
| I-II | 19 | |||||
| III-IV | 191 | |||||
| I-II vs III-IV | 0.019 | 0.022 | 0.001 | 0.149 | 0.001 | |
| Follow-up | ||||||
| Follow-up available | 23 |
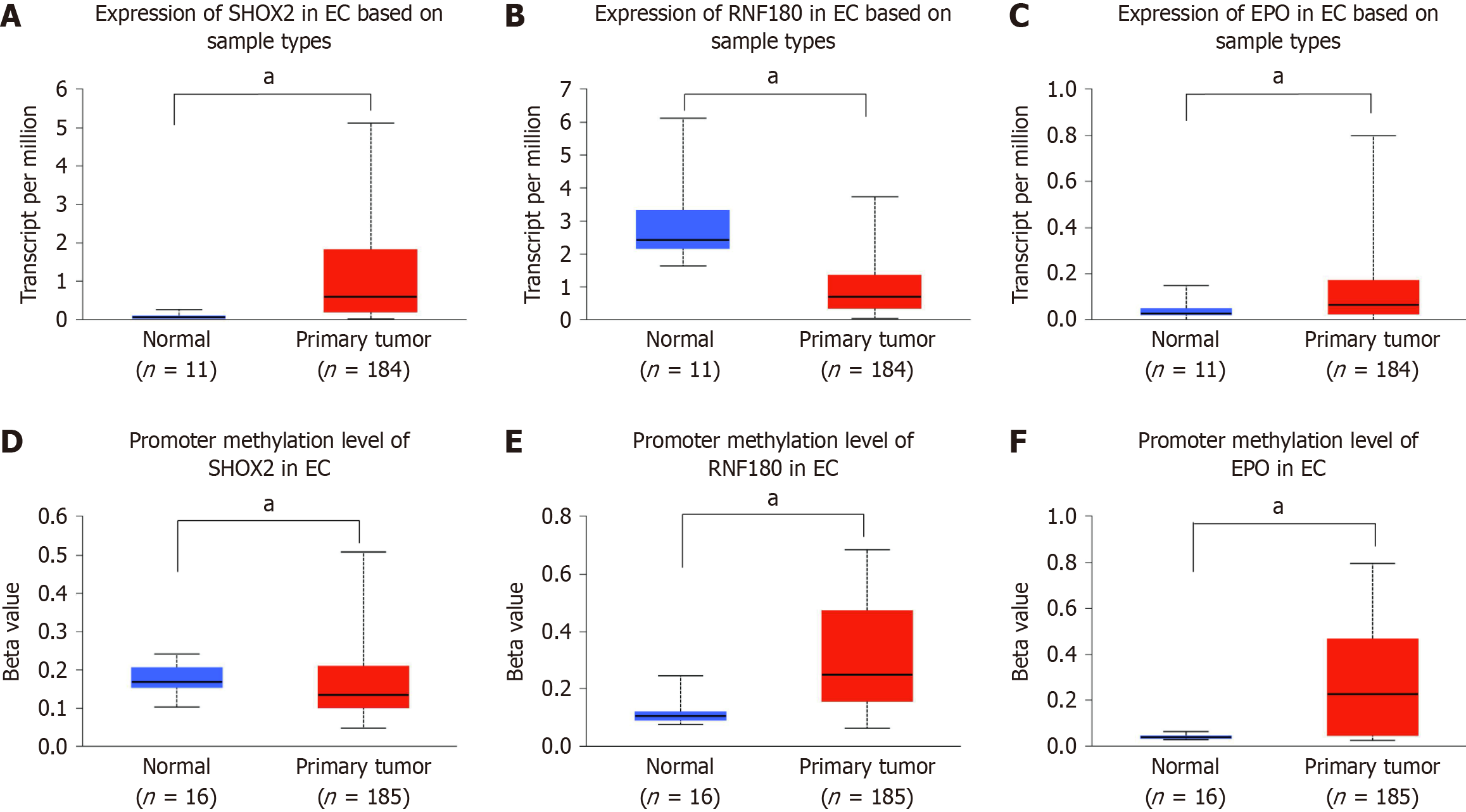
According to TCGA data analysis, EPO and SHOX2 expression in EC tissue samples was significantly higher than that in normal tissue samples, while RNF180 expression in EC tissue samples was lower (P < 0.05). Promoter methylation levels of EPO, SHOX2, and RNF180 in normal (n = 16) and EC (n = 185) tissue samples were indicated by beta values ranging from 0 (unmethylated) to 1 (fully methylated). Beta values between 0.7 and 0.5 indicated hypermethylation and those between 0.3 and 0.25 indicated hypomethylation (Figure 1).
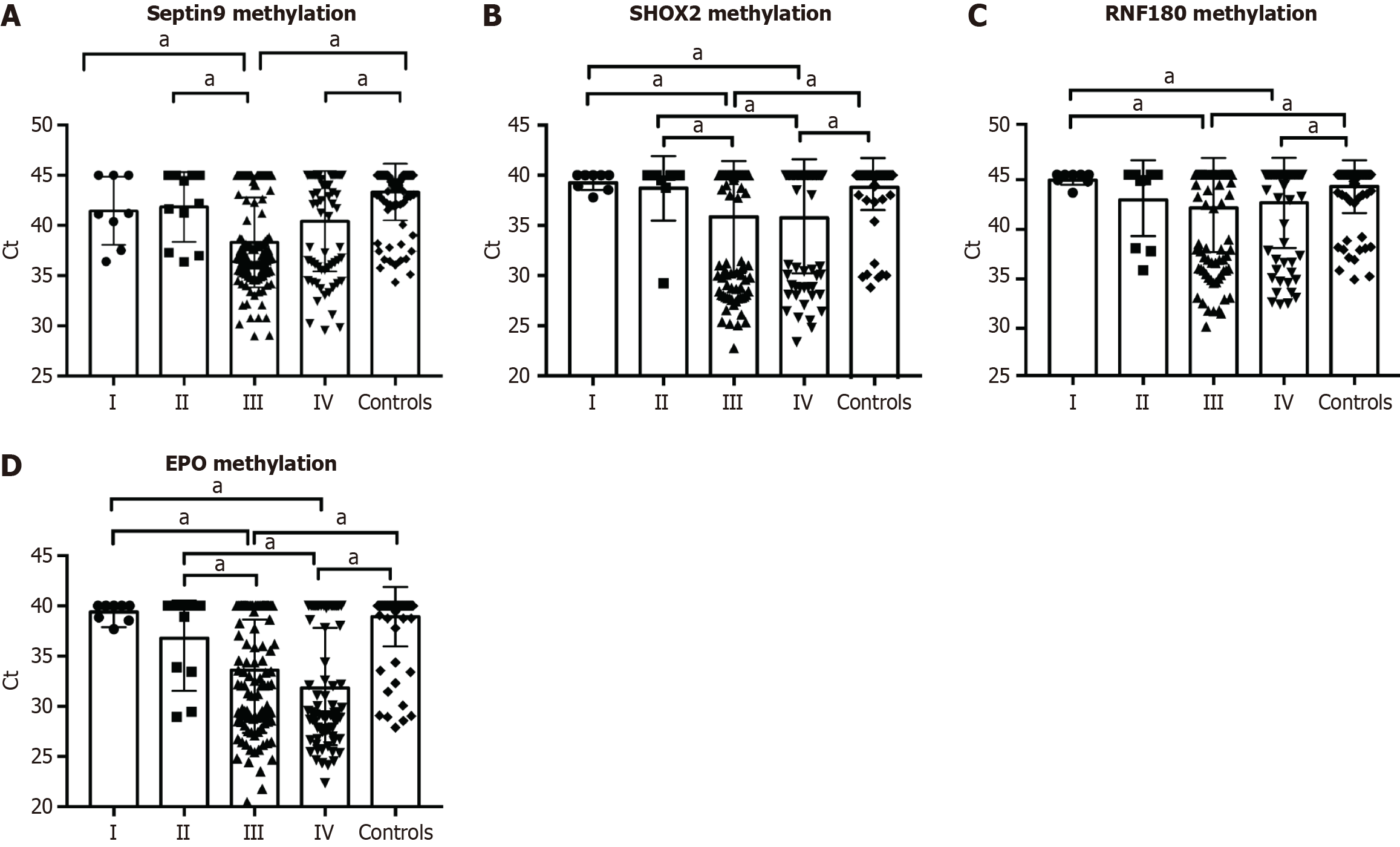
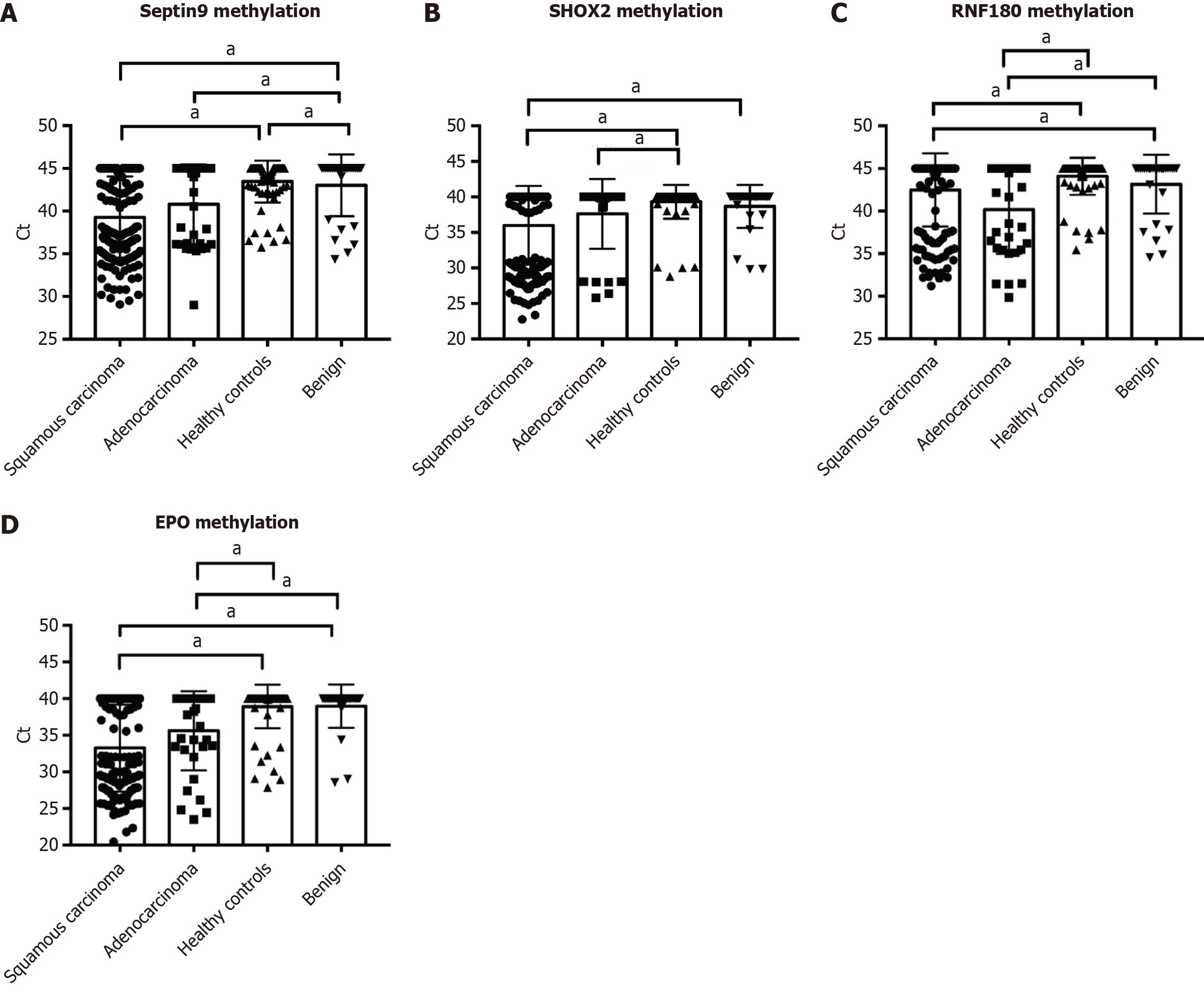
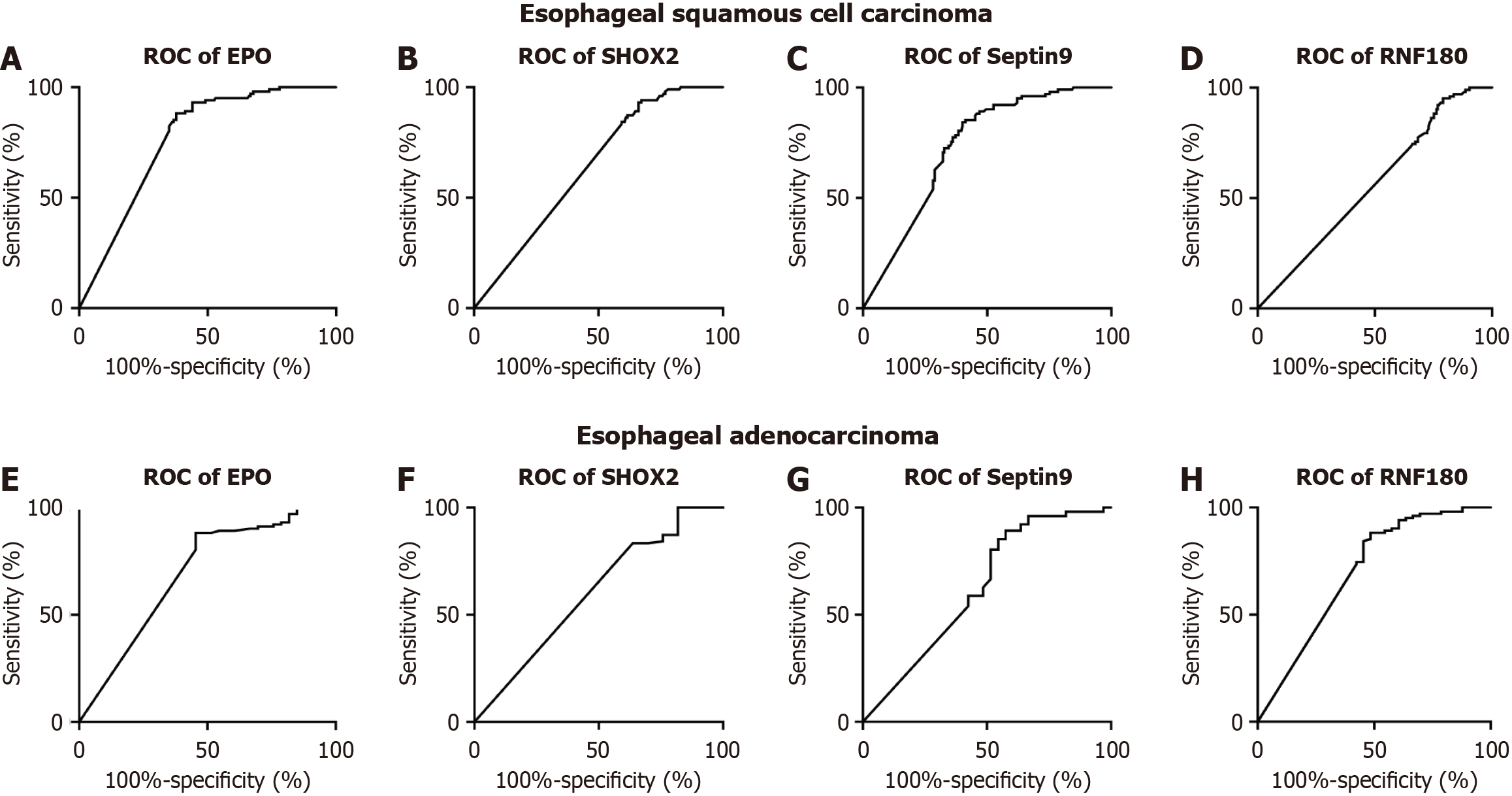
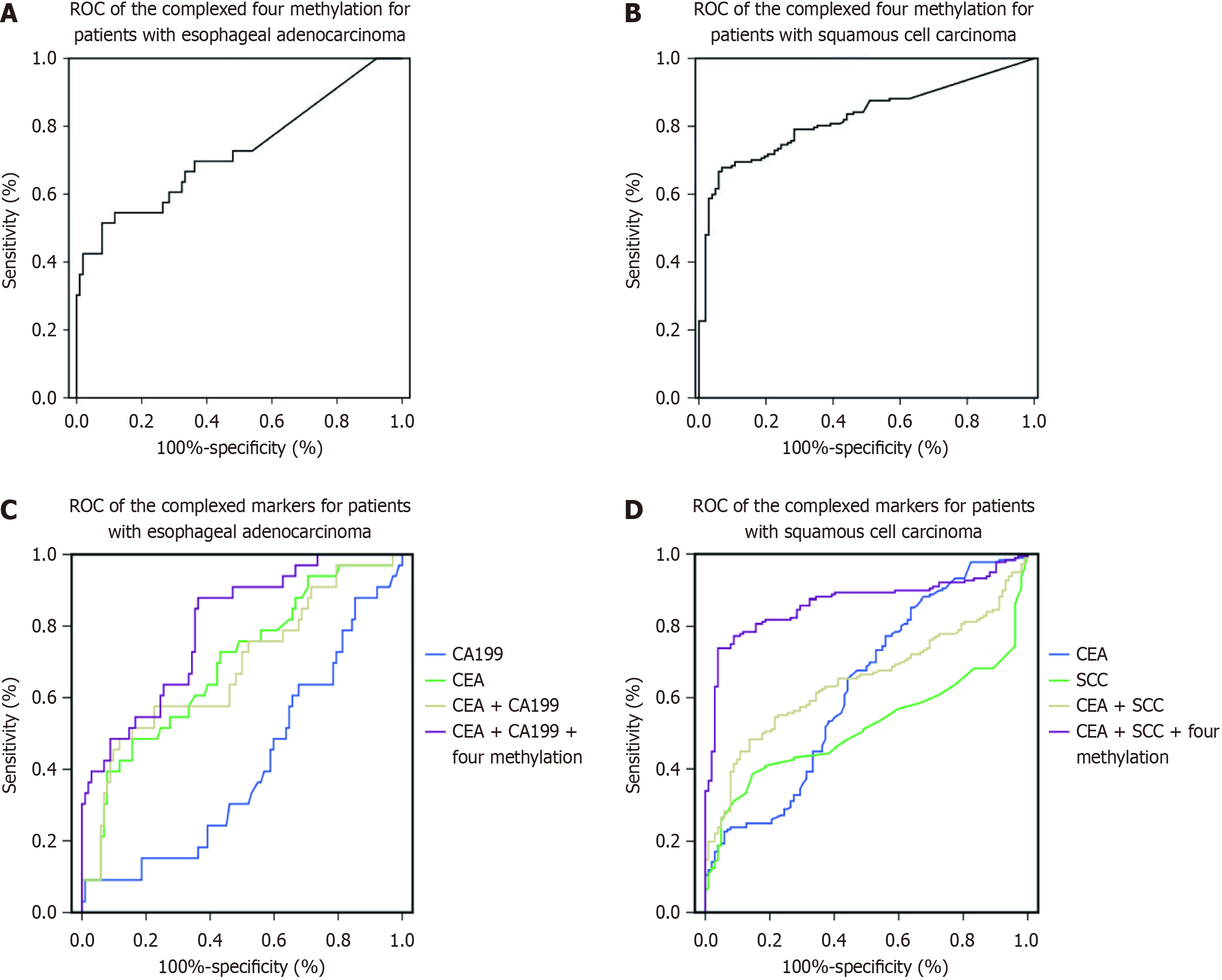
Figure 2 and Figure 3 depict the frequency distribution of methylation for the four aforementioned genes for detecting esophageal adenocarcinoma and SCC. In EC patients with advanced stage disease, SHOX2, RNF180, SEPTIN9, and EPO DNA methylation was more prevalent than in EC patients with early stage disease (Figure 2). However, there were no significant differences in the positive percentages of SHOX2, RNF180, SEPTIN9, and EPO methylation in EC patients with SCC and adenocarcinoma (Figure 3). The sensitivity and specificity of SHOX2, RNF180, SEPTIN9, and EPO methylation for diagnosing EC were 18%–56% and 80%–90%, respectively (Figure 4). Figure 4 depicts the ROC curves for methylation of these genes. The sensitivity and specificity of SHOX2, EPO, SEPTIN9, and RNF180 methylation for diagnosing esophageal adenocarcinoma were 18.18% and 94.12%, 42.42% and 88.24%, 42.42% and 86.27%, and 45.45% and 88.24%, respectively, while the sensitivity for diagnosing esophageal SCC was 33.90%, 55.93%, 54.80%, and 23.73%, respectively. When combining the methylation of the four genes, the sensitivity and specificity for diagnosing adenocarcinoma, SCC, and EC were 66.67% and 86.27%, 77.40% and 85.29%, and 76.19% and 86.27%, respectively. The AUC values of SHOX2, EPO, SEPTIN9, and RNF180 methylation for diagnosing esophageal SCC were 0.639 (95%CI: 0.575–0.703), 0.765 (95%CI: 0.710–0.820), 0.731 (95%CI: 0.673–0.789), and 0.559 (95%CI: 0.491–0.626), and the corresponding values for diagnosing esophageal adenocarcinoma were 0.601 (95%CI: 0.484-0.719), 0.691 (95%CI: 0.577-0.805), 0.626 (95%CI: 0.504-0.749), 0.697 (95%CI: 0.582-0.813), respectively (Figure 4). The AUC values for the combined evaluation to diagnose adenocarcinoma, SCC, and EC were 0.737 (95%CI: 0.584–0.89), 0.824 (95%CI: 0.775–0.891), and 0.864 (95%CI: 0.809–0.92), respectively (Figure 5).
Of the 210 patients with EC, 19 had stage I-II and 191 had stage III-IV disease (Table 1). The sensitivity and specificity of SHOX2, EPO, SEPTIN9, and RNF180 methylation for diagnosing stage I-II EC were 52.63% and 86.54%, respectively.
Serum SCCA demonstrated a 28.81% sensitivity and 90.20% specificity for detecting esophageal SCC, while CEA demonstrated a 19.2% sensitivity. CA199 and CEA demonstrated a 18.18% and 21.21% sensitivity and 87.25% and 86.27% specificity, respectively, for diagnosing adenocarcinoma. The AUC values of SCCA, CEA, and complex makers to diagnose esophageal SCC were 0.516 (95%CI: 0.448–0.583), 0.626 (95%CI: 0.557–0.695), and 0.648 (95%CI: 0.584-0.711), respectively (Figure 5). For diagnosing adenocarcinoma, the AUC value of the CEA, CA199, and complex markers were 0.683 (95%CI: 0.572–0.793), 0.408 (95%CI: 0.297-0.519), and 0.693 (95%CI: 0.588-0.798), respectively. The AUC value of the combined tumor markers (SCCA and CEA) and SHOX2, EPO, SEPTIN9, and RNF180 methylation was 0.864 (0.820–0.908) for diagnosing esophageal SCC, while it was 0.798 (95%CI: 0.714-0.883) for diagnosing adenocarcinoma (Figure 5).
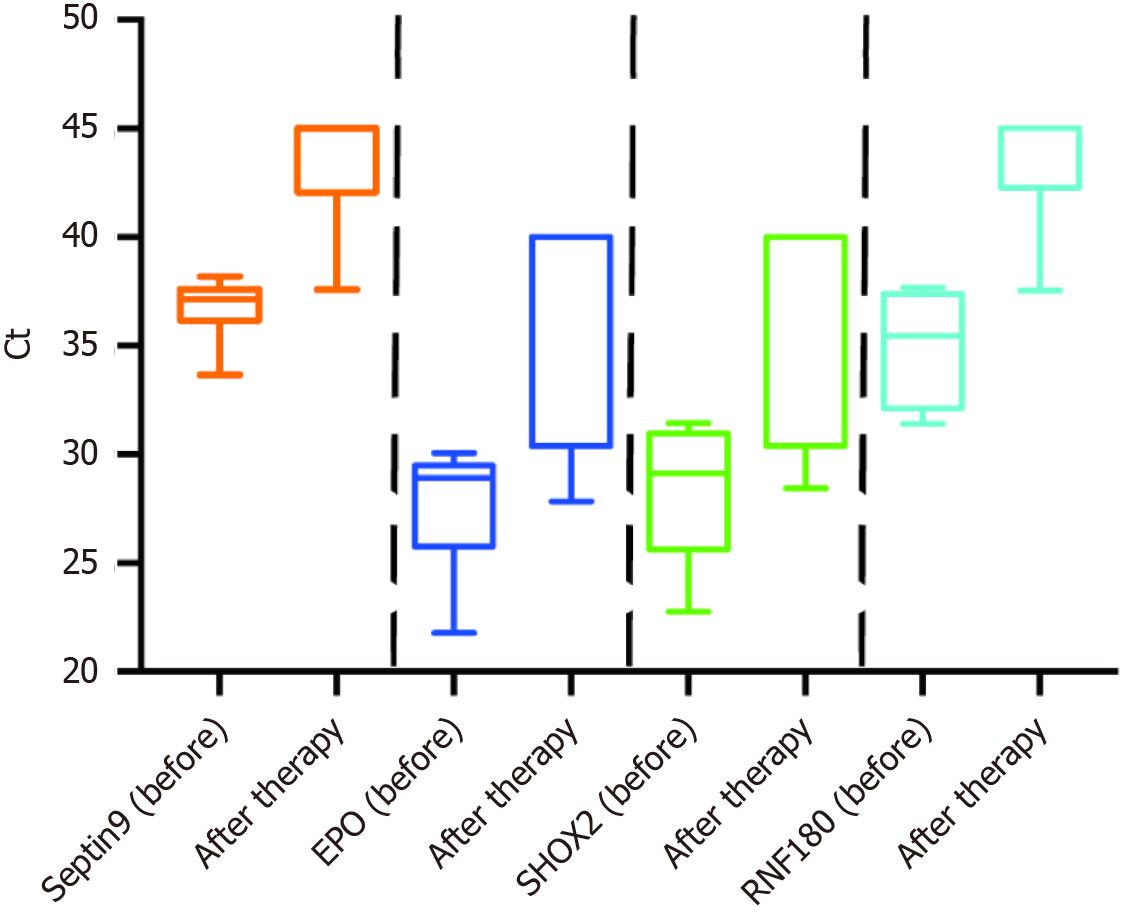
Figure 6 depicts the frequency distribution of plasma DNA methylation before and after therapy in 23 patients. DNA methylation in the plasma of 10 patients exhibited a considerable negative change post-therapy.
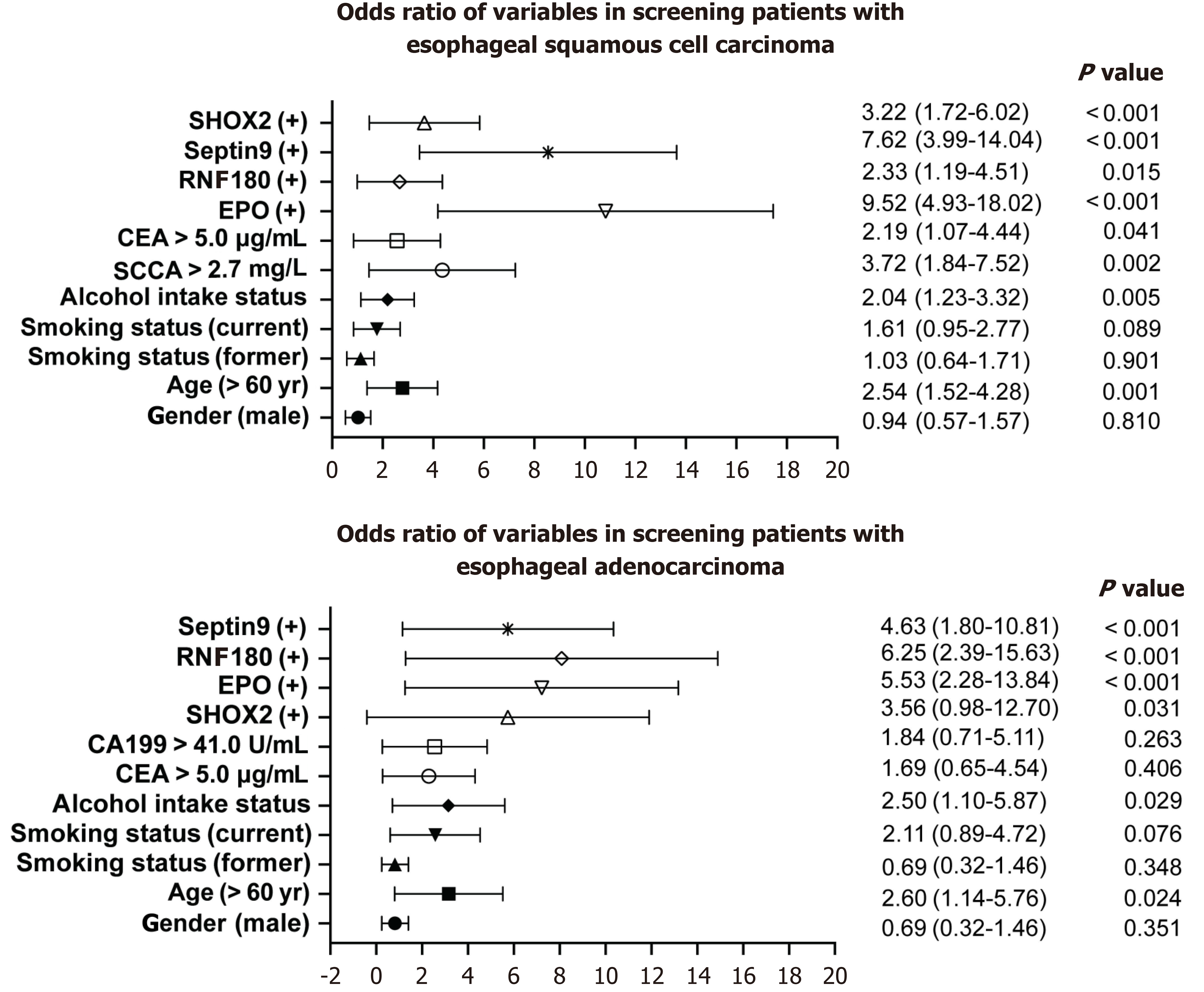
The multivariate logistic regression model revealed that methylation of the four genes [SHOX2 methylation: odds ratio (OR) = 3.22, 95%CI: 1.76–6.02; EPO methylation: OR = 9.52, 95%CI: 4.93–18.02; RNF180 methylation: OR = 2.33, 95%CI: 1.19–4.51; and SEPTIN9 methylation: OR = 7.62, 95%CI: 3.99–14.04), age (> 60 years: OR = 2.54, 95%CI: 1.52–4.28), smoking status (current smoker: OR = 1.61, 95%CI: 0.95–2.77), alcohol intake status (OR = 2.04, 95%CI: 1.23–3.32), high SCCA levels (> 2.7 mg/L: OR = 3.72, 95%CI: 1.84–7.52), and high CEA levels (> 5.0 μg/mL: OR = 2.19, 95%CI: 1.07–4.44)] were positively associated with an esophageal SCC risk (Figure 7). While the independent risk factors for adenocarcinoma were SHOX2 methylation (OR = 3.56, 95%CI: 0.98–12.7) EPO methylation (OR = 5.53, 95%CI: 2.28–13.84), RNF180 methylation (OR = 6.25, 95%CI: 2.39–15.63), SEPTIN9 methylation (OR = 4.63, 95%CI: 1.80–10.81), age (> 60 years: OR = 2.60, 95%CI: 1.14–5.76), and alcohol intake status (OR = 2.50, 95%CI: 1.10–5.87) (Figure 7). In addition, age-adjusted logistic regression analysis indicated a positive association between EC risk and EPO and SEPTIN9 methylation as well as SCCA levels (Table 2).
| Univariate analysis | Number of patients | OR (95%CI) | P value | |
| Gender | (Male vs female) | 210 | 0.65 (0.32-1.34) | 0.568 |
| Age at diagnosis | (≤ 60 vs > 60 years) | 210 | 2.32 (1.45-6.61) | 0.021 |
| SCCA | (≤ 3.5 vs > 3.5 U/mL) | 210 | 3.01 (1.54-6.45) | 0.023 |
| EPO methylation | (EPO− vs EPO+) | 210 | 7.89 (3.28-18.21) | 0.001 |
| SEPT9 methylation | (SEPT9− vs SEPT9+) | 210 | 7.15 (2.86-17.63) | 0.001 |
In this study, we employed two sets of models: The epidemiological model, which included four established predictors of EC (age, alcohol intake status, tumor markers, and ctDNA methylation), and the full model, which, through stepwise logistic regression, further incorporated two metabolic markers—SCCA and ctDNA methylation.
Esophagoscopy plays a crucial role in the early screening of EC[21]. However, EC diagnosis is often delayed due to occult symptoms in the early stages of tumor development. As esophagoscopy is not a routine option in normal physical examinations, and a significant number of patients with EC are diagnosed at advanced stages in China, leading to a poor prognosis[23], there is a critical need to identify an ideal screening method for EC that is cost-effective, well-tolerated, and applicable in primary care. Plasma samples offer an easily obtainable, less invasive, and highly acceptable alternative.
ctDNA methylation is a potentially valuable tumor marker for EC. Herein we explored the clinical significance of SHOX2, EPO, SEPTIN9, and RNF180 methylation for the early screening, clinical diagnosis, and monitoring of esophageal adenocarcinoma and SCC. Our results indicated that the combined analysis of SHOX2, SEPTIN9, RNF180, and EPO promoter methylation effectively distinguished esophageal adenocarcinoma and SCC, and EC and controls without any tumors. For EC, the area under the operating characteristic curve value of combined detection of methylation of the four genes was 0.864, which was significantly higher than those for SCCA, CA199, and CEA. For esophageal adenocarcinoma and SCC, the area under the operating characteristic curve values of the combined methylation detection were 0.737 and 0.824, respectively. Moreover, the sensitivity and specificity of the combined methylation detection for early EC diagnosis were 40% and 80%, respectively. Our study was the first study on ctDNA methylation in detection of early-stage disease. Moreover, the incorporation of SCCA, CA199, CEA, and ctDNA methylation as metabolic markers satisfactorily improved the discriminatory performance of ctDNA methylation alone (C-statistic for the full model, 0.735). As all the indicators in this model can be easily acquired from general clinical or screening assays, the potential for translation into practical use is considerable. Internal validation indicated that the model may perform well with regard to model discrimination when applied to other populations.
Clearly, the role of ctDNA methylation in screening early EC is superior (> 50%). A blood-based test could be an ideal screening platform for implementation in primary care settings. ctDNA methylation holds greater value in diagnosing advanced EC and shows promise for evaluating efficacy and monitoring disease progression. Plasma DNA methylation, utilized to predict the recurrence of EC, reflects transient changes in treatment response and tumor load, enabling accurate tumor staging and early diagnosis of recurrence. The combined detection of multiple methylation targets can enhance diagnostic performance in EC. The combined methylation of EPO, SHOX2, SEPTIN9, and RNF180, as jointly detected in our study, effectively distinguished EC and benign esophageal diseases from healthy subjects with high sensitivity and specificity. These promising results suggest the potential use of these DNA methylation markers in plasma to complement current EC detection methods and monitor EC in patients with primary sclerosing cholangitis. However, we found no significant differences in the methylation of SHOX2, EPO, SEPTIN9, and RNF180 between adenocarcinoma and SCC. It is presumable that the common four methylation markers may be applicable for all EC types, not limited to a specific subtype at a particular anatomic location.
Over the years, investigators have explored esophageal cell collection devices that can be deployed either via a catheter or swallowed by patients[24]. Unfortunately, the accuracy for detecting Barrett’s esophagus was deemed too low for clinical utility, primarily due to a low cell yield and reliance on standard cytology for identifying cell atypia[24-26]. Cytology samples can undergo additional testing, such as DNA methylation assays, so as to increase diagnostic accuracy and facilitate risk stratification[26,27]. In this study, the methylation status of EC cytology samples was positive, while that of tissue samples from benign esophageal diseases was negative. Combining cytology with DNA methylation assays appears to be superior to plasma methylation for early EC screening.
EC remains relatively concealed, necessitating patient selection for screening based on symptoms, family history, and other risk factors to enhance cost-effectiveness. It is imperative to conduct studies in relevant populations to avoid misleading estimates of sensitivity and specificity in detection. Monitoring ctDNA methylation in addition to traditional tumor markers seems effective for EC diagnosis and treatment. Herein the combined detection of methylation of EPO, SHOX2, SEPTIN9, and RNF180 could effectively distinguish EC and benign esophageal diseases. We believe that cytology combined with DNA methylation assays may prove superior for early EC screening.
We are grateful to the China Scholarship Council for generously sponsoring Dr. Jing Yu in pursuing studies abroad.
| 1. | Ma L, Li X, Wang M, Zhang Y, Wu J, He Y, Fan X, Zhang B, Zhou X. The Incidence, Mortality, and DALYs Trends Associated with Esophageal Cancer - China, 1990-2019. China CDC Wkly. 2022;4:956-961. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 2. | Zeng H, Chen W, Zheng R, Zhang S, Ji JS, Zou X, Xia C, Sun K, Yang Z, Li H, Wang N, Han R, Liu S, Li H, Mu H, He Y, Xu Y, Fu Z, Zhou Y, Jiang J, Yang Y, Chen J, Wei K, Fan D, Wang J, Fu F, Zhao D, Song G, Chen J, Jiang C, Zhou X, Gu X, Jin F, Li Q, Li Y, Wu T, Yan C, Dong J, Hua Z, Baade P, Bray F, Jemal A, Yu XQ, He J. Changing cancer survival in China during 2003-15: a pooled analysis of 17 population-based cancer registries. Lancet Glob Health. 2018;6:e555-e567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 795] [Cited by in RCA: 970] [Article Influence: 138.6] [Reference Citation Analysis (2)] |
| 3. | Domper Arnal MJ, Ferrández Arenas Á, Lanas Arbeloa Á. Esophageal cancer: Risk factors, screening and endoscopic treatment in Western and Eastern countries. World J Gastroenterol. 2015;21:7933-7943. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 554] [Cited by in RCA: 746] [Article Influence: 74.6] [Reference Citation Analysis (15)] |
| 4. | Li H, Teng Y, Yan X, Cao M, Yang F, He S, Zhang S, Li Q, Xia C, Li K, Chen W. Profiles and Findings of Population-Based Esophageal Cancer Screening With Endoscopy in China: Systematic Review and Meta-analysis. JMIR Public Health Surveill. 2023;9:e45360. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 5. | Gao QY, Fang JY. Early esophageal cancer screening in China. Best Pract Res Clin Gastroenterol. 2015;29:885-893. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 45] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 6. | Li R, Sun J, Wang T, Huang L, Wang S, Sun P, Yu C. Comparison of Secular Trends in Esophageal Cancer Mortality in China and Japan during 1990-2019: An Age-Period-Cohort Analysis. Int J Environ Res Public Health. 2022;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 7. | Shields MD, Chen K, Dutcher G, Patel I, Pellini B. Making the Rounds: Exploring the Role of Circulating Tumor DNA (ctDNA) in Non-Small Cell Lung Cancer. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 32] [Reference Citation Analysis (0)] |
| 8. | Pellini B, Szymanski J, Chin RI, Jones PA, Chaudhuri AA. Liquid Biopsies Using Circulating Tumor DNA in Non-Small Cell Lung Cancer. Thorac Surg Clin. 2020;30:165-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 9. | Nunes SP, Moreira-Barbosa C, Salta S, Palma de Sousa S, Pousa I, Oliveira J, Soares M, Rego L, Dias T, Rodrigues J, Antunes L, Henrique R, Jerónimo C. Cell-Free DNA Methylation of Selected Genes Allows for Early Detection of the Major Cancers in Women. Cancers (Basel). 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 53] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 10. | Nebgen DR, Lu KH, Bast RC Jr. Novel Approaches to Ovarian Cancer Screening. Curr Oncol Rep. 2019;21:75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 70] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 11. | Hoque MO, Feng Q, Toure P, Dem A, Critchlow CW, Hawes SE, Wood T, Jeronimo C, Rosenbaum E, Stern J, Yu M, Trink B, Kiviat NB, Sidransky D. Detection of aberrant methylation of four genes in plasma DNA for the detection of breast cancer. J Clin Oncol. 2006;24:4262-4269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 176] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 12. | Kneip C, Schmidt B, Seegebarth A, Weickmann S, Fleischhacker M, Liebenberg V, Field JK, Dietrich D. SHOX2 DNA methylation is a biomarker for the diagnosis of lung cancer in plasma. J Thorac Oncol. 2011;6:1632-1638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 190] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 13. | Vo TTL, Nguyen TN, Nguyen TT, Pham ATD, Vuong DL, Ta VT, Ho VS. SHOX2 methylation in Vietnamese patients with lung cancer. Mol Biol Rep. 2022;49:3413-3421. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 14. | Steinmann K, Richter AM, Dammann RH. Epigenetic silencing of erythropoietin in human cancers. Genes Cancer. 2011;2:65-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 15. | Ren M, Wang C, Sheng D, Shi Y, Jin M, Xu S. Methylation analysis of SHOX2 and RASSF1A in bronchoalveolar lavage fluid for early lung cancer diagnosis. Ann Diagn Pathol. 2017;27:57-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 16. | Zhou X, Lu X, Wu H, Liu J, Huang H. Diagnostic performance of SHOX2 promoter methylation as biomarker for lung cancer identification: A meta-analysis update. Thorac Cancer. 2021;12:3327-3332. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 17. | Li Q, Jiang W, Zhang Y, Yang X, Huang T, Huang Y, Yang S, Wang Q. Methylation of Septin9, SRSF1, and PAX8 in Early Screening of Colorectal Cancer in the Population Undergoing Physical Examinations. Clin Lab. 2023;69. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 18. | Imperiale TF, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, Lidgard GP, Ahlquist DA, Berger BM. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;370:1287-1297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1015] [Cited by in RCA: 1236] [Article Influence: 112.4] [Reference Citation Analysis (1)] |
| 19. | Zhao L, Liu Y, Zhang S, Li M. Plasma Methylated RNF180 for Noninvasive Diagnosis of Gastric Cancer. Biomed Res Int. 2022;2022:6548945. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Reference Citation Analysis (0)] |
| 20. | Nie Y, Gao X, Cai X, Wu Z, Liang Q, Xu G, Liu N, Gao P, Deng J, Xu H, Shen Z, Cao C, Chen F, Zhang N, Song Y, Sun M, Liu C, Zhou G, Han W, Dou J, Xie H, Yao L, Liu Z, Ji G, Wang X, Zhao Q, Shang L, Fan D, Han X, Ren J, Liang H, Wang Z, Wang J, Wu Q, Yu J, Wu K; MAGIS Study Group. Combining methylated SEPTIN9 and RNF180 plasma markers for diagnosis and early detection of gastric cancer. Cancer Commun (Lond). 2023;43:1275-1279. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 4] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 21. | Kim JA, Shah PM. Screening and prevention strategies and endoscopic management of early esophageal cancer. Chin Clin Oncol. 2017;6:50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 22. | Zhang L, Yang X, Tian Y, Yu Q, Zhou D, Wu Z, Zhao X. Noninvasive Detection of Esophageal Cancer by the Combination of mSEPT9 and SNCG. Genet Test Mol Biomarkers. 2022;26:8-16. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 23. | Zhu H, Ma X, Ye T, Wang H, Wang Z, Liu Q, Zhao K. Esophageal cancer in China: Practice and research in the new era. Int J Cancer. 2023;152:1741-1751. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 96] [Article Influence: 48.0] [Reference Citation Analysis (0)] |
| 24. | Sharma P. Diagnostic Testing for Barrett Esophagus. Gastroenterol Hepatol (N Y). 2020;16:92-94. [PubMed] |
| 25. | Codipilly DC, Iyer PG. Novel Screening Tests for Barrett's Esophagus. Curr Gastroenterol Rep. 2019;21:42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 26. | Krishna Chandar A, Sharma A, Chak A. Novel Screening Alternatives for Barrett Esophagus. Gastroenterol Hepatol (N Y). 2020;16:238-245. [PubMed] |
| 27. | Raut JR, Guan Z, Schrotz-King P, Brenner H. Whole-blood DNA Methylation Markers for Risk Stratification in Colorectal Cancer Screening: A Systematic Review. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |