Published online Jan 28, 2024. doi: 10.3748/wjg.v30.i4.381
Peer-review started: October 31, 2023
First decision: November 24, 2023
Revised: December 5, 2023
Accepted: January 10, 2024
Article in press: January 10, 2024
Published online: January 28, 2024
Processing time: 86 Days and 21.4 Hours
Radiomics is a promising tool that may increase the value of magnetic resonance imaging (MRI) for different tasks related to the management of patients with hepatocellular carcinoma (HCC). However, its implementation in clinical practice is still far, with many issues related to the methodological quality of radiomic studies.
To systematically review the current status of MRI radiomic studies concerning HCC using the Radiomics Quality Score (RQS).
A systematic literature search of PubMed, Google Scholar, and Web of Science databases was performed to identify original articles focusing on the use of MRI radiomics for HCC management published between 2017 and 2023. The methodological quality of radiomic studies was assessed using the RQS tool. Spearman’s correlation (ρ) analysis was performed to explore if RQS was correlated with journal metrics and characteristics of the studies. The level of statistical signi-ficance was set at P < 0.05.
One hundred and twenty-seven articles were included, of which 43 focused on HCC prognosis, 39 on prediction of pathological findings, 16 on prediction of the expression of molecular markers outcomes, 18 had a diagnostic purpose, and 11 had multiple purposes. The mean RQS was 8 ± 6.22, and the corresponding percentage was 24.15% ± 15.25% (ranging from 0.0% to 58.33%). RQS was positively correlated with journal impact factor (IF; ρ = 0.36, P = 2.98 × 10-5), 5-years IF (ρ = 0.33, P = 1.56 × 10-4), number of patients included in the study (ρ = 0.51, P < 9.37 × 10-10) and number of radiomics features extracted in the study (ρ = 0.59, P < 4.59 × 10-13), and time of publication (ρ = -0.23, P < 0.0072).
Although MRI radiomics in HCC represents a promising tool to develop adequate personalized treatment as a noninvasive approach in HCC patients, our study revealed that studies in this field still lack the quality required to allow its introduction into clinical practice.
Core Tip: This systematic review aimed at evaluating the status of magnetic resonance imaging (MRI) radiomic studies related to hepatocellular carcinoma (HCC) using the Radiomics Quality Score (RQS) to assess methodological quality. A systematic literature search identified 127 articles covering various steps of HCC management. The mean RQS was 8 ± 6.22, with significant variation. RQS was significantly correlated with journal impact factor (IF), 5-year IF, the number of patients involved, the number of radiomic features extracted, and the publication year. Despite the potential of MRI radiomics in HCC, its clinical implementation is hindered by a lack of quality in studies in this field.
- Citation: Brancato V, Cerrone M, Garbino N, Salvatore M, Cavaliere C. Current status of magnetic resonance imaging radiomics in hepatocellular carcinoma: A quantitative review with Radiomics Quality Score. World J Gastroenterol 2024; 30(4): 381-417
- URL: https://www.wjgnet.com/1007-9327/full/v30/i4/381.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i4.381
Medical imaging has progressed over the last few decades from a simple diagnostic tool for diseases to a massive supply of quantitative data free of the normal subjective interpretation that characterizes conventional clinical practice. The introduction of technological advances and the quest for precision medicine have given rise to a new potential branch of research known as "radiomics". Radiomics is a quantitative technique that turns digitized medical pictures into high-dimensional mineable features that may be correlated with clinical endpoints such as pathological findings, treatment response, and survival. Radiomics can also be integrated with other quantitative data, such as genomics and pathomics data, to provide a comprehensive approach to disease[1-4]. As a quantitative analysis of digital images, radiomics has the potential to reveal specific disease characteristics that are otherwise inaccessible to the naked eye using conventional imaging modalities. This method may increase the quantity of clinically relevant data that may be extracted from medical images, offering the possibility of discovering innovative imaging biomarkers for the diagnosis, characterization, and prediction of outcomes in a wide range of diseases, including oncologic diseases[5]. In the field of oncology, the rationale behind radiomics is that biological tumor characteristics might be mirrored by quantifying medical image heterogeneity using extracted radiomic features, encompassing aspects of tumor progression, response to therapeutic interventions, and clinical outcomes. Quantitative imaging has garnered significant interest in the non-invasive detection of tumor heterogeneity, and recent radiomics studies across various oncological fields have shown a strong association between imaging heterogeneity and the characteristics of solid tumors[6].
Hepatocellular carcinoma (HCC) is one of the leading causes of cancer-related deaths worldwide and poses serious challenges for screening, early diagnosis and treatment firstly because most HCC is diagnosed at an advanced stage when curative treatment options are limited, and also because of its complex heterogeneity at multiple levels: heterogeneity between tumor nodules from the same patient (intertumor heterogeneity), within the same tumor nodule (intratumor heterogeneity) and between patients (interpatient heterogeneity)[7,8]. Furthermore, current clinical practice based on single bioptic or tumor tissue section fails to discover useful biomarkers, and many existing staging systems for HCC are based on postoperative pathological examinations, which cannot aid in preoperative decision-making[9]. In contrast to numerous other solid tumors, HCC can be diagnosed by using distinctive enhancement patterns on dynamic multiphasic CT or magnetic resonance imaging (MRI), without additional histopathologic confirmation[10,11]. Although imaging plays an important role in the screening, early identification, and management of HCC patients, the imaging evaluation of HCC is still based on subjective interpretation of qualitative imaging descriptors and tumor size estimate, both of which are prone to variability[10,12,13]. Of note, although CT is more generally available, faster, and needs less exper-ience to administer and interpret pictures than MRI, its downsides include radiation exposure and low soft tissue contrast, which demands the use of iodinated contrast agents. The increased soft tissue contrast of MRI, on the other hand, enables for the examination of a range of tissue features that may be relevant in HCC therapy[14,15]. In this context, recent advantages in MRI radiomics can potentially address the urgent need for noninvasive, radiation-free strategies that can aid in the early detection of HCC and preoperative prediction of tumor behavior, as well as address the inherent variability of qualitative imaging descriptors and provide previously unavailable information to obtain a better stratification of HCC patients for a more precise treatment decision making.
Over the last decade, there has been a significant increase in radiomics studies in the field of HCC. Many of these studies have demonstrated the effectiveness of radiomic features for differential diagnosis, grading, predicting microvascular invasion, overall survival, recurrence, and treatment response[16-19]. Nevertheless, radiomics is presently limited to academic literature in the context of HCC, as physicians question its utility due to the absence of a translation from research studies to clinical application. This is attributed, at least in part, to the overall deficiency of streamlined and productive methods for integrating imaging biomarkers into clinical practice[20-22]. Lambin et al[2] developed the Radiomics Quality Score (RQS) to provide a standardized evaluation of the radiomics performance, reproducibility, and clinical. The RQS metric system determines the validity and comprehensiveness of radiomics investigations. This tool is modality-independent tool and was designed to assess the methodological quality of radiomics studies. The methodology and analyses of a radiomics study are evaluated based on 16 criteria that reward or penalize, promoting the best scientific practice[2]. Recent research tried to examine the current state of the art in HCC radiomics, stressing the major concepts, clinical applications, and limitations[23-25]. However, it is clear from these research that the bulk of radiomic investigations on HCC have been conducted on CT, with only a few looking into MRI. Furthermore, the quality of science and reporting in HCC MRI radiomics research investigations is mainly unknown.
Hence, the objective of this study was to provide a comprehensive overview of the existing state of MRI radiomic investigations related to HCC. Simultaneously, we aimed to evaluate the methodological quality of each study using the RQS to assess the radiomics analyses conducted in prior publications. The study’s goal is to promote the quality of MRI radiomics research studies in HCC as a diagnostic, prognostic, and/or predictive tool, to allow radiomics to become an appropriate medical decision-making tool by facilitating the combined analysis of clinical data and high-throughput imaging features, while taking advantage of the benefits arising from the MRI technique.
A systematic search was conducted for all published studies exploring the role of MRI radiomics in the field of HCC. PubMed, Web of Science and Google Scholar electronic databases were comprehensively explored and used to build the search. Only studies published in the last six years were selected. The last search was performed on June 1, 2023. The search terms consisted in: (“radiomics” OR “texture” OR “histogram”) AND (“MRI” OR “Magnetic Resonance Imaging”) AND (“Hepatocellular Carcinoma” OR “HCC”). The literature search was limited to English language publications and studies of human subjects. Two reviewers, after having independently screened the identified titles and abstracts, assessed the full text of articles aiming at exploring MRI radiomics in the field of HCC and that were not review articles. For articles meeting these criteria with full text available, the following further selection criteria had to be fulfilled: Involvement of adult patients (age > 18 years); involvement of patients with HCC confirmed by pathology and/or surgery and/or overall analysis combined with medical history, clinical symptoms, and imaging data; presence of information about MRI protocol. Moreover, studies were excluded if they performed analyses on mixed patients (e.g., groups of patients with multiple hepatic malignant diseases) that did not allow conclusions to be drawn only about HCC patients; if they did not evaluate an outcome measure; if they were focused only on semantic imaging features (radiologist-dependent). After selecting the studies that met the inclusion and exclusion criteria, reference lists of these studies were also searched in order to recruit any potential eligible studies. In addition, pre-existing reviews/systematic reviews/meta-analyses were also searched in order to recruit any other potentially eligible studies from their reference lists.
After the above-mentioned selection procedure, selected articles were analysed by two reviewers, and data useful for conducting the systematic review were collected in a predesigned sheet. Extracted data will include the following: first author name, publication year, Journal name, scientometric indexes [impact factor (IF), 5-years IF, CiteScore, H-index, first author IF with and without self-citations], study design, in particular prospective/retrospective, clinical purpose, specific output measured in the study, number and type of patients, imaging modalities used for radiomic feature extraction, information on region of interest (ROI)/volume of interest (VOI) placement (segmentation technique and ROI/VOI type), software used for radiomic feature extraction, number and features type, feature selection methods (if used), classification methods, information on if models were applied to a separate dataset, highest accuracy/most important results and main findings.
This systematic review was conducted according to the PRISMA statement[26].
The methodological quality of each radiomics study was assessed by two reviewers using the RQS tool[2]. The assessment was performed independently, and any disagreement was resolved by consensus. RQS tool is composed of 16 items structured to assess various crucial steps in the workflow of radiomics analyses (see Supplementary Table 1). In particular, a maximum of 36 points can be assigned to each study: up to 2 points for the first RQS checkpoint (a single item, namely “Image protocol quality”), up to 3 points for the second RQS checkpoint (3 items, specifically on multiple segmentation strategies, the use of phantoms and multiple imaging time points) and up to 31 points for the third RQS checkpoint (12 items, encompassing feature extraction, exploratory analysis design as well as model building and validation). The total score ranges between −8 and 36 and can be translated into a final 0–100 RQS percentage, with −8 to 0 defined as 0%, indicating the lowest quality, and 36 as 100%, indicating the highest quality in terms of the methodology and reporting standards of the radiomics study[2].
Spearman’s correlation (ρ) analysis was performed to explore if there was a correlation between RQS and journal metrics, comprising IF of the journal at the year of publication, 5-year IF, CiteScore, and H-index at the year of publication. Additionally, Spearman’s correlation was used to explore the correlation between RQS and H-index of the first author at the year of publication of the study (both with and without self-citations), time of publication (calculated as time between the publication date and the date of last literature research, in months), as well as the association with the number of patients involved and the number of radiomic features extracted in the study. Finally, to explore if there was a difference in RQS according to clinical purpose of the study, a subgroup analysis using Kruskal-Wallis H test was performed. In case of significance, Wilcoxon rank-sum post hoc tests with Bonferroni correction were carried out on each pair of groups. The significance level was set at 0.05. All statistical analysis was performed using SPSS (version 27).
A total of 537 articles were identified from scientific electronic scientific databases. Only 211 articles were retained after the removal of duplicates.
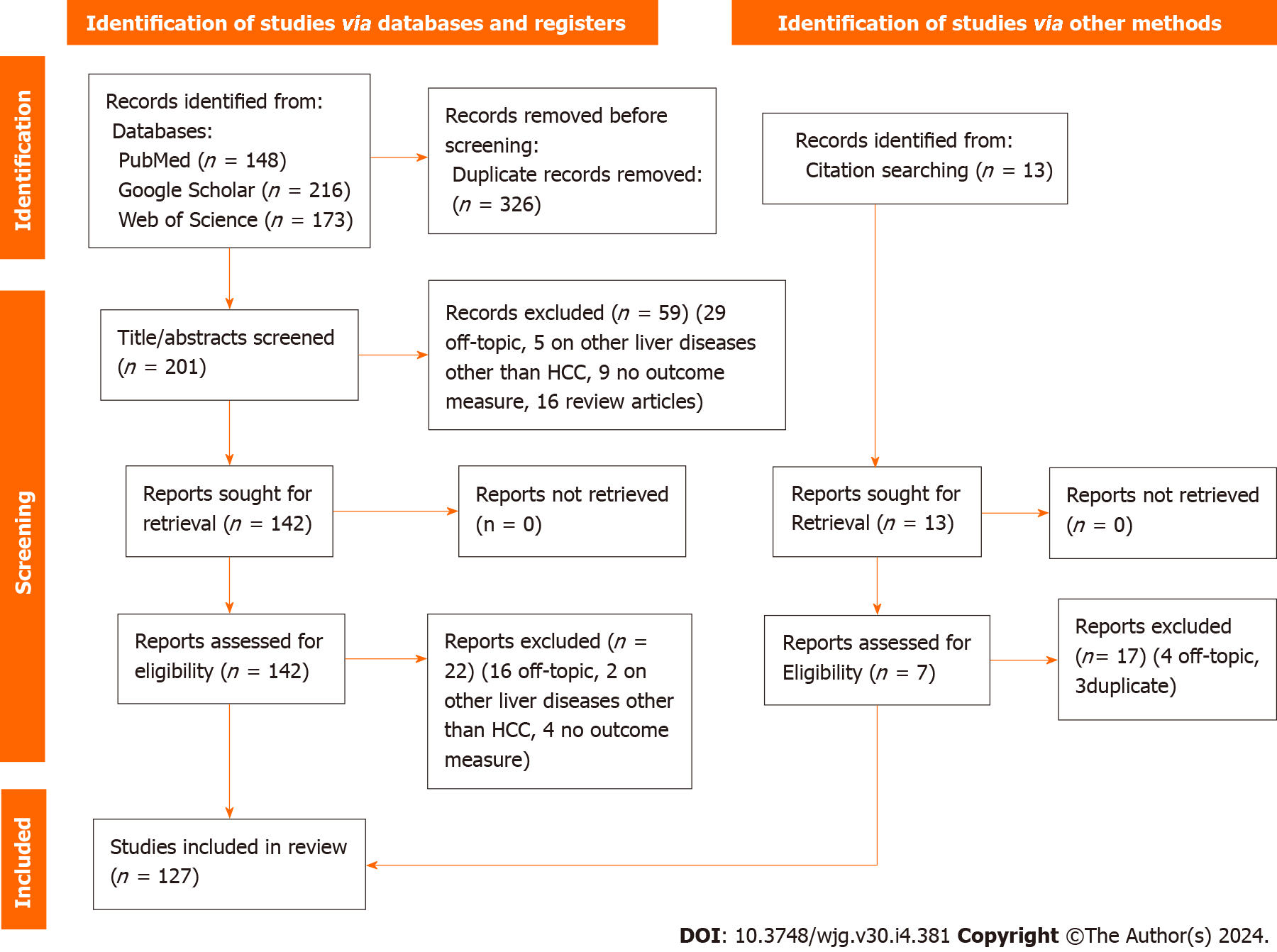
We reviewed the titles and abstracts of these records, excluding 59 due to non-compliance with inclusion criteria (29 unrelated to the topic, 16 were reviews, 5 conducted analyses on mixed patients, and 9 did not assess an outcome measure). The full text of 149 articles was assessed, leading to the exclusion of 16 off-topic articles. Additionally, four studies were excluded for not evaluating an outcome measure, and two for analyzing mixed patients. Thirteen more articles were found through references in selected articles or existing reviews/systematic reviews/meta-analyses, and seven of these were incorporated into the review. A total of 127 data sets were included in the review. Figure 1 shows the PRISMA flow diagram of the included studies based on the inclusion and exclusion criteria.
The details regarding the characteristics of the 127 studies chosen for this review are presented in Table 1. Approximately half of these studies (51 out of 127) were published in the last two years, and only 9 studies deviated from a retrospective design. Most of the selected studies (43 out of 127) explored radiomic approaches for HCC prognosis after surgical, radiofrequency ablation and/or trans-arterial chemo embolization treatment. Forty studies investigated the ability of radiomics in predicting pathological findings [e.g., microvascular invasion (MVI), vessels encapsulating tumor clusters, histologic grade], of which 27 aimed at investigating the performance of radiomics analysis for MVI prediction. Sixteen studies aimed at exploring if MRI radiomics could infer the expression of molecular markers (e.g., CK19, Ki67, GPC3) outcomes. Among the remaining studies, 24/127 aimed to evaluate the power of radiomics for distinguishing HCC from other solid hepatic lesions, while 11 had multiple aims.
| Ref. | ST | CP | Specific outcome | NP (type) | Modalities used for feature extraction | Seg | Software used for feature extraction | Features number (type) | FS | CM | Model applied to a separate dataset? | Most important result | Main findings |
| Liu et al[42], 2023 | R | PPF | MVI | 104 (HCC) | T2WI | M, 3D | AK SOFTWARE | 851 (first order, shape, GLCM, GLSZM, GLRLM, NGTDM, and GLDM) | LASSO, LR | LR | Yes | AUC = 0.867 in the TS, 0.820 in the VS | A prediction model using radiomic features from single T2WI can predict MVI in HCC |
| Wang et al[43], 2023 | R | PR | LRT | 100 (HCC) | AP, PVP, T2WI | M, 3D | 3D SLICER | 851 (first-order, shape, GLCM, GLDM, GLSZM, GLRLM, NGTDM and wavelet) | t-test/Mann Whitney, LASSO | ROC | Yes | AUC = 0.867 | MRI-based radiomics analysis may serve as a promising and noninvasive tool to predict outcome of locoregional treatment in HCC patients |
| Gong et al[44], 2023 | R | MC | PD-1/PD-L1 | 108 (HCC) | T2WI FS, AP, PVP | M, 3D | NS | 352 (GLCM, GLRLM, intensity histogram, and shape) | ICC, t-test/ MANN WHYTNEY, LASSO | LR | Yes | AUC = 0.946 in the TS and 0.815 in the VS | A radiomics model based on multisequence MRI has the potential to predict the preoperative expression of PD-1 and PD-L1 in HCC |
| Zhang et al[45], 2023 | R | MC | CK 19+/-HCC | 311 (HCC) | T1WI, T2WI, DWI, AP, VP, and DP | M, 3D | uRP | 2286 (first order, wavelet) | ICC, LASSO | LR | Yes | in the TS (C-index, 0.914), internal (C-index, 0.855), and external VS (C-index, 0.795) | The combined model based on clinic-radiological radiomics features can be used for predicting CK19+ HCC preoperatively |
| Zhang et al[46], 2023 | R | PPF | MTM HCC | 232 (HCC) | DCE-MRI | M, 3D | Pyradiomics | 1037 (first order, shape GLRLM, GLSZM, NGTDM, GLCM, GLDM LoG and wavelet) | ICC, GBDT | LR, KNN, Naive-Bayes, Decision Tree, SVM | Yes | AUCs of 0.896 and 0.805 in the TS e VS | The nomogram containing radiomics, age, alpha-fetoprotein, tumour size, and tumour-to-liver ADC ratio revealed excellent predictive ability in preoperatively identifying the MTM-HCC Subtype |
| Dong et al[47], 2024 | R | D, PR | VETC | 221 (HCC) | DCE-MRI | M, 3D | Pyradiomics | 1218 (FIRST ORDER) | ICC | LR, decision tree, RF, SVM, KNN, and Bayes | Yes | AUC = 0.844 | The DLR model provides a noninvasive method to discriminate VETC status and prognosis of HCC patients preoperatively |
| Tabari et al[48], 2023 | R | PPF | Pre-ablation tumor radiomics | 97 (HCC) | AP, DCE-MRI | M, 3D | NS | 112 first-order, (GLCM, GLDM, GLRLM, GLSZM, NGTDM) | mRMR | RF | Yes | AUC = 0.83 | Pre-ablation MRI radiomics could act as a valuable imaging biomarker for the prediction of tumor pathologic response in patients with HCC |
| Cao et al[49], 2023 | R | PR | RFS | 249 (HCC) | T2WI FS, T1WI FS, DCE-MRI | M, 3D | Pyradiomics | NS (first-order, shape, and texture, wavelet, Laplacian) | LASSO | Cox regression | Yes | C-index = 0.893 TS, 0.851 (test set), 0.797 (external) | The combined radiomic model provides superior ability to discern the possibility of recurrence-free survival in HCC over the total radiomic and the clinical–radiological models |
| İnce et al[50], 2023 | R | PPF | TARE | 82 (HCC) | DCE-MRI | S, 3D | Pyradiomics | 1128 (first-order, GLCM, GLDM, GLRLM, GLSZM, and NGTDM) | ICC, PCA, SFS | SVM, LR, RM, LightGBM | No | AUC = 0.94 | Machine learning–based clinicoradiomic models demonstrated potential to predict response to TARE |
| Chen et al[51], 2023 | R | PR | TACE | 144 (HCC) | T2WI, AP, PVP, DP | M, 3D | Pyradiomics | 110 (NS) | mRMR, LASSO, DNN | SVM, LR | Yes | AUC = 0.974 | DNN model performs better than other classifiers in predicting TACE response. Integrating with clinically significant factors, the CD model may be valuable in pre-treatment counseling of HCC patients who may benefit the most from TACE intervention |
| Jiang et al[52], 2023 | R | PPF | MVI | 102 (HCC) | T1_in, T1_A, T2W, DWI | M, 3D | Pyradiomics | 1967 (first-order, shapes, textures, GLCM, GLSZM, GLDM, GLRLM, and filter-transformed) | LASSO | ULR | Yes | AUC = 0.901, 0.923 for TS and VS | The multiparametric MRI-based radiomics nomogram is a promising tool for the preoperative diagnosis of peritumoral MVI in HCCs |
| Hu et al[53], 2023 | R | D, MC | CK19+ | 110 (HCC) | AP, VP, HBP | M, 3D | PyRadiomics | 1130 (shape, first order, GLCOM, GLRLM, GLSZ, GLDM) | ICC | RFE | No | AUC = 0.92 | The established radiomics signature based on preoperative gadoxetic acid-enhanced MRI could be an accurate and potential imaging biomarker for HCC CK19 (+) prediction |
| Chong et al[54], 2023 | R | MC, PR | Glypican 3-Positive HCC | 259 (HCC) | T2WI, DWI, PRE, AP, PVP, TP and HBP | M, 3D | PyRadiomics | 749 (first order statistics, shape and size) and textural property types (GLSZM, GLCM, GLDM, GLRLM, and NGTDM) | Test-retest procedure, ICC, LASSO, RF, SVM | LR, RF, SVM | Yes | AUC = 0.943 vs 0.931 TS and VS respectively | Preoperative EOB-MRI radiomics-based nomogram satisfactorily distinguished GPC3 status and outcomes of solitary HCC 5 cm |
| Hu et al[55], 2023 | R | D, PPF | Functional liver reserve | 403 (HCC) | DCE MRI | M, 3D | Pyradiomics | 851 (shape, first-order GLCM, GLRLM, GLSZ, GLDM, NGTDM, wavelet) | ICC, Spearman’s correlation | LR, SVM | No | AUC = 0.71 | A radiomics model based on gadoxetic acid-enhanced MRI was constructed in this study to discriminate HCC with different histopathologic grades |
| Tao Y et al[56], 2023 | R | MC | PD-L2 | 108 (HCC) | T2WI, AP, PV | M, 3D | R | 1130 | ICC, LASSO | ROC | No | AUC = 0.871 | Prediction based on the radiomic characteristics of MRI could noninvasively predict the expression of PD-L2 in HCC |
| Yang et al[57], 2022 | R | PR | ER | 181 (HCC) | T1WI, T2WI | M, 3D | LIFEx | 34 (Histogram, Shape) | LASSO | ROC | Yes | AUC = 0.79 | The model for early recurrence of HCC after ablation based on the clinical, imaging, and radiomics features presented good predictive performance |
| Liu et al[58], 2023 | R | PPF | MVI | 161 (HCC) | AP, PVP, DP | M, 3D | 3D Slicer, Pyradiomics | 321 (shape, first-order histogram, GLCM, GLDM, GLRLM, GLSZM, NGTDM) | LASSO, ICC | LR | Yes | AUC = 0.87 | The nomogram model can effectively predict MVI in patients with HCC |
| Zhang et al[59], 2022 | R | PPF | MVI | 189 (HCC) | HBP | M, 3D | IBEX SOFTWARE | 1768 | LASSO, ICC | nomogram | Yes | AUC = 0.884 | Depending on the clinicoradiological factors and radiological features, nomograms can effectively predict MVI status in HCC patients |
| Sim et al[60], 2022 | R | PPF | MVI | 50 (HCC) | T1 AP, T1PVP | M, 2D | MaZda | 290 (area, histogram, gradient, GLCM, GLRLM, autoregressive, and wavelet) | Mutual Information, recursive pruning | SVM | No | Accuracy = 0.878 | Texture analysis of tumours on pre-operative MRI can predict presence of MVI in HCC |
| Zhang et al[61], 2022 | R | PR | RFA, ER | 90 (HCC) | T1WI, T2WI, CE-MRI | M, 2D | AK Software | 1316 (first-order histogram, shape, texture, GLCM, GLRLM, GLSZM, NGTDM, GLDM, and local binary pattern, high-order, and wavelet) | ANOVA | RF, LASSO | Yes | AUC of 0.822 in the TS and 0.812 in the VS | The multi-parametric MRI-based radiomics nomogram has a high predictive value for ER of small HCC after RFA |
| Zhao et al[62], 2023 | R | PR | HAIC | 112 (HCC) | T2WI | M, 3D | AK software | 396 (histogram, form factor, texture, GLZSM, GLCM, GLRLM, and Haralick) | LASSO | ROC | Yes | Accuracy = 0.81 | The nomogram based on the combined model consisting of MRI radiomics and ALBI score could be used as a biomarker to predict the therapeutic response of unresectable HCC after HAIC |
| Lu et al[63], 2022 | R | PPF | MVI | 165 (HCC) | T2WI, DWI (b = 800 s/mm2), T1WI, AP, PP, TP, and HBP | M, 3D | Pyradiomics | 1227 (shape, first-order, texture, GLSZM, GLRLM, GLCM, NGTDM, and GLDM) | LASSO | multivariate LR | Yes | AUC = 0.826 | The combined model based on radiomics features of Gd-EOB-DTPA enhanced MRI, tumour margin, and peritumoural hypointensity was valuable for predicting HCC MVI |
| Yang et al[64], 2022 | R | PPF | MVI | 110 (HCC) | DCE-MRI | M, 3D | A.K. Software | 11 (Grey Histogram, GLCM) | NO | ROC | No | AUC = 0.797 | The combination of MR image features and texture analysis may improve the efficiency in prediction of MVI |
| Ameli et al[65], 2022 | R | D | Degree of tumor differentiation | 129 (HCC) | ADC, VE MAPS | S, 3D | MATLAB R2017B | 95 (global, histogram, GLCM, GLRLM, GLSZM, NGTDM) | multi-class classification algorithm | RF | Yes | AUC = 0.832 | The addition of radiomics-based texture analysis improved HCC grading over that of ADC or venous enhancement values alone |
| Li et al[66], 2022 | R | PR | ER | 302 (HCC) | T2WI, DWI (800 s/mm2), AP, and PVP | M, 3D | Pyradiomics | 853 (shape, first order, texture, and wavelet) | SPSS, LASSO, ICC | ROC | Yes | AUCs of 0.91 and 0.87 in the TS and VS | The proposed predictive model incorporating clinico-radiological factors and the fusion radiomics signature derived from multiparametric MR images may be an effective tool for the individualized prediction of postoperative ER in patients with HCC |
| Zeng et al[67], 2022 | R | PPF | BETA-CATENIN MUTATION | 98 (HCC) | AP, PVP, DP, HBP | M, 3D | Pyradiomics | 1674 (first order, GLCOM, GLSZM, GLRLM, GLDM) | T-test, fisher's exact test | LSVC | Yes | AUC = 0.86 | The RHBP radiomics model may be used as an effective model indicative of HCCs with b-catenin mutation preoperatively |
| Aujay et al[68], 2022 | R | PR | TARE | 22 (HCC) | AP, PVP | M, 3D | Pyradiomics | 107 (Shape, first- and second- order) | Mann-Whitney U test | LR | No | AUC = 0.92 | Radiomics could aid in the prediction of early treatment response following TARE in patients with HCC |
| Chen et al[69], 2022 | R | PPF | MVI | 415 (HCC) | T1WI, T2WI, DWI, AP, PVP, HBP | M, 3D | R | 1409 (First order, shape, two order texture, Laplacian, wavelet, logarithmic, and exponential filters) | LASSO | SVM, XGBoost, RF, LR | Yes | AUC = 0.979 | Machine learning with an LR classifier yielded the best radiomics score for HBP and DWI. The radiomics nomogram developed as a noninvasive preoperative prediction method showed favorable predictive accuracy for evaluating MVI in sHCC |
| Wu et al[70], 2023 | R | D | DP-HCC | 179 (DPHCC, non DPHCC) | DCE-MRI | M, 3D | PyRadiomics | 1781 (first-order statistics, shape, and texture) | PCC, RFE | SVM, LR, LR-LASSO | Yes | AUC = 0.908 | MRI radiomics models may be useful for discriminating DPHCC from non-DPHCC before surgery |
| Li et al[71], 2022 | R | PPF | MVI | 113 (HCC) | T2WI, T1WI, DCE MRI | M, 2D | MaZda | 101 (histogram, GLCOM, GLRLM) | t-test, Mann-whitney U test | ROC | No | AUC = 0.939 | Noninvasive MRI radiomic model based on MDF values and imaging biomarkers may be useful to make preoperative prediction of MVI in patients with primary HCC |
| Wang et al[72], 2022 | R | PR | ER | 190 (HCC) | T2WI, T2WI FS, DCE MRI | M, 3D | PyRadiomics | 1316 (first-order histogram, texture, shape, GLZSM, GLRLM, GLCM, GLDM, and NGTDM, wavelet, local binary pattern, and Laplacian of Gaussian) | ICC, LASSO | LASSO, ICC, LR | Yes | AUC = 0.90 | The predictive model incorporated the clinical–radiological risk factors and radiomics features that could adequately predict the individualized ER risk in patients with solitary HCC ≤ 5 cm |
| Zhang et al[73], 2023 | P | PPF | MVI | 602 (HCC) | T1WI, T2WI, AP, VP, HBP and ADC | M, 3D | Radcloud platform | 1409 (First order, second order, shape, texture) | LASSO | LR, RF, SVM | Yes | AUC = 0.824 E 0.821 in the TS and VS | The combination of clinicoradiological factors and fusion radiomics signature of AP and VP images based on Gd-EOB-DTPA-enhanced MRI can effectively predict MVI |
| Brancato et al[74], 2022 | R | PPF | IABR | 38 (HCC) | T2WI, DCE-MRI | M, 3D | Pyradiomics | 386 (shape, first-order, and texture) | correlation filter, Wilcoxon-rank sum test, MI | LR | No | AUC = 0.96 | Radiomics MRI based on T2 and DCE-MRI revealed promising results concerning both HCC detection and grading |
| Fan et al[75], 2022 | R | PR | VEGF | 202 (HCC) | AP, PV, HBP, BP, DP | M, 3D | PyRadiomics | 1906 (first order, shape) | ICC, ANOVA | LR | Yes | AUC = 0.892 in the TS, 0.800 in the VS | The combined model acquired from Gd-EOB-DTPA enhanced MRI could be considered as a credible prognostic marker for the level of VEGF in HCC |
| Gao et al[76], 2022 | R | PPF | MVI | 115 (HCC) | T2WI, T1WI, AP, PVP, DP, and HBP | M, 3D | Pyradiomics | 107 (shape, first-order, and textural) | LR, SVC, RFC, and AdaBoost | LR | Yes | AUCs of 0.866 in the TS and 0.855 in the VS | The fusion model of multi-region radiomics achieves an enhanced prediction of the individualized risk estimation of MVI in HCC patients |
| Hu et al[77], 2022 | R | PPF | MVI | 501 (HCC) | T1WI, AP, PVP, HBP | M, 3D | Pyradiomics | 2600 (first order, shape, GLCM, GLRLM, GLSZM, GLDM and NGTDM) | LASSO | ROC | Yes | AUC = 0.962 | The radiomics signatures of the dual regions for tumor and peritumor on AP and PVP images are of significance to predict MVI |
| He et al[78], 2022 | R | PR | DFS, OS | 103 (HCC) | DCE MRI | M, 2D | AK software | 1217 (First order, Morphological, GLCM, GLRLM, GLSZM, GLDM, LOG) | ICC, Lasso, cox regression | LASSO | Yes | AUC = 0.884 | Multimodal radiomics models can serve as effective visual tools for predicting prognosis in patients with liver cancer |
| Ren et al[79], 2023 | R | PR | HCC grade | 270 (HCC) | T2WI | M, 3D | Pyradiomics | 1197 (first-order and shape, GLCM, GLRLM, GLRM, and spatial gray scale corre-lation matrix) | MIC, Spearman’s correlation, LR | LR | Yes | AUC = 0.864 | The clinical–radiomics model integrating radiomics features and clinical factors can improve recurrence predictions beyond predictions made using clinical factors or radiomics features alone |
| Luo et al[80], 2022 | R | PR | TACE | 61 (HCC) | T1WI, T1WI AP, T1WI PP, T2WI, DWI (b = 800), ADC | M, 3D | Pyradiomics | 1782 (shape, GLCM, GLRLM, GLSZM, NGTDM) | RF, single cox regression | ROC | No | AUC = 0.71 | Radiomic signatures derived from pretreatment MRIs could predict response to combined Lenvatinib and TACE therapy. Furthermore, it can increase the accuracy of a combined model for predicting disease progression |
| Wang et al[81], 2022 | R | PPF | MVI | 113 (HCC) | AP | M, 3D | MATLAB | 12 (first order) | NO | Mann-Whitney U test, LR | No | AUC = 0.741 | Peritumoral AP enhanced degree on MRI showed an encouraging predictive performance for preoperative prediction of MVI |
| Mao et al[82], 2022 | R | PPF | HCC GRADE | 122 (HCC) | T2WI (AP, HBP phases) | M, 3D | Image Analyzer | 121 (histogram, shape, texture, GLRLM and GLCM) | ICC | ANN, LR | Yes | AUC = 0.889 | Prediction models consisting of clinical parameters and Gd-EOB-DTPA-enhanced MRI radiomic features could distinguish between high-grade HCCs and low-grade HCCs |
| Anderson et al[83], 2023 | P | PR | IVIM | 17 (HCC) | DWI-MRI | M, 2D | Matlab | 3 (10th, 50th, and 90th percentiles) | NO | Wilcoxon signed-rank test | No | NS | DW-MRI with IVIM and histogram analysis revealed significant reductions of D* early after treatment as well as an association between D at baseline and smaller tumor growth at three months |
| Li et al[84], 2022 | R | PPF | SEV, MVI | 43 (HCC) | DWI, DCE-MRI | M, 2D | Matlab, SPSS, Medcalc | 8 (Histogram) | NO | ROC | No | AUC = 0.863 | Histogram parameters DDC and ADC, but not the α value, are useful predictors of MVI. The fifth percentile of DDC was the most useful value to predict MVI of HCC |
| Li et al[85], 2022 | R | PPF | MVI | 301 (HCC) | T1WI, T2WI | M, 3D | MITK SOFTWARE | 328 (first-order, GLCM, GLRLM, form factor) | LASSO, ANOVA, MANN-WHITNEY TEST | LASSO | Yes | AUC = 0.914 | The preoperative MRI-based radiomic-clinical model predicted the MVI of HCC effectively and was more efficient compared with the radiomic model or clinical model alone |
| Wang et al[86], 2022 | R | D | DD (cCC-HCC, HCC, CC) | 196 (33 cHCC-CC, 88 HCC and 75 CC) | DCE (ART, PVP, DP) | M; 3D | Pyradiomics | 1316 (shape, first-order, texture -GLCM, GLSZM, GLRLM, GLDM, NGTDM- from original, LoG and wavelet filtered images) | MI, F-test, Chi2-test, LASSO | SVM | No | AUC = 0.91 | The classification ability of cHCC-CC, HCC and CC can be further improved by extracting MRI high- order features and using a two-level feature selection method |
| Yang et al[87], 2021 | R | PPF | MVI | 201 (HCC) | DCE (Pre-T1WI, AP, PVP, DP and HBP) | S; 3D | AK software | 851 (shape, first-order, texture-GLCM, GLSZM, GLRLM, GLDM, NGTDM-, wavelet-transformed) | mRMR, LASSO | ROC; LR | Yes | Radiomics: AUC = 0.896 (TS), 0.788 (VS); Radiomics + clinical: AUC = 0.932 (TS), 0.917 (VS) | The preoperative nomogram integrating clinicoradiological risk factors and the MR radiomics signature showed favourable predictive efficiency for predicting MVI |
| Lv et al[88], 2021 | R | PR | AIR of RFA-treated HCC | 58 (HCC) | DCE | S; 3D | AK software | 396 (histogram, GLCM, GRLM, GLSZM, formfactor) | LASSO | LASSO, ROC | Yes | AUC = 0.941 and 0.818 in the TS and VS | The predictive nomogram integrated with clinical factors and CE-T1WI -based radiomics signature could accurately predict the occurrence of AIR after RFA |
| Yu et al[89], 2022 | R | PPF, PR | VECT, PFS in VETC + and VETC-patients | 182 (HCC) | HBP | M; 3D | Pyradiomics | 1316 (shape, first-order, texture-GLCM, GLRLM, GLSZM, GLDM, NGTDM-) | LASSO | Multivariate LR; forest, SVM; DT | Yes | AUC = 0.972 (peritumoral radiomics model), AUC = 0.91 (intratumoral model) | The intratumoral or peritumoral radiomics model may be useful in predicting VETC and patient prognosis preoperatively. The peritumoral radiomics model may yield an incremental value over intratumoral model |
| Fang et al[90], 2021 | R | PR | PFS of TACE + RFA treated HCC | 113 (HCC) | DCE (HAP, PVP, SPP, and DP) | S; 3D | AK software | 396 (histogram, GLCM, GLSZM GRLM) | LASSO | Cox regression; ROC | Yes | C-index radiomics: 0.646 and 0.669 in TS and VS; C-index combined model: 0.772 and 0.821 in TS and VS | A nomogram combining radiomics and clinical factors predicted the PFS of intermediate and advanced HCC treated with TACE plus RFA |
| Yang et al[91], 2021 | R | MC | CK19+ HCC | 257 (HCC) | T2WI; DWI | M; 3D | MATLAB | 968 (shape, first-order, texture-GLCM, GLRLM, GLSZM, NGTDM-, wavelet) | Univariate analysis, mRMR | Multiple LR; SVM; RF; ANN | Yes | ANN-model: AUROCs = 0.857, 0.726, and 0.790 in the TS and VS A and B | The combined model based on mpMRI-radiomics accurately classify CK19+ HCC |
| Chen et al[92], 2021 | R | MC | CK19+ HCC | 141 (HCC) | HBP | S; 3D | Python (U-Net) | 1024 (Deep semantic) | grid search | GBDT | Yes | AUC = 0.820 and 0.781 in TS and VS | DCE-MRI-based radiomics DLR model can preoperatively predict CK19-positive HCCs |
| Horvat et al[93], 2021 | R | PR | Sustained complete response in RFA-treated HCC | 34 (HCC) | DCE (AP and EP) | M; 3D | Pyradiomics | 107 (shape, first-order, texture-GLDM, NGTDM, GLSZM, GLCM-) | NO | ROC | No | AUC > 0.7 | Second-order features extracted from equilibrium phase obtained highest discriminatory performance |
| Alksas et al[94], 2021 | R | D | DD (types and grades of liver tumors) | 95 (38 benign tumors, 19 intermediate tumors, 38 HCC) | DCE (Pre-T1WI, LAP, PVP, and DP) | M; 3D | NS | 249 (morphological, functional, first-order, texture-GLCM, GLRLM-) | Wrapper approach, and Gini impurity-based selection | RF; SVM; NB, KNN; LDA | No | Accuracy = 0.88 | The identified imaging markers and CAD system can early and accurately detect and grade liver cancer |
| Chong et al[95], 2021 | R | PR | 2 yr RFS after hepatectomy | 23 (HCC) | DCE (AP, PVP, TP, HBP) | M; 3D | Pyradiomics | 2950 (shape, first-order, texture-GLCM, GLRLM, GLSZM, GLDM, NGTDM- from original and filtered images -Wavelets, Gaussian, Laplacian Sharpening-) | Inter-correlation, LASSO | LR, RF, SVM | Yes | AUC = 0.93 and 0.84 in TS and VS | DCE-MRI-based peritumoral dilation radiomics is a potential preoperative biomarker for early recurrence of HCC patients without MVI |
| Ding et al[96], 2021 | R | D | DD (HCC vs FNH) | 224 (149 HCC, 75 FNH) | AP and PVP | M; 3D | Pyradiomics | 2260 (shape, first-order, texture -GLDM, GLCM, GLRLM, GLSZM, NGTDM-, from original LoG and wavelet filtered images) | mRMR, RF, correlation, LASSO | LR | Yes | AUC combined model = 0.984 and 0.972 in TS and VS | The combined model can differentiate HCC from FNH in non-cirrhotic liver with higher accuracy than the clinical model |
| Fan et al[97], 2021 | R | MC | Ki67+ HCC | 51 (HCC) | DCE (AP, PVP, HPB); T2WI | M; 3D | Pyradiomics | 1300 (shape, first-order, texture -GLCM, GLSZM, GLRLM, GLDM, NGTDM- from original, LoG and wavelet filtered images) | LASSO | LR | Yes | Combined model: AUC = 0.922 (TS) and 0.863 (VS) | Combined AP-Rad-score-serum AFP model can preoperatively predict Ki-67 expression in HCC and outperforms AP-based radiomics model |
| Gao et al[98], 2021 | R | PPF | MVI | 225 (HCC) | T2WI | M; 3D | Matlab, SE-DenseNet | 180 low level (intensity, shape, GLCM, GLRLM) + high-level semantic with CNN | LASSO | LR, KNN, RF, SVM, CNNs | Yes | AUC = 0.826 | The proposed ensemble learning algorithm is proved to be an effective method for MVI prediction |
| Li et al[99], 2022 | R | MC | GOLM1, SETD7, and RND1 expression levels | 92 (HCC) | T2WI | M; 2D | MATLAB | 307 (first-order statistics, GLCM, GLRLM, GLSZM, NGTDM), with five, LBP, fractal analysis, shape metrics, FOS, variance, power) | Correlation, RELIEFF | SVM | Yes | r = 0.67 | MRI radiomics features could help quantify GOLM1, SETD7, and RND1 expression levels and predict the recurrence risk for early-stage HCC patients |
| Shi et al[100], 2022 | R | PR | Functional liver reserve | 60 (HCC) | HBP | M; 3D | QTIELAB | 165 (shape, histogram, texture-GLCM, GLRLM, GLZSM-) | Boruta algorithm | RF | No | AUC = 0.90, 0.95, 0.99 for ICG R15 < 10%, < 15%, and < 20% | Radiomics analysis of Gd-EOB-DTPA enhanced hepatic MRI can be used for assessment of functional liver reserve in HCC patients |
| Dai et al[101], 2021 | R | PPF | MVI | 69 (HCC) | DCE (Pre-T1WI, AP, PVP or HBP) | M; 3D | Matlab | 106 (texture -GLCM, GLRLM, GLSZM, SGLDM, NGTDM, and NGLDS-) | LASSO, SVM-RFE, mRMR, LASSO-RFE | GBDT; SVM; LR; RF | No | AUC = 0.792 for HBP model | The radiomics model based on the HBP had better predictive performance than those based on the AP, PVP, and pre-enhanced T1W |
| Fan et al[102], 2021 | R | PPF | VECT+ HCC | 81 (HCC) | DCE (AP and HBP) | M; 3D | Pyradiomics | 1316 (first-order, texture -GLCM, GLSZM, GLRLM, GLDM, NGTDM- from original, wavelet and LoG filtered images) | ICC, LASSO | ROC; LR | No | AUC = 0.84 | Texture analysis based on Gd-EOB-DTPA-enhanced MRI can help identify VETC-positive HCC |
| Yang et al[103], 2021 | R | PPF | Poorly differentiated HCC | 188 (HCC) | T1WI, T2WI, DCE (AP, PP and DP) | M; 3D | LIFEx | 200 (shape, histogram, texture -GLCM, NGLDM, GLRLM, GLZLM-) | LASSO | LASSO | Yes | Model1: AUC = 0.623 and 0.576 in TS and VS, while it is 0.576 in the validation cohort. Model2: AUC = 0.721, and 0.681 in TS and VS | The MRI-based radiomics signature and clinical model can distinguish HCC patients that belong in a low differentiation group fromother patients |
| Chen et al[104], 2021 | R | PPF | MVI | 269 (HCC) | T2WI; DWI, DCE (AP, PVP, and HBP) | M; 3D | Pyradiomics | 1395 (first-order, GLRLM, GLCM from original, Laplacian, logarithmic, exponential, and wavelet filtered images) | Variance threshold, LASSO | KNN SVM, XGBoost, RF, LR, DT | Yes | For HBP model: AUC = 0.942 (SVM), 0.938 (XGBoost), and 0.936 (LR) | Radiomics signatures with machine learning can further improve the ability to predict MVI and are best modeled during HBP |
| Kong et al[16], 2021 | R | PR | Response to TACE | 99 (HCC) | T2WI | M; 3D | AK software | 396 (histogram, texture-GLSZM, GLCM, GLRLM-) | LASSO, correlation | ROC | Yes | AUC = 0.861 and 0.884 in TS and VS | The radiomics and clinical-based nomogram can well predict TR in intermediate-advanced HCC |
| Zhao et al[105], 2021 | R | MC | GPC3 | 143 (HCC) | DCE-MRI, DWI | M; 3D | MedCalc, R | 6 (Histogram) | NO | Mann-Whitney U test | No | C-INDEX = 0.804 | Elevated serum AFP levels and lower 75th percentile ADC values were helpful in differentiating GPC3-positive and GPC3-negative HCCs. The combined nomogram achieved satisfactory preoperative risk prediction of GPC3 expression in HCC patients |
| Song et al[106], 2021 | R | PPF | MVI | 601 (HCC) | T2WI FS; DWI; ADC; DCE (AP, PVP, and DP) | M; 3D | PyRadiomics | 110 (shape, first-order, texture) | PCA, ANOVA | SVM, AE, LDA, RF, LR, LASSO, AdaBoost, DT, Gaussian process, NB, DL | Yes | DLC model: AUC = 0.931 for MVI prediction; AUC = 0.793 for MVI-grade stratification | DLC model predicts and grades MVI better than DL model |
| Zhong et al[107], 2021 | R | D | DD (small HCC 3 cm vs benign nodules) | 150 (112 HCC, 44 benign nodules) | in phase sequence; T2WI FS; ADC | M; 2D | MaZda | 837 (histogram, GLCM, RLM, wavelet, absolute gradient, autoregressive model) | ICC, Mann-Whitney, LASSO | LR; ROC | No | AUC = 0.917 | MRI-based radiomics analysis showed additive value to the LI-RADS v 2018 algorithm for differentiating small HCCs from benign nodules in the cirrhotic liver |
| Zhao et al[105], 2021 | R | MC | GPC3 expression | 143 (HCC) | ADC | M;3D | MR Multiparametric Analysis software | 6 (histogram) | Univariate analysis (t-test, Mann-Whitney, Pearson, χ2, Fisher) | LR | No | C-index = 0.804 | The combined nomogram achieved satisfactory preoperative risk pre-diction of GPC3 expression in HCC patients |
| Chen et al[108], 2021 | R | PR | Post hepatectomy liver failure | 144 (HCC) | HBP | M; 2D | AK software | 1,044 (shape, first-order, texture-GLSZM, GLCM, GLRLM-) | Correlation, RFE | LR; ROC; liver failure model | Yes | AUC = 0.956 and 0.844 in TS and VS | The LF model is able to predict PHLF in HCC patients |
| Liang et al[109], 2021 | R | PPF | MVD | 30 (HCC) | DCE | M; 2D | AK software | 376 (histogram, texture-GLSZM, GLCM, GLRLM-) | Mann-Whitney | LR | No | AUC = 0.83 and 0.94 | DITET model provides a better indication of the microcirculation of HCC than SITET |
| Zhang et al[110], 2021 | R | PR | RFS of HCC patients treated with surgical resection | 153 (HCC) | T2WI FS; DCE (AP, PVP, and HBP) | M; 3D | Pyradiomics | 107 (shape, first-order, texture -GLCM, GLSZM, GLRLM, GLDM, NGTDM-) | LASSO | LASSO Cox regression | Yes | C-index 0.725 | The prediction model combining MRI radiomics signatures with clinical factors predicts the prognosis of surgically resected HCC patients |
| Zhang et al[111], 2021 | R | PR | RFS after curative ablation | 132 (HCC) | T2WI FS; T1WI FS; DCE (AP, PVP, and HBP) | M; 3D | Pyradiomics | 1316 (shape, first-order, texture -GLCM, GLSZM, GLRLM, GLDM, NGTDM-, LoG, wavelet) | RandomForestSRC | Cox regression; random survival forest; ROC | Yes | C-index = 0.706 | The radiomics model combining DCE-MRI with clinical characteristics could predict HCC recurrence after curative ablation |
| Zhang et al[112], 2021 | R | PPF | MVI | 195 (HCC) | T2WI FS; DWI; ADC; DCE (AP, PP, DP) | M; 3D | AK software | 396 (histogram, GLCM, GLSZM, RLM, formfactor, haralick) | ANOVA, Mann-Whitney U-test, correlation, LASSO | Multivariate LR | Yes | AUC = 0.901 and 0.840 in the TS and VS | The combined radiomics-clinical model can preoperatively and noninvasively predict MVI in HCC |
| Zhao et al[113], 2021 | R | PR | Response to TACE | 122 (HCC) | DCE (AP, PVP, and DP) | M; 3D | AK software | 789 (histogram,GLCM, GLRLM, GLZSM, Haralick,Gaussian transform) | ICC, Spearman's correlation, univariate LR, LASSO | LR; ROC | Yes | AUC = 0.838 and 0.833 in TS and VS | The combined model (radiomics score + clinical-radiological risk factors) showed better performance than the clinical-radiological model in predicting TACE efficacy in HCC patients |
| Kuang et al[114], 2021 | R | PR | Predict short-term response after TACE in HCC | 153 (HCC) | T2WI; DCE (AP) | A; 3D | AK software | 396 (shape, histogram, GLSZM, GLCM, RLM) | mRMR, LASSO | LR | Yes | AUC = 0.83 and 0.81 in TS and VS | MRI-based nomogram has greater predictive efficacy to predict the response after TACE than radiomics and clinics models alone |
| Meng et al[115], 2021 | R | PPF | MVI | 402 (HCC) | T1WI, T2WI, DWI, CE-CT | M, 3D | Pyradiomics | 1288 | ICC, MANN-WHITNEY, LASSO | LR | Yes | AUC = 0.804 | CT and MRI had a comparable predictive performance for MVI in solitary HCC. The RS of MRI only hadsignificant added value for predicting MVI in HCC of 2–5 cm |
| Zhu et al[116], 2021 | R | D | DD (MTM-HCC vs HCC) | 88 (32 MTM-HCCs, 56 Non-MTM-HCC) | T2WI FS; in-phase and out-of-phase sequences; DCE (AP, PVP, and DP) | M; 2D | MaZda | 101 (histogram, the absolute gradient, GLRLM, GLCM, autoregressive model and wavelet transform) | Fisher, MI, POE + ACC, LASSO | LR; ROC | No | AUC = 0.785 | A DCE-MRI-based radiomics nomogram can predict MTM-HCC |
| Liu et al[117], 2021 | R | PR | TACE, MWA | 102 (HCC) | T1WI, T2WI, PVP | M, 2D | MaZda | 20 (First order, GLCM) | NS | ROC | No | AUC = 0.876 | MR imaging texture features may be used to predict the prognosis of HCC treated with TACE combined with MWA |
| Chong et al[118], 2021 | R | PR | MVI, RFS after curative surgery (HCC≤ 5 cm) | 356 (HCC) | DWI, DCE (Pre-T1WI, AP, PVP, TP, HBP) | M; 3D | Pyradiomics | 854 (shape, first-order, texture -GLCM, GLDM, GLRLM, NGTDM, GLSZLM- from original and wavelet filtered images) | LASSO | RF; LR | Yes | AUC = 0.920 with RF, 0.879 with LR in validation cohort | Preoperative radiomics-based nomogram using random forest is a potential biomarker of MVI and RFS prediction for solitary HCC ≤ 5 cm |
| Gu et al[119], 2020 | R | MC | GPC3+ HCC | 293 (HCC) | DCE (DP) | M; 3D | Pyradiomics | 853 (shape, histogram, texture -GLCM, GLSZM, GLRLM, GLDM, NGTDM-, wavelet) | ICC, Mann-Whitney, Fisher | LR; SVM | Yes | AUC = 0.926 and 0.914 in TS and VS | The combined AFP + radiomics nomogram may provide an effective tool for noninvasive and individualized prediction of GPC3-positive in HCC patients |
| Zhao et al[120], 2021 | R | PR | ER after partial hepatectomy | 113 (HCC) | T2WI; in-phase and out-of-phase sequences; DWI; DCE (AP, PVP, and DP) | M; 3D | AK software | 1146 (shape, histogram, texture -GLCM, GLRLM, GLSZM-) | Spearman's correlation, LASSO, stepwise LR | Multivariate LR | Yes | Radiomics: AUC = 0.771 in the VS. Combined nomogram: AUC = 0.873 | A combined nomogram incorporating the mpMRI radiomics score and clinicopathologic-radiologic characteristics can predict ER (≤ 2 yr) in HCC |
| Ai et al[121], 2020 | R | D | DD (HCC, HH, HC) | 89 (33 HH, 22 HC, 34 HCC) | IVIM | M; 3D | MITK-DI | 13 (histogram) | Kruskal-Wallis | ROC | No | AUC = 0.883 | A multiparametric histogram from IVIM is an effective means of identifying HH, HC, and HCC |
| Shaghaghi et al[122], 2021 | R | PR | Post-TACE OS and TFS | 104 (HCC) | ADC | S; 3D | NS | 3 (mean, skewness, and kurtosis) | NO | NO | No | Significant results for changes in ADC mean and Kurtosis | Changes in mean ADC and ADC kurtosis can be used to predict post-TACE OS and TFS in well-circumscribed HCC |
| Li et al[123], 2020 | R | D | DD (HCC vs HMRC) | 75 (41 HCC, 34 HMRC) | DCE | M; 2D | OmniKinetic | 67 (First order, histogram, GLCM, Haralick, RLM) | t-test, ROC | FDA | No | AUC = 0.86 (radiomics + pharmacokinetic) and 0.89 (DA based on radiomics) | A model based on DCEMRI radiomics and pharmacokinetic parameters was useful for differentiating HCC from HMRC |
| Geng et al[124], 2021 | R | PPF, MC | MVI; GRADE; CK-7, CK-19, GPC3 expression status | 53 (HCC) | SWI | M;3D | PyRadiomics | 107 (first-order, shape, GLCM, GLRLM, GLSZM, NGTDM) | ICC | LR | No | AUC = 0.905 (CK-19+), 0.837 (CK-7+), 0.800 (high histopathologic grade) and 0.760 (GPC-3+) | Extracting the radiomics features from SWI images was feasible to evaluate multiple histo-pathologic indexes of HCC |
| Zhang et al[125], 2020 | R | PR | OS after surgical resection | 136 (44 MCC, 59 HCC, 33 CHCC) | DCE (EP); DWI | M; 3D | AK software | 384 (histogram, GLCM, GLSZM, RLM, formfactor, haralick) | mRMR method and the elastic network algorithm | Multivariable cox regression | No | Parameters independently associated with OS (P < 0.05) | Clinicopathological and radiomics features are independently associated with the OS of patients with primary liver cancer |
| Zhang et al[126], 2020 | P | PR | OS after surgical resection | 120 (HCC) | T2WI FS; DCE (AP, PVP, TP, and HBP) | S; 2D | AK software | 350 (histogram, form factor, GLCM, GLRLM) | ICC, LASSO | LASSO Cox regression | Yes | C-index = 0.92 | Radiomics + clinic-radiological predictors can efficiently aid in preoperative HCC prognosis prediction after surgical resection with respect to clinic-radiological model |
| Hectors et al[127], 2020 | P | PR | 6- and 12- week response to 90 yr | 24 (HCC) | DCE-MRI, IVIM-DWI | M; 3D | Matlab | 40 DCE MRI histogram parameters and 20 IVIM DWI histo-gram parameters | Stepwise feature selection | LR | No | AUC = 0.92 | Diffusion and perfusion MRI can be combied to evaluate the response of HCC to radioembolization |
| Shi et al[128], 2020 | P | PPF, MC | HCC GRADE, KI67+ HCC, CAPSULE FORMATION+ | 52 (HCC) | IVIM | M; 3D | ImageJ, Mazda | 15 (histogram) | t-test | LR | No | AUC = 0.92 (grading), 0.86 (Ki67+) and 0.84 (capsule formation) | Multiple prognostic factors can be accurately predicted with assistance of histogram metrics sourced from a single IVIM scan |
| Feng et al[129], 2020 | R | D, MC | DD | 104 (HCC) | Gd-EOB-DTPA-enhanced MRI and T2WI | M, 3D | Mazda | 262 (Histogram, GLCOM, GLRLM, WAVELET TRANSFORM) | PCA, LDA, NDA, RDA | ROC | No | AUC = 0.879 | Texture analysis of Gd-EOB-DTPA-enhanced MRI and T2WI was valuable and might be a promising method in identifying the HCC grade |
| Nebbia et al[130], 2020 | R | PPF | MVI | 99 (HCC) | T2WI; DCE (AP and PP); DWI | M; 3D | Pyradiomics | 100 (shape, first-order, texture -GLCM, GLDM, GLSZM-) | LASSO | SVM; DT; KNN, NB | No | AUC = 0.867 | Information from mpMRI sequences is complementary in identifying MVI |
| Schobert et al[131], 2020 | R | PR | Response to DEB-TACE, PFS | 46 (HCC) | DCE (HAP, PVP, and DP) | M; 3D | Pyradiomics | 14 (shape, first-order) | Univariate analysis, stepwise forward selection | LinearRegression; Cox regression; Kaplan–Meier analysis | No | High NLR and PLR correlated with non-spherical tumor growth (P = 0.001 and P < 0.001) | This study establishes the prognostic value of quantitative inflammatory biomarkers associated with aggressive nonspherical tumor growth and predictive of poorer tumor response and shorter PFS after DEB-TACE |
| Sun et al[132], 2020 | R | PR | Early progression of unresectable HCC after TACE | 84 (HCC) | T2WI; DWI; ADC | M; 3D | Pyradiomics | 1597 (first-order, shape, texture -GLCM, GLRLM, GLSZM, NGTDM, GLDM-) | Variance threshold, Pearson's correlation, LASSO | LR | Yes | AUC = 0.800 | mpMRI-based radiomic model predicts the outcome of TACE therapy for unresectable HCC outperforms monomodality radiomic models |
| Wilson et al[133], 2020 | R | PPF, PR | MVI, OS, DFS after surgery | 38 (HCC) | T2WI; in-phase and out-of-phase sequences; DCE (HAP, and PVP) | M; 2D | TexRAD | 7 (histogram) | NO | LR | No | AUC = 0.83 | Tumor entropy and mean are both associated with MVI. Texture analysis on preoperative imaging correlates with microscopic features of HCC |
| Hectors et al[134], 2020 | R | MC, PR | Immuno-oncological markers (CD3, CD68, CD31), recurrence at 12 m | 48 (HCC) | DCE (Pre-T1WI, AP, PVP, LVP, and HBP); ADC | M; 2D | MATLAB | 36 (Haralick, qualitative and quantitative) | NO | LR | No | AUC = 0.76–0.80 | MRI radiomics features may serve as noninvasive predictors of HCC immuno-oncological characteristics and tumour recurrence |
| Wang et al[135], 2020 | R | MC | CK19+ HCC | 227 (HCC) | DWI; ADC; T2WI; DCE (Pre-T1WI, AP, PVP, DP, and HBP) | M; 3D | Pyradiomics | 647 (shape, histogram, texture, wavelet) | ICC, LASSO | Logistic model; ROC | Yes | AUC = 0.95 | The combined model based on a fusion radiomics signature derived from AP and HBP can be a reliable biomarker for CK19 status of HCC |
| Wang et al[136], 2020 | R | PR | 5 yr survival after curative hepatectomy | 201 (HCC) | T1WI; T2WI; DWI; ADC; DCE (AP, PVP, and EP) | S; 3D | Precision Medicine Open Platform | 3144 (histogram, texture, wavelet, statistical) | Gini index | Random Forest | Yes | AUC = 0.9804 and 0.7578 in the TS and VS | This radiomics model is a valid method to predict 5-year survival in HCC patients |
| Song et al[137], 2020 | R | PR | RFS after c-TACE | 184 (HCC) | DCE (AP, and PVP) | S; 3D | AK software | 396 (histogram, GLCM, GRLM, GLSZM) | ICC, LASSO | LASSO Cox regression | Yes | C-index = 0.802 | The combined model is more valuable than the clinical-radiological model or radiomics model alone for evaluating the RFS of HCC patients after c-TACE |
| Zhang et al[138], 2019 | R | PR | ER (1 yr after hepatectomy) | 100 (HCC) | DCE (AP, PVP, and DP) | S; 3D | Omni Kinetic | 6 (skewness, kurtosis, uniformity, energy, entropy, and correlation) | NO | LR | No | AUC = 0.867 | Texture analysis based on preoperative MRI are potential quantitative predictors of ER in HCC patients after hepatectomy |
| Huang al[139], 2019 | R | D, PR | DD (HCC vs DPHCC), DFS, OS after surgery | 100 (HCC) | DCE (AP, PVP, DP, and HBP) | M; 3D | Huiying Medical Technology | 1029 (First-order, shape, texture -GLCM, GLRLM, GLSZM-) | LASSO | Multi-layer perceptron; SVM; LR; K-nearestneighbor; ROC | No | Accuracy of LR in PVP (0.77), DP (0.798), HBP (0.756) and of multi-layer perceptron in PVP (0.798) | The radiomics features extracted from DCE-MRI can be used to diagnose preoperative DPHCC |
| Ye et al[140], 2019 | P | MC | Ki67 expression | 89 (HCC) | T2WI FS; DCE (Pre-T1WI, AP, PVP, TP, and HBP) | M; 3D | AK software | 396 (histogram, texture, GLCM, GLRLM) | LASSO | LR | No | C-index = 0.936 | The combination of DCE-MRI texture signature and clinical factors demonstrated the potential to preoperatively predict Ki-67 status of HCC after curative resection |
| Zhang et al[141] 2019 | R | PPF | MVI | 267 (HCC) | T2WI FS; in-phase and out-of-phase sequences; T1WI; DWI; DCE (AP, PVP, and EP) | M; 3D | MATLAB | 484 (intensity, texture, wavelet) | mRMR | LR | Yes | AUC = 0.784 and 0.820 in TS and VS | The radiomics nomogram can serve as a visual predictive tool for MVI in HCC and outperformed clinico-radiological model |
| Chen et al[142], 2020 | R | MC | CK19+, EpCAM | 115 (HCC) | T2WI, pre-T1WI, DCE (AP, PVP, HBP), ADC | M; 3D | AK software | 23 (histogram) | Univariate analysis | LR | No | Accuracy = 0.86, C-index = 0.94 | Noninvasive prediction of HCCs with progenitor phenotype can be achieved with high accuracy by integrated interpretation of biochemical and radiological information |
| Xu et al[143], 2019 | R | PPF | HCC GRADE | 51 (HCC) | ADC | M; 3D | SPSS | 27 (histogram) | NO | NO | No | ρ = −0.397 for ADC 25th percentile; AUC = 0.76 for ADC min | The 25th percentile ADC showed a stronger correlation with the histological grade of HCC than other ADC parameters, and the minimum ADC value might be an optimal metric for determining poor and fair diferentiations of HCC in DWI |
| Li et al[144], 2019 | R | MC | Ki67 expression | 83 (HCC) | DCE (HAP, PVP, EP, and HBP); T2WI FS | M; 3D | MaZda | 30 (histogram, GLCM, GLRLM, absolute gradient, the autoregressive model, wavelet transform) | Fisher coefficient, MI, POE + ACC, correlation | ROC (accuracy) | No | Lowest misclassification rates: PCA-PVP = 40.96%; LDA-PVP = 9.64%; NDA-AP = 6.02%. | Texture analysis of HBP, arterial phase, and portal venous phase are helpful for predicting Ki67 expression |
| Oyama et al[145], 2019 | R | D | DD (HCC, MT, HH) | 93 (50 HCCs, 50 MTs, 50 HHs) | T1WI | M; 3D | MATLAB | 43 (GLCM, GLRLM, GLSZM, NGTDM) | correlation | LDA | No | Accuracy = 92% (texture analysis) and 85% (persistence imges analyses) | Texture analysis or topological data analysis support the classifcation of HCC, MT, and HH with considerable accuracy, solely based on non-contrast-enhanced T1WI 3D |
| Wang et al[146], 2019 | R | MC | CK19+ HCC | 48 (HCC) | T2WI FS; in-phase and out-of-phase sequences; DCE (AP, PVP, DP); DWI (b values 0 and 500 s/mm²); ADC | M; 2D | In-house software | 2415 (intensity, gradient, Gabor wavelet, local binary pattern histogram Fourier, GLCM, GLGCM) | LDA (AUC) | LR | No | AUC = 0.765 | The StdSeparation 3D texture character may be a reliable imaging biomarker which can improve the diagnostic performance. |
| Zhu et al[147], 2019 | R | PPF | MVI | 142 (HCC) | DCE (AP, PVP) | M; 3D | Omni-kinetics software | 58 (histogram, GLCM, Haralick, GRLM) | Kruskal-Wallis, univariate LR, Pearson's correlation | LR | Yes | AUC = 0.81 | The combined model of arterial phase radiomic features with clinical-radiological features could improve MVI prediction ability |
| Zhang et al[148], 2019 | P | PR | ER (1 yr after surgical resection) | 155 (HCC) | T2WI FS, DCE (AP, PVP, TP, and HBP) | M; 3D | AK software | 385 (histogram, texture, GLCM, GLRLM) | LASSO | LR; ROC | Yes | AUC = 0.844 | The radiomics nomogram integrating the radiomics score with clinical-radiological risk factors showed better discriminative performance than the clinical-radiological nomogram |
| Gordic et al[149], 2019 | R | PR | CR, PR, SD | 22 (HCC) | volumetric ADC | M; 3D | MATLAB | 7 (histogram) | Wald test | LR | No | AUC = 0.91 | Diffusion histogram parameters obtained at 6w and early changes in ADC from baseline are predictive of subsequent response of HCCs treated with RE |
| Jansen et al[150], 2019 | R | D | DD (adenomas, cysts, hemangiomas, HCC, metastases) | 211 (40 adenomas, 29 cysts, 56 hemangiomas, 30 HCC, 56 metastases) | DCE-MRI, T2WI | M; 2D | NS | 164 (contrast curve, histogram, and GLCM texture) | ANOVA F-SCORE | Randomized tree classifier | No | Accuracy = 0.77 | The proposed classification system using features derived from clinical DCE-MR and T2WI, with additional risk factors is able to differentiate five common types of lesions and is a step forward to a clinically useful aid for focal liver lesion diagnosis |
| Ma et al[151], 2019 | R | PR | Post RFA progression | 64 (HCC) | ADC | M; 3D | Volume View | 8 (histogram) | NO | Cox-regression | No | C-index = 0.62 | Pre-RFA ADC histogram analysis might serve as a useful biomarker for predicting tumor progression and survival in patients with HCC treated with RFA |
| Wu et al[152], 2019 | R | D | DD (HCC, HH) | 369 (222 HCCs, 224 HHs) | In-phase and out-of-phase sequences; T2WI; DWI | M; 3D | PyRadiomics | 1029 (shape, first-order, texture -GLCM, GLRLM, GLSZM-, exponential, square, square root, logarithm, and wavelet) | Variance threshold, select k best, LASSO | Decision tree; random forest; K nearest neighbours; LR; ROC | Yes | AUC = 0.86 and 0.89 in TS and VS | mpMRI radiomics signature is an adjunct tool to distinguish HCC and HH, outperformed a less experienced radiologist, and is nearly equal to an experienced radiologist |
| Kim et al[153], 2019 | R | PR | ER (< = 2 yr), LR (> 2 yr) after curative resection | 167 (HCC) | DCE (AP, PP, HBP, AP-PP, AP-HBP, PP-HBP, and AP-PP-HBP) | S; 3D | PyRadiomics | 1301 (first-order, shape, texture -GLCM, GLRLM, GLSZM, NGTDM, GLDM-, LoG, wavelet) | RF minimal depth algorithm | random survival forest | Yes | C-index = 0.716 | The clinicopathologic-radiomic model showed best performances, suggesting the importance of including clinicopathologic information in the radiomic analysis of HCC |
| Lewis et al[154], 2019 | R | D | DD (HCC, ICC, HCC-ICC) | 63 (36 HCC; 17 ICC; 12 HCC-ICC) | ADC | M; 3D | MATLAB | 11 (histogram) | Wald criteria | Binary LR and AUROC | No | AUC = 0.9 | The combination of quantitative ADC histogram parameters and LI-RADS categorization yielded the best prediction accuracy for distinction of HCC vs ICC and combined HCC-ICC |
| Chen et al[155], 2019 | R | MC | Immunoscore (CD3+ and CD8+) | 207 (HCC) | HBP | M; 3D | AK software | 1044 (histogram, texture, factor parameters, GLCM, GLRLM, GLSZM) | Recursive elimination | LR | Yes | AUC = 0.92 | The combined MRI-radiomics-based clinical nomogram is effective in predicting immunoscore in HCC |
| Feng et al[156], 2019 | R | PPF | MVI | 160 (HCC) | HBP | M; 3D | AK software | 1044 (histogram, texture, wavelet transformed, filter transformed texture) | LASSO | LR | Yes | AUC = 0.85 and 0.83 in TS and VS | A combined intratumoural and peritumoural radiomics model based on DCE-MRI is able to pre-operatively predict MVI in primary HCC patients |
| Wu et al[157], 2019 | R | PPF | HCC grade | 170 (HCC) | T1WI; T2WI FS | M; 3D | MATLAB | 656 (histogram, shape, GLCM, wavelet) | LASSO | LR | Yes | AUC = 0.8 | The combination of the radiomics signatures with clinical factors may be helpful for the preoperative prediction of HCC grade |
| Yang et al[158], 2019 | R | PPF | MVI | 208 (HCC) | T2WI FS; DWI; DCE (AP, PVP, DP, and HBP) | M; 3D | MATLAB | 647 (shape, intensity, textur-GLCM, GLRLM, GLZLM, NGLDS-) | LASSO, AIC | LR | Yes | AUC = 0.94 and 0.86 | The nomogram incorporating clinicoradiological risk factors and radiomic features derived from HBP images achieved satisfactory preoperative prediction of the individualized risk of MVI in HCC patients |
| Stocker et al[159], 2018 | R | D | DD (HCC vs FNH vs HA) | 108 (55 HCC, 24 HA, 29 FNH) | T1WI FS; T2WI; DCE (AP, PVP, and HBP) | M; 2D | MATLAB | 19 (histogram, GLCM, GLRLM) | LR | LR; ROC | No | AUC = 0.92 | 2D-TA of MR images may help to distinguish HCC from benign hepatocellular tumors in the non-cirrhotic liver, with most promising results were found in TA features in the AP images |
| Ahn et al[160], 2019 | R | PR | ER (1y after surgical resection) | 179 (HCC) | HBP | M; 3D | In-house software program | 13 (histogram, GLCM) | Univariate analysis | LR | No | AUC = 0.83 | When added texture variables to MRI findings, the diagnostic performance for predicting early recurrence is improved |
| Hui et al[161], 2018 | R | PR | ER (1yr), LNR (late or no recurrence) after surgery | 50 (HCC) | T2WI; DCE (AP, PVP and EP) | M; 2D | MaZda | 290 (histogram, texture, autoregressive model, GRLM, GLCM, wavelet) | PRTools | ROC | No | Accuracy 78%-84% | Texture analysis of preoperative MRI has the potential to predict ER of HCC with up to 84% accuracy using an appropriate, single texture analysis parameter |
| Zou et al[162], 2019 | R | D | DD (IMCC and HCC) | 33 IMCC, 98 HCC | volumetric ADC, DCE-MRI | M; 3D | SPSS | 9 (histogram) | NO | ROC | No | AUC = 0.79 | Volumetric ADC histogram analysis provides additional value to dynamic enhanced MRI in differentiating IMCC from HCC |
| Li et al[163], 2018 | P | PPF | MVI | 41 (HCC) | IVIM-DWI | M; 3D | MATLAB | 10 (histogram) | Univariate analysis | ROC | No | AUC = 0.87 | Histogram analysis of IVIM based on whole tumor volume can be useful for predicting MVI. The 5th percentile of D was most useful value to predict MVI of HCC |
| Wu et al[164], 2019 | P | PR | TTP after TACE | 55 (HCC) | IVIM-DWI | S; 3D | MR OncoTreat | 8 histogram parameters | NO | Cox-regression | No | AUC = 0.82 | Pre-TACE kurtosis of ADCtotal is the best independent predictor for TTP |
| Li et al[165], 2017 | R | D | DD (HH vs HM vs HCC) | 162 (55 HH, 67 HM, 40 HCC) | SPAIR T2WI | M; 2D | MATLAB | 233 (histogram, GLCM, GLGCM, GLRLM, GWTF, ISZM) | CCC, DR, R2 | ROC, KNN, BP-ANN, SVM, LR | Yes | Misclassification rates: 11.7% (HH vs HM), 9.6% (HM vs HCC) and 9.7% (HH vs HCC) | Texture features of T2WI SPAIR can classify HH, HM and HCC |
| Moriya et al[166], 2017 | R | D | HCC grade | 53 (HCC) | DWI, ADC | A; 3D | SPSS | 11 (First Level) | ANOVA | ROC | No | sensitivity: 100%, specificity: 54% | Minimum ADC was most useful to differentiate poorly differentiated HCC in 3D analysis of ADC histograms |
The number of total included patients was 18.949, with a sample size varying from 17 to 602 patients (median: 309.5). Most studies (96 out of 127) explored more than one phase/sequence to perform radiomic analysis. Most studies (106 out of 127) performed 3D segmentation. In 114 of them, segmentation was manually performed, while in the remaining studies was used a semiautomatic (12 studies) or automatic (2 study) segmentation approach. Concerning software used for feature extraction, PyRadiomics was the most popular (used in 42 out of 127 studies), followed by AK software (used in 23 out of 127 studies) and Matlab (used in 19 out of 127studies). The number of radiomics features extracted from each phase/sequence ranged from 3 to 3144 (mean: 68 ± 206). Shape features were extracted in 55 out of 127 studies, first-order features in all but three studies, textural features in 82/127 studies, and features from filtered images (e.g., wavelet, Laplacian of gaussian) in 34 out of 127 studies. Concerning feature selection algorithms, the Least Absolute Shrinkage and Selection Operator regression was the most widely used (used in 55 out of 127 studies). Other frequently used algorithms for feature selection were intra-class correlation coefficient (used in 25 studies), correlation (used in 12 studies) and minimum redundancy maximum relevancy (used in 9 studies). The performance metrics of the studies, when present, corresponded to accuracy in 9 out of 127 studies, area under the receiver operating characteristic curve (AUC) in 99 out of 127 studies and to C-index in 12 out of 127 studies. Most studies involved machine learning techniques for radiomic analysis, of which 51 splitted the subjects into training and test cohort to test the prediction models performance. Further details on these characteristics can be found in Table 1 and Supplementary Table 2.
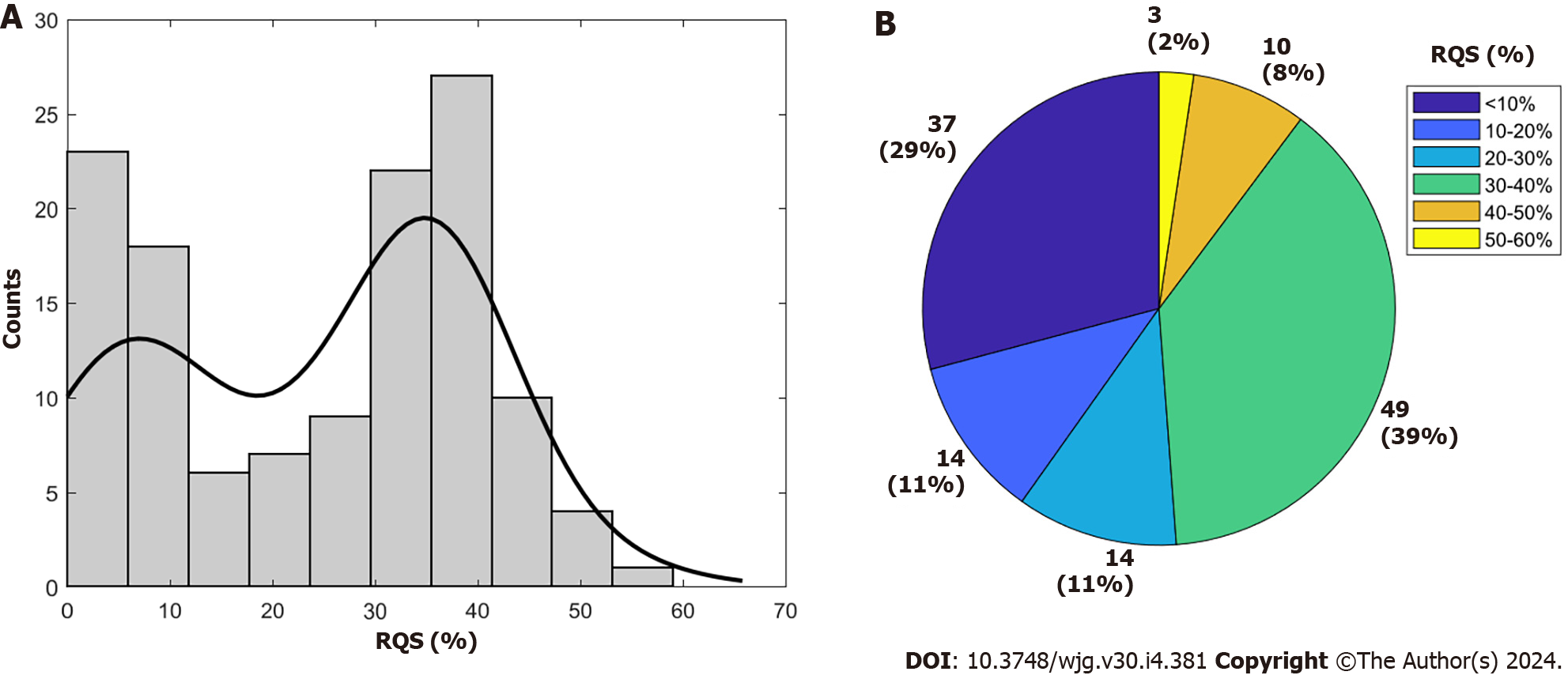
Supplementary Table 3 provides the RQS details of all included studies. The average total RQS score was 8 ± 6.22, corresponding to a percentage of 24.15% ± 15.25%, with a range from 0.0% to 58.33% (Figure 2). Concerning the first RQS checkpoint, nearly all studies, excluding ten, provided thorough documentation of the imaging protocol, yet none achieved the maximum points for utilizing a public protocol. In relation to the second RQS checkpoint (items 2 to 4), a majority of studies (84.25%, 107 out of 127) employed multiple segmentation, mainly by different radiologists, but none of the articles met the requirement for 'imaging at multiple time points' and only one article met the requirement for a 'phantom study'. With respect to the third RQS checkpoint (items 5 to 16), feature reduction techniques were applied in all but 15 studies (88.28%). Multivariable analysis with non-radiomics features was performed in 85 studies (66.92%) of the 128 included articles. However, only 40 (31.25%) identified and discussed biological correlates and only 50 (39.06%) provided cut-off analysis.
Of the 127 studies included, almost all (123) reported discrimination statistics and their statistical significance. About a quarter of these studies used resampling techniques. However, only 58 studies reported calibration statistics, and none of them applied resampling techniques.
A significant proportion (39.37%) of the studies (50 out of 127) did not provide any validation of their results. Only three studies validated their results using one external validation cohort and five studies used two external validation cohorts. Furthermore, only 47 out of 127 research examined the clinical utility of the produced model using decision curve analysis, while 42 out of 127 studies compared radiomics models with the particular gold standard (based on the study purpose).
Lastly, no study disclosed code and data to the public or performed a cost-effectiveness analysis.
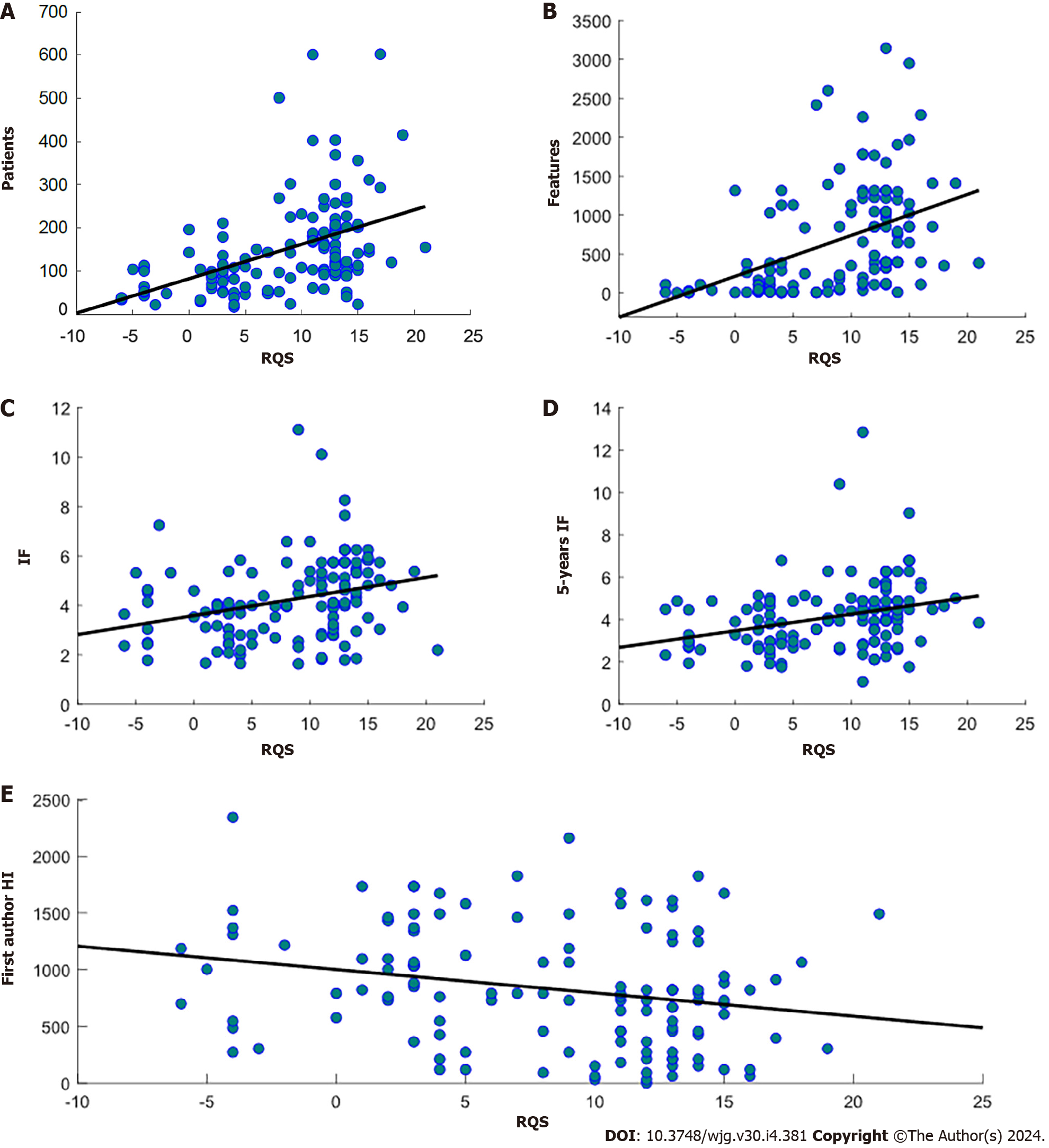
A significant positive correlation was found between RQS and journal IF (ρ = 0.36, P = 2.98 × 10-5), 5-years IF (ρ = 0.33, P = 1.56 × 10-4), number of patients involved (ρ = 0.51, P < 9.37 × 10-10) and number of radiomics features (ρ = 0.59, P < 4.59 × 10-13) extracted in the study. On the other hand, there was a significant negative correlation between RQS and time between the publication and the performed literature research (ρ = -0.23, P = 0.0072) and there were no statistically significant differences identified in the RQS among studies with different objectives. Scatterplots with regression lines showing significant correlations between RQS and journal metrics are shown in Figure 3.
In this systematic review, we aimed at summarizing the current status of the fast-growing research on MRI radiomics for the management of HCC. We explored whether it could offer diagnostic, prognostic, and predictive information about pathological outcomes and molecular expression. Additionally, we assessed the quality of the science and reporting across the studies using the RQS tools. 127 studies from November 2017 onwards were examined in our study. Despite promising results obtained from each of them (with best AUC and C-indexes reaching 0.98 and 0.94, respectively), our study revealed that the methodological variability of the research is considerable, and the reporting quality is insufficient.
Mean RQS was 8 out of 36, with a mean percentage RQS of 24.15%. These results are consistent with previously published data on a variety of tumors, including prostate, breast, lung, renal, and brain cancer[27-31]. Recent studies evaluating research quality in HCC radiomics also align with our findings[25,32]. However, direct comparison with our study is not possible due to differences in purpose and inclusion criteria.
The results of our analysis showed that the poor RQS scores of the included studies were mostly caused by the absence of rigorous procedures pertaining to radiomics workflow.
Regarding RQS checkpoint 1, practically all investigations have a thorough documentation of the imaging methodology. Nevertheless, the lack of public image methods in the investigations negatively impacts the radiomic studies' repeatability and reproducibility. Notably, the CE-T1WI MRI sequence emerged as the most extensively explored, given its primary role in preoperative HCC assessment. Nevertheless, there exists variability in MRI acquisition due to differences in manufacturers, scanning protocols, contrast media, and phases employed. A significant diversity across the included studies was also noted in terms of RQS checkpoint 2. Specifically, 108 out of 127 studies adopted multiple segmentations to mitigate bias arising from segmentation variability. It's crucial to highlight, however, the lack of consistency among studies regarding the type of ROI (2D/3D) and the segmentation method used (manual, semi-automatic, automatic). It is worth mentioning that the majority of studies used manual or semi-automated image segmentation with manual correction, which restricts the studies included. Both manual and semi-automatic segmentation can introduce significant observation bias, which may affect studies on intra- and interobserver variation in ROI/VOI delineation[1].
None of the studies determined scanner/manufacturer variability or collected images at multiple time points, making it difficult to detect potential feature variability between scanners and manufacturers, as well as temporal variability. Positively, all but twelve studies performed feature reduction, which is consistent with the third RQS checkpoint. In fact, excessive dimensionality of features can negatively affect model performance and lead to overfitting[33]. The RQS showed high variability in items 6, 7 and 8. However, it is important to note that these items are highly dependent on the aim of the study.
Another notable finding from our review was that only nine of the studies in the review were prospective studies, which is the highest weighting in the RQS tool. This constitutes a significant drawback in radiological studies since a meticulously planned prospective trial serves to diminish and control potential confounding factors, thereby offering a superior level of evidence regarding the trial's quality. This elucidates the rationale behind assigning the highest weight (7 points) to studies with a prospective design in the RQS tool, representing approximately 20% of the total score. Thus, this limitation highlights the importance of conducting well-designed prospective studies.
It is noteworthy that nearly half of the examined papers lacked outcome validation which increases the risk of false-positive results and hinders the implementation of radiomics in clinical practice. However, approximately half of the studies that did not validate their results with an independent cohort chose to perform cross-validation.
The majority of the studies did not provide open access to their data sets, segmentations or codes, which limits the ability to verify and reproduce their results[34,35].
Cost-effectiveness analyses that evaluate radiomic prediction models from a health economic perspective when applied in clinical practice have the same limitation. The assumption is that a new predictor should be no more costly than existing predictors, given comparable accuracy. In addition, the health impact of a radiomics predictor is compared to a condition in which no radiomic predictor[2,32]. However, this criterion of RQS is not as important as the need to standardize and validate the models.
As far as we are aware, this is the first systematic review that looks into the possibility of employing MRI radiomics to gather information regarding the management of HCC and to assess studies using the RQS tool.
Previous studies evaluated the quality of radiomic analysis in different studies for different oncologic applications[27-31,36]. Similar to our study, Wakabayashi et al[25] assessed whether radiomics is a valuable and reproducible method for clinical management of HCC using RQS. However, their work included studies up to 2018 and was not focused on MRI modality. In addition, Wang et al[32] also aimed to assess the methodological quality of radiomics studies for HCC management. However, although similar findings with respect to our study were found (mean RQS of 10), their study was focused on the prediction of MVI in HCC patients and also included studies involving other imaging modalities than MRI.
I contrast to most studies that focus on assessing the quality of radiomic studies by means of RQS, our approach involved exploring the potential correlation between RQS and scientometric indixes. Our findings revealed that publications that have higher RQS were published in journals with higher IF and 5-years IF. However, studies with high/low RQS and low/high IF and 5-years IF were also found. Moreover, although no significant correlation was found, it was observed that RQS tended to increase with time (decreasing number of months passed from literature research). Interestingly, we discovered that the quality of included studies increased as the number of included patients and extracted attributes grew.
It is crucial to underscore that only 45 out of 127 studies referenced the Image Biomarker Standardization Initiative (IBSI) guidelines or utilized software for radiomic feature extraction compliant with IBSI standards (e.g., PyRadiomics). Emphasizing the importance of adhering to standardized radiomic features nomenclature and calculation according to IBSI, our study highlights the need for future research to align with these standards, thus enhancing the reproducibility of scientific researches[37].
Despite the insights gained, our study is not without limitations. The RQS scoring system, as acknowledged in prior research, is not a definitive standard for evaluating radiomics studies and requires ongoing refinement for widespread acceptance in radiology. The existing research is limited by issues including conducting phantom studies across all scanners, applying imaging at multiple time points, and lacking definition for a particular study purpose[38,39]. Additionally, the predominantly retrospective nature of the included studies introduces bias, compounded by the absence of external validation cohorts and comparisons with reference standards, hindering conclusive remarks on the efficacy of MRI radiomics in HCC[40,41]. Variability in sample size, inclusion criteria, and methodological settings across studies precluded a meta-analysis aligned with study objectives. Furthermore, the study did not explore specific shared radiomic features among different studies, considering the wide-ranging variability in imaging protocols and software for feature extraction.
In summary, despite the potential of recent developments in MRI radiomics to fulfill the urgent requirement for noninvasive, radiation-free, and quantitative approaches to support decision-making in HCC treatment, the current studies in this domain lack the requisite quality for integration into clinical practice. Emphasizing the significance of external validation, addressing concerns related to feature reproducibility, conducting clinical utility analyses, and fostering scientific openness are crucial steps that need to be addressed. This endeavor aims to provide fresh perspectives and contribute to the establishment of a consensus regarding the application of the radiomic method in assessing HCC.
Radiomics is a promising tool that may increase the value of Magnetic Resonance Imaging (MRI) for different tasks linked to the management of patients with hepatocellular carcinoma (HCC).
Over the last decade, there has been a substantial increase in radiomics studies in the field of HCC. Many of these studies have demonstrated the power of radiomic features for differential diagnosis, grading, predicting microvascular invasion, overall survival, recurrence, and treatment response. However, the use of radiomics in HCC is currently limited to academic literature, and no studies have yet been translated into clinical applications. This has led to doubts among clinicians about the radiomics validity. This is in part due to many issues related to the methodological quality of radiomic studies.
To summarize the status of MRI radiomic studies concerning HCC, using the radiomics quality score (RQS) to assess the quality of the methodology used in each study.
We systematically reviewed PubMed, Google Scholar, and Web of Science databases to identify original articles focused on using MRI radiomics for HCC management published between 2017 and 2023. The RQS tool was employed to evaluate the methodological quality of radiomic studies. Spearman’s correlation (ρ) analysis was conducted to investigate the association between RQS and journal metrics, as well as the characteristics of the studies. The threshold for statistical significance was established at P < 0.05.
127 articles were included, of which 43 focused on HCC prognosis, 39 on prediction of pathological findings, 16 on prediction of the expression of molecular markers outcomes, 18 had a diagnostic purpose, and 11 had multiple aims. Mean RQS was 8 ± 6.22, with the corresponding percentage of 24.15% ± 15.25% (ranging from 0.0 to 58.33%). RQS was positively correlated with journal impact factor (IF; ρ = 0.36, P =2.98 × 10-5), 5-years IF (ρ = 0.33, P = 1.56 × 10-4), number of patients involved (ρ = 0.51, P < 9.37 × 10-10) and number of radiomics features (ρ = 0.59, P < 4.59 × 10-13) extracted in the study, and time of publication (ρ = -0.23, P = 0.0072).
Although the MRI radiomics in HCC represents an auspicious tool for developing adequate personalized treatment as a noninvasive approach in HCC patients, our study revealed that studies in this field still lack the quality required to allow its introduction in clinical practice.
Although recent advantages in MRI radiomics can potentially satisfy the urgent need for noninvasive, radiation-free and quantitative strategies that can aid in HCC treatment decision making, studies in this field still lack the quality required to allow its introduction in clinical practice. Future studies including external validation and adhering to the standardization of radiomics features are necessary. Moreover, limitations and challenges related to feature reproducibility, analysis of the clinical utility, and openness of science need to be addressed. This work may provide new insights and contribute to a common understanding of the use of radiomics in the assessment of HCC.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Italy
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Wang W, China; Wang SM, China S-Editor: Lin C L-Editor: A P-Editor: Yu HG
| 1. | van Timmeren JE, Cester D, Tanadini-Lang S, Alkadhi H, Baessler B. Radiomics in medical imaging-"how-to" guide and critical reflection. Insights Imaging. 2020;11:91. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 588] [Cited by in RCA: 771] [Article Influence: 154.2] [Reference Citation Analysis (0)] |
| 2. | Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, Sanduleanu S, Larue RTHM, Even AJG, Jochems A, van Wijk Y, Woodruff H, van Soest J, Lustberg T, Roelofs E, van Elmpt W, Dekker A, Mottaghy FM, Wildberger JE, Walsh S. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749-762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1825] [Cited by in RCA: 3570] [Article Influence: 446.3] [Reference Citation Analysis (0)] |
| 3. | Lu C, Shiradkar R, Liu Z. Integrating pathomics with radiomics and genomics for cancer prognosis: A brief review. Chin J Cancer Res. 2021;33:563-573. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 4. | Bodalal Z, Trebeschi S, Nguyen-Kim TDL, Schats W, Beets-Tan R. Radiogenomics: bridging imaging and genomics. Abdom Radiol (NY). 2019;44:1960-1984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 190] [Article Influence: 31.7] [Reference Citation Analysis (0)] |
| 5. | Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology. 2016;278:563-577. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4541] [Cited by in RCA: 5559] [Article Influence: 617.7] [Reference Citation Analysis (3)] |
| 6. | Limkin EJ, Sun R, Dercle L, Zacharaki EI, Robert C, Reuzé S, Schernberg A, Paragios N, Deutsch E, Ferté C. Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann Oncol. 2017;28:1191-1206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 345] [Cited by in RCA: 553] [Article Influence: 79.0] [Reference Citation Analysis (0)] |
| 7. | Barcena-Varela M, Lujambio A. The Endless Sources of Hepatocellular Carcinoma Heterogeneity. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 8. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64698] [Article Influence: 16174.5] [Reference Citation Analysis (177)] |
| 9. | Vogel A, Cervantes A, Chau I, Daniele B, Llovet JM, Meyer T, Nault JC, Neumann U, Ricke J, Sangro B, Schirmacher P, Verslype C, Zech CJ, Arnold D, Martinelli E; ESMO Guidelines Committee. Hepatocellular carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018;29:iv238-iv255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 595] [Cited by in RCA: 723] [Article Influence: 103.3] [Reference Citation Analysis (0)] |
| 10. | Osho A, Rich NE, Singal AG. Role of imaging in management of hepatocellular carcinoma: surveillance, diagnosis, and treatment response. Hepatoma Res. 2020;6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 11. | European Association For The Study Of The Liver; European Organisation For Research And Treatment Of Cancer. EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56:908-943. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4059] [Cited by in RCA: 4521] [Article Influence: 347.8] [Reference Citation Analysis (2)] |
| 12. | Navin PJ, Venkatesh SK. Hepatocellular Carcinoma: State of the Art Imaging and Recent Advances. J Clin Transl Hepatol. 2019;7:72-85. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 13. | Chernyak V, Fowler KJ, Kamaya A, Kielar AZ, Elsayes KM, Bashir MR, Kono Y, Do RK, Mitchell DG, Singal AG, Tang A, Sirlin CB. Liver Imaging Reporting and Data System (LI-RADS) Version 2018: Imaging of Hepatocellular Carcinoma in At-Risk Patients. Radiology. 2018;289:816-830. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 356] [Cited by in RCA: 785] [Article Influence: 112.1] [Reference Citation Analysis (0)] |
| 14. | Choi JY, Lee JM, Sirlin CB. CT and MR imaging diagnosis and staging of hepatocellular carcinoma: part I. Development, growth, and spread: key pathologic and imaging aspects. Radiology. 2014;272:635-654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 371] [Cited by in RCA: 351] [Article Influence: 31.9] [Reference Citation Analysis (1)] |
| 15. | Choi JY, Lee JM, Sirlin CB. CT and MR imaging diagnosis and staging of hepatocellular carcinoma: part II. Extracellular agents, hepatobiliary agents, and ancillary imaging features. Radiology. 2014;273:30-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 324] [Cited by in RCA: 394] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 16. | Kong C, Zhao Z, Chen W, Lv X, Shu G, Ye M, Song J, Ying X, Weng Q, Weng W, Fang S, Chen M, Tu J, Ji J. Prediction of tumor response via a pretreatment MRI radiomics-based nomogram in HCC treated with TACE. Eur Radiol. 2021;31:7500-7511. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 86] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 17. | Harding-Theobald E, Louissaint J, Maraj B, Cuaresma E, Townsend W, Mendiratta-Lala M, Singal AG, Su GL, Lok AS, Parikh ND. Systematic review: radiomics for the diagnosis and prognosis of hepatocellular carcinoma. Aliment Pharmacol Ther. 2021;54:890-901. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 91] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 18. | Tang YY, Zhao YN, Zhang T, Chen ZY, Ma XL. Comprehensive radiomics nomogram for predicting survival of patients with combined hepatocellular carcinoma and cholangiocarcinoma. World J Gastroenterol. 2021;27:7173-7189. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 5] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 19. | Chen W, Zhang T, Xu L, Zhao L, Liu H, Gu LR, Wang DZ, Zhang M. Radiomics Analysis of Contrast-Enhanced CT for Hepatocellular Carcinoma Grading. Front Oncol. 2021;11:660509. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 20. | Vallières M, Zwanenburg A, Badic B, Cheze Le Rest C, Visvikis D, Hatt M. Responsible Radiomics Research for Faster Clinical Translation. J Nucl Med. 2018;59:189-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 153] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 21. | Shaikh F, Franc B, Allen E, Sala E, Awan O, Hendrata K, Halabi S, Mohiuddin S, Malik S, Hadley D, Shrestha R. Translational Radiomics: Defining the Strategy Pipeline and Considerations for Application-Part 2: From Clinical Implementation to Enterprise. J Am Coll Radiol. 2018;15:543-549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 22. | Shaikh F, Franc B, Allen E, Sala E, Awan O, Hendrata K, Halabi S, Mohiuddin S, Malik S, Hadley D, Shrestha R. Translational Radiomics: Defining the Strategy Pipeline and Considerations for Application-Part 1: From Methodology to Clinical Implementation. J Am Coll Radiol. 2018;15:538-542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 23. | Sagir Kahraman A. Radiomics in Hepatocellular Carcinoma. J Gastrointest Cancer. 2020;51:1165-1168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 24. | Miranda Magalhaes Santos JM, Clemente Oliveira B, Araujo-Filho JAB, Assuncao-Jr AN, de M Machado FA, Carlos Tavares Rocha C, Horvat JV, Menezes MR, Horvat N. State-of-the-art in radiomics of hepatocellular carcinoma: a review of basic principles, applications, and limitations. Abdom Radiol (NY). 2020;45:342-353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 25. | Wakabayashi T, Ouhmich F, Gonzalez-Cabrera C, Felli E, Saviano A, Agnus V, Savadjiev P, Baumert TF, Pessaux P, Marescaux J, Gallix B. Radiomics in hepatocellular carcinoma: a quantitative review. Hepatol Int. 2019;13:546-559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 110] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 26. | Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, Shamseer L, Tetzlaff JM, Akl EA, Brennan SE, Chou R, Glanville J, Grimshaw JM, Hróbjartsson A, Lalu MM, Li T, Loder EW, Mayo-Wilson E, McDonald S, McGuinness LA, Stewart LA, Thomas J, Tricco AC, Welch VA, Whiting P, Moher D. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372:n71. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44932] [Cited by in RCA: 40624] [Article Influence: 10156.0] [Reference Citation Analysis (2)] |
| 27. | Stanzione A, Gambardella M, Cuocolo R, Ponsiglione A, Romeo V, Imbriaco M. Prostate MRI radiomics: A systematic review and radiomic quality score assessment. Eur J Radiol. 2020;129:109095. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 88] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 28. | Granzier RWY, van Nijnatten TJA, Woodruff HC, Smidt ML, Lobbes MBI. Exploring breast cancer response prediction to neoadjuvant systemic therapy using MRI-based radiomics: A systematic review. Eur J Radiol. 2019;121:108736. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 61] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 29. | Ursprung S, Beer L, Bruining A, Woitek R, Stewart GD, Gallagher FA, Sala E. Radiomics of computed tomography and magnetic resonance imaging in renal cell carcinoma-a systematic review and meta-analysis. Eur Radiol. 2020;30:3558-3566. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 106] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 30. | Park JE, Kim HS, Kim D, Park SY, Kim JY, Cho SJ, Kim JH. A systematic review reporting quality of radiomics research in neuro-oncology: toward clinical utility and quality improvement using high-dimensional imaging features. BMC Cancer. 2020;20:29. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 83] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 31. | Chen Q, Zhang L, Mo X, You J, Chen L, Fang J, Wang F, Jin Z, Zhang B, Zhang S. Current status and quality of radiomic studies for predicting immunotherapy response and outcome in patients with non-small cell lung cancer: a systematic review and meta-analysis. Eur J Nucl Med Mol Imaging. 2021;49:345-360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 47] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 32. | Wang Q, Li C, Zhang J, Hu X, Fan Y, Ma K, Sparrelid E, Brismar TB. Radiomics Models for Predicting Microvascular Invasion in Hepatocellular Carcinoma: A Systematic Review and Radiomics Quality Score Assessment. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 36] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 33. | Park JE, Park SY, Kim HJ, Kim HS. Reproducibility and Generalizability in Radiomics Modeling: Possible Strategies in Radiologic and Statistical Perspectives. Korean J Radiol. 2019;20:1124-1137. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 185] [Cited by in RCA: 241] [Article Influence: 40.2] [Reference Citation Analysis (0)] |
| 34. | Vesteghem C, Brøndum RF, Sønderkær M, Sommer M, Schmitz A, Bødker JS, Dybkær K, El-Galaly TC, Bøgsted M. Implementing the FAIR Data Principles in precision oncology: review of supporting initiatives. Brief Bioinform. 2020;21:936-945. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 35] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 35. | Hasselbring W, Carr L, Hettrick S, Packer H, Tiropanis T. From FAIR research data toward FAIR and open research software. it – Inf Technol. 2020;62:39-47. [RCA] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 36. | Wang H, Zhou Y, Li L, Hou W, Ma X, Tian R. Current status and quality of radiomics studies in lymphoma: a systematic review. Eur Radiol. 2020;30:6228-6240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 44] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 37. | Zwanenburg A, Vallières M, Abdalah MA, Aerts HJWL, Andrearczyk V, Apte A, Ashrafinia S, Bakas S, Beukinga RJ, Boellaard R, Bogowicz M, Boldrini L, Buvat I, Cook GJR, Davatzikos C, Depeursinge A, Desseroit MC, Dinapoli N, Dinh CV, Echegaray S, El Naqa I, Fedorov AY, Gatta R, Gillies RJ, Goh V, Götz M, Guckenberger M, Ha SM, Hatt M, Isensee F, Lambin P, Leger S, Leijenaar RTH, Lenkowicz J, Lippert F, Losnegård A, Maier-Hein KH, Morin O, Müller H, Napel S, Nioche C, Orlhac F, Pati S, Pfaehler EAG, Rahmim A, Rao AUK, Scherer J, Siddique MM, Sijtsema NM, Socarras Fernandez J, Spezi E, Steenbakkers RJHM, Tanadini-Lang S, Thorwarth D, Troost EGC, Upadhaya T, Valentini V, van Dijk LV, van Griethuysen J, van Velden FHP, Whybra P, Richter C, Löck S. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology. 2020;295:328-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2090] [Cited by in RCA: 2321] [Article Influence: 464.2] [Reference Citation Analysis (0)] |
| 38. | Sanduleanu S, Woodruff HC, de Jong EEC, van Timmeren JE, Jochems A, Dubois L, Lambin P. Tracking tumor biology with radiomics: A systematic review utilizing a radiomics quality score. Radiother Oncol. 2018;127:349-360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 172] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 39. | Park JE, Kim D, Kim HS, Park SY, Kim JY, Cho SJ, Shin JH, Kim JH. Quality of science and reporting of radiomics in oncologic studies: room for improvement according to radiomics quality score and TRIPOD statement. Eur Radiol. 2020;30:523-536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 182] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 40. | Norvell DC. Study types and bias-Don't judge a study by the abstract's conclusion alone. Evid Based Spine Care J. 2010;1:7-10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 41] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 41. | Tripepi G, Jager KJ, Dekker FW, Zoccali C. Selection bias and information bias in clinical research. Nephron Clin Pract. 2010;115:c94-c99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 268] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 42. | Liu J, Cheng D, Liao Y, Luo C, Lei Q, Zhang X, Wang L, Wen Z, Gao M. Development of a magnetic resonance imaging-derived radiomics model to predict microvascular invasion in patients with hepatocellular carcinoma. Quant Imaging Med Surg. 2023;13:3948-3961. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 43. | Wang Y, Liu Z, Xu H, Yang D, Jiang J, Asayo H, Yang Z. MRI-based radiomics model and nomogram for predicting the outcome of locoregional treatment in patients with hepatocellular carcinoma. BMC Med Imaging. 2023;23:67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 44. | Gong XQ, Liu N, Tao YY, Li L, Li ZM, Yang L, Zhang XM. Radiomics models based on multisequence MRI for predicting PD-1/PD-L1 expression in hepatocellular carcinoma. Sci Rep. 2023;13:7710. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Reference Citation Analysis (0)] |
| 45. | Zhang L, Zhou H, Zhang X, Ding Z, Xu J. A radiomics nomogram for predicting cytokeratin 19-positive hepatocellular carcinoma: a two-center study. Front Oncol. 2023;13:1174069. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 46. | Zhang Y, He D, Liu J, Wei YG, Shi LL. Preoperative prediction of macrotrabecular-massive hepatocellular carcinoma through dynamic contrast-enhanced magnetic resonance imaging-based radiomics. World J Gastroenterol. 2023;29:2001-2014. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 47. | Dong X, Yang J, Zhang B, Li Y, Wang G, Chen J, Wei Y, Zhang H, Chen Q, Jin S, Wang L, He H, Gan M, Ji W. Deep Learning Radiomics Model of Dynamic Contrast-Enhanced MRI for Evaluating Vessels Encapsulating Tumor Clusters and Prognosis in Hepatocellular Carcinoma. J Magn Reson Imaging. 2024;59:108-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 23] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 48. | Tabari A, D'Amore B, Cox M, Brito S, Gee MS, Wehrenberg-Klee E, Uppot RN, Daye D. Machine Learning-Based Radiomic Features on Pre-Ablation MRI as Predictors of Pathologic Response in Patients with Hepatocellular Carcinoma Who Underwent Hepatic Transplant. Cancers (Basel). 2023;15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 49. | Cao X, Yang H, Luo X, Zou L, Zhang Q, Li Q, Zhang J, Li X, Shi Y, Jin C. A Cox Nomogram for Assessing Recurrence Free Survival in Hepatocellular Carcinoma Following Surgical Resection Using Dynamic Contrast-Enhanced MRI Radiomics. J Magn Reson Imaging. 2023;58:1930-1941. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 50. | İnce O, Önder H, Gençtürk M, Cebeci H, Golzarian J, Young S. Prediction of Response of Hepatocellular Carcinoma to Radioembolization: Machine Learning Using Preprocedural Clinical Factors and MR Imaging Radiomics. J Vasc Interv Radiol. 2023;34:235-243.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 51. | Chen M, Kong C, Qiao E, Chen Y, Chen W, Jiang X, Fang S, Zhang D, Chen M, Ji J. Multi-algorithms analysis for pre-treatment prediction of response to transarterial chemoembolization in hepatocellular carcinoma on multiphase MRI. Insights Imaging. 2023;14:38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 52. | Jiang T, He S, Yang H, Dong Y, Yu T, Luo Y, Jiang X. Multiparametric MRI-based radiomics for the prediction of microvascular invasion in hepatocellular carcinoma. Acta Radiol. 2023;64:456-466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 53. | Hu X, Wang Q, Huang G, He X, Sparrelid E, Brismar TB, Fan Y. Gadoxetic Acid-Enhanced MRI-Based Radiomics Signature: A Potential Imaging Biomarker for Identifying Cytokeratin 19-Positive Hepatocellular Carcinoma. Comput Math Methods Med. 2023;2023:5424204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 54. | Chong H, Gong Y, Zhang Y, Dai Y, Sheng R, Zeng M. Radiomics on Gadoxetate Disodium-enhanced MRI: Non-invasively Identifying Glypican 3-Positive Hepatocellular Carcinoma and Postoperative Recurrence. Acad Radiol. 2023;30:49-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 55. | Hu X, Li C, Wang Q, Wu X, Chen Z, Xia F, Cai P, Zhang L, Fan Y, Ma K. Development and External Validation of a Radiomics Model Derived from Preoperative Gadoxetic Acid-Enhanced MRI for Predicting Histopathologic Grade of Hepatocellular Carcinoma. Diagnostics (Basel). 2023;13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 56. | Tao YY, Shi Y, Gong XQ, Li L, Li ZM, Yang L, Zhang XM. Radiomic Analysis Based on Magnetic Resonance Imaging for Predicting PD-L2 Expression in Hepatocellular Carcinoma. Cancers (Basel). 2023;15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 16] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 57. | Yang X, Yuan C, Zhang Y, Li K, Wang Z. Predicting hepatocellular carcinoma early recurrence after ablation based on magnetic resonance imaging radiomics nomogram. Medicine (Baltimore). 2022;101:e32584. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 58. | Liu HF, Zhang YZ, Wang Q, Zhu ZH, Xing W. A nomogram model integrating LI-RADS features and radiomics based on contrast-enhanced magnetic resonance imaging for predicting microvascular invasion in hepatocellular carcinoma falling the Milan criteria. Transl Oncol. 2023;27:101597. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 59. | Zhang S, Duan C, Zhou X, Liu F, Wang X, Shao Q, Gao Y, Duan F, Zhao R, Wang G. Radiomics nomogram for prediction of microvascular invasion in hepatocellular carcinoma based on MR imaging with Gd-EOB-DTPA. Front Oncol. 2022;12:1034519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 60. | Sim JZT, Hui TCH, Chuah TK, Low HM, Tan CH, Shelat VG. Efficacy of texture analysis of pre-operative magnetic resonance imaging in predicting microvascular invasion in hepatocellular carcinoma. World J Clin Oncol. 2022;13:918-928. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 4] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 61. | Zhang X, Wang C, Zheng D, Liao Y, Wang X, Huang Z, Zhong Q. Radiomics nomogram based on multi-parametric magnetic resonance imaging for predicting early recurrence in small hepatocellular carcinoma after radiofrequency ablation. Front Oncol. 2022;12:1013770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 11] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 62. | Zhao Y, Huang F, Liu S, Jian L, Xia X, Lin H, Liu J. Prediction of therapeutic response of unresectable hepatocellular carcinoma to hepatic arterial infusion chemotherapy based on pretherapeutic MRI radiomics and Albumin-Bilirubin score. J Cancer Res Clin Oncol. 2023;149:5181-5192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 15] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 63. | Lu XY, Zhang JY, Zhang T, Zhang XQ, Lu J, Miao XF, Chen WB, Jiang JF, Ding D, Du S. Using pre-operative radiomics to predict microvascular invasion of hepatocellular carcinoma based on Gd-EOB-DTPA enhanced MRI. BMC Med Imaging. 2022;22:157. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 64. | Yang WL, Zhu F, Chen WX. Texture analysis of contrast-enhanced magnetic resonance imaging predicts microvascular invasion in hepatocellular carcinoma. Eur J Radiol. 2022;156:110528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 65. | Ameli S, Venkatesh BA, Shaghaghi M, Ghadimi M, Hazhirkarzar B, Rezvani Habibabadi R, Aliyari Ghasabeh M, Khoshpouri P, Pandey A, Pandey P, Pan L, Grimm R, Kamel IR. Role of MRI-Derived Radiomics Features in Determining Degree of Tumor Differentiation of Hepatocellular Carcinoma. Diagnostics (Basel). 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 66. | Li W, Shen H, Han L, Liu J, Xiao B, Li X, Ye Z. A Multiparametric Fusion Radiomics Signature Based on Contrast-Enhanced MRI for Predicting Early Recurrence of Hepatocellular Carcinoma. J Oncol. 2022;2022:3704987. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 67. | Zeng F, Dai H, Li X, Guo L, Jia N, Yang J, Huang D, Zeng H, Chen W, Zhang L, Qin G. Preoperative radiomics model using gadobenate dimeglumine-enhanced magnetic resonance imaging for predicting β-catenin mutation in patients with hepatocellular carcinoma: A retrospective study. Front Oncol. 2022;12:916126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 68. | Aujay G, Etchegaray C, Blanc JF, Lapuyade B, Papadopoulos P, Pey MA, Bordenave L, Trillaud H, Saut O, Pinaquy JB. Comparison of MRI-based response criteria and radiomics for the prediction of early response to transarterial radioembolization in patients with hepatocellular carcinoma. Diagn Interv Imaging. 2022;103:360-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 29] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 69. | Chen YD, Zhang L, Zhou ZP, Lin B, Jiang ZJ, Tang C, Dang YW, Xia YW, Song B, Long LL. Radiomics and nomogram of magnetic resonance imaging for preoperative prediction of microvascular invasion in small hepatocellular carcinoma. World J Gastroenterol. 2022;28:4399-4416. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 16] [Cited by in RCA: 19] [Article Influence: 6.3] [Reference Citation Analysis (1)] |
| 70. | Wu Q, Yu YX, Zhang T, Zhu WJ, Fan YF, Wang XM, Hu CH. Preoperative Diagnosis of Dual-Phenotype Hepatocellular Carcinoma Using Enhanced MRI Radiomics Models. J Magn Reson Imaging. 2023;57:1185-1196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 11] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 71. | Li YM, Zhu YM, Gao LM, Han ZW, Chen XJ, Yan C, Ye RP, Cao DR. Radiomic analysis based on multi-phase magnetic resonance imaging to predict preoperatively microvascular invasion in hepatocellular carcinoma. World J Gastroenterol. 2022;28:2733-2747. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 2] [Cited by in RCA: 7] [Article Influence: 2.3] [Reference Citation Analysis (1)] |
| 72. | Wang L, Ma X, Feng B, Wang S, Liang M, Li D, Zhao X. Multi-Sequence MR-Based Radiomics Signature for Predicting Early Recurrence in Solitary Hepatocellular Carcinoma ≤5 cm. Front Oncol. 2022;12:899404. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 73. | Zhang HD, Li XM, Zhang YH, Hu F, Tan L, Wang F, Jing Y, Guo DJ, Xu Y, Hu XL, Liu C, Wang J. Evaluation of Preoperative Microvascular Invasion in Hepatocellular Carcinoma Through Multidimensional Parameter Combination Modeling Based on Gd-EOB-DTPA MRI. J Clin Transl Hepatol. 2023;11:350-359. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 74. | Brancato V, Garbino N, Salvatore M, Cavaliere C. MRI-Based Radiomic Features Help Identify Lesions and Predict Histopathological Grade of Hepatocellular Carcinoma. Diagnostics (Basel). 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 75. | Fan T, Li S, Li K, Xu J, Zhao S, Li J, Zhou X, Jiang H. A Potential Prognostic Marker for Recognizing VEGF-Positive Hepatocellular Carcinoma Based on Magnetic Resonance Radiomics Signature. Front Oncol. 2022;12:857715. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 76. | Gao L, Xiong M, Chen X, Han Z, Yan C, Ye R, Zhou L, Li Y. Multi-Region Radiomic Analysis Based on Multi-Sequence MRI Can Preoperatively Predict Microvascular Invasion in Hepatocellular Carcinoma. Front Oncol. 2022;12:818681. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 77. | Hu F, Zhang Y, Li M, Liu C, Zhang H, Li X, Liu S, Hu X, Wang J. Preoperative Prediction of Microvascular Invasion Risk Grades in Hepatocellular Carcinoma Based on Tumor and Peritumor Dual-Region Radiomics Signatures. Front Oncol. 2022;12:853336. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 78. | He Y, Hu B, Zhu C, Xu W, Ge Y, Hao X, Dong B, Chen X, Dong Q, Zhou X. A Novel Multimodal Radiomics Model for Predicting Prognosis of Resected Hepatocellular Carcinoma. Front Oncol. 2022;12:745258. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 79. | Ren Y, Bo L, Shen B, Yang J, Xu S, Shen W, Chen H, Wang X, Cai X. Development and validation of a clinical-radiomics model to predict recurrence for patients with hepatocellular carcinoma after curative resection. Med Phys. 2023;50:778-790. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 80. | Luo J, Huang Z, Wang M, Li T, Huang J. Prognostic role of multiparameter MRI and radiomics in progression of advanced unresectable hepatocellular carcinoma following combined transcatheter arterial chemoembolization and lenvatinib therapy. BMC Gastroenterol. 2022;22:108. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 81. | Wang X, Sun Y, Zhou X, Shen Z, Zhang H, Xing J, Zhou Y. Histogram peritumoral enhanced features on MRI arterial phase with extracellular contrast agent can improve prediction of microvascular invasion of hepatocellular carcinoma. Quant Imaging Med Surg. 2022;12:1372-1384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 82. | Mao Y, Wang J, Zhu Y, Chen J, Mao L, Kong W, Qiu Y, Wu X, Guan Y, He J. Gd-EOB-DTPA-enhanced MRI radiomic features for predicting histological grade of hepatocellular carcinoma. Hepatobiliary Surg Nutr. 2022;11:13-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 30] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 83. | Andersson M, Jalnefjord O, Montelius M, Rizell M, Sternby Eilard M, Ljungberg M. Evaluation of response in patients with hepatocellular carcinoma treated with intratumoral dendritic cell vaccination using intravoxel incoherent motion (IVIM) MRI and histogram analysis. Acta Radiol. 2023;64:32-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 84. | Li H, Wang L, Zhang J, Duan Q, Xu Y, Xue Y. Evaluation of microvascular invasion of hepatocellular carcinoma using whole-lesion histogram analysis with the stretched-exponential diffusion model. Br J Radiol. 2022;95:20210631. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 85. | Li L, Su Q, Yang H. Preoperative prediction of microvascular invasion in hepatocellular carcinoma: a radiomic nomogram based on MRI. Clin Radiol. 2022;77:e269-e279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 86. | Wang X, Wang S, Yin X, Zheng Y. MRI-based radiomics distinguish different pathological types of hepatocellular carcinoma. Comput Biol Med. 2022;141:105058. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (61)] |
| 87. | Yang Y, Fan W, Gu T, Yu L, Chen H, Lv Y, Liu H, Wang G, Zhang D. Radiomic Features of Multi-ROI and Multi-Phase MRI for the Prediction of Microvascular Invasion in Solitary Hepatocellular Carcinoma. Front Oncol. 2021;11:756216. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 88. | Lv X, Chen M, Kong C, Shu G, Meng M, Ye W, Cheng S, Zheng L, Fang S, Chen C, Wu F, Weng Q, Tu J, Zhao Z, Ji J. Construction of a novel radiomics nomogram for the prediction of aggressive intrasegmental recurrence of HCC after radiofrequency ablation. Eur J Radiol. 2021;144:109955. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 89. | Yu Y, Fan Y, Wang X, Zhu M, Hu M, Shi C, Hu C. Gd-EOB-DTPA-enhanced MRI radiomics to predict vessels encapsulating tumor clusters (VETC) and patient prognosis in hepatocellular carcinoma. Eur Radiol. 2022;32:959-970. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 58] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 90. | Fang S, Lai L, Zhu J, Zheng L, Xu Y, Chen W, Wu F, Wu X, Chen M, Weng Q, Ji J, Zhao Z, Tu J. A Radiomics Signature-Based Nomogram to Predict the Progression-Free Survival of Patients With Hepatocellular Carcinoma After Transcatheter Arterial Chemoembolization Plus Radiofrequency Ablation. Front Mol Biosci. 2021;8:662366. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 91. | Yang F, Wan Y, Xu L, Wu Y, Shen X, Wang J, Lu D, Shao C, Zheng S, Niu T, Xu X. MRI-Radiomics Prediction for Cytokeratin 19-Positive Hepatocellular Carcinoma: A Multicenter Study. Front Oncol. 2021;11:672126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 92. | Chen Y, Chen J, Zhang Y, Lin Z, Wang M, Huang L, Huang M, Tang M, Zhou X, Peng Z, Huang B, Feng ST. Preoperative Prediction of Cytokeratin 19 Expression for Hepatocellular Carcinoma with Deep Learning Radiomics Based on Gadoxetic Acid-Enhanced Magnetic Resonance Imaging. J Hepatocell Carcinoma. 2021;8:795-808. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 93. | Horvat N, Araujo-Filho JAB, Assuncao-Jr AN, Machado FAM, Sims JA, Rocha CCT, Oliveira BC, Horvat JV, Maccali C, Puga ALBL, Chagas AL, Menezes MR, Cerri GG. Radiomic analysis of MRI to Predict Sustained Complete Response after Radiofrequency Ablation in Patients with Hepatocellular Carcinoma - A Pilot Study. Clinics (Sao Paulo). 2021;76:e2888. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 94. | Alksas A, Shehata M, Saleh GA, Shaffie A, Soliman A, Ghazal M, Khelifi A, Khalifeh HA, Razek AA, Giridharan GA, El-Baz A. A novel computer-aided diagnostic system for accurate detection and grading of liver tumors. Sci Rep. 2021;11:13148. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 95. | Chong H, Gong Y, Pan X, Liu A, Chen L, Yang C, Zeng M. Peritumoral Dilation Radiomics of Gadoxetate Disodium-Enhanced MRI Excellently Predicts Early Recurrence of Hepatocellular Carcinoma without Macrovascular Invasion After Hepatectomy. J Hepatocell Carcinoma. 2021;8:545-563. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 37] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 96. | Ding Z, Lin K, Fu J, Huang Q, Fang G, Tang Y, You W, Lin Z, Pan X, Zeng Y. An MR-based radiomics model for differentiation between hepatocellular carcinoma and focal nodular hyperplasia in non-cirrhotic liver. World J Surg Oncol. 2021;19:181. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 97. | Fan Y, Yu Y, Wang X, Hu M, Hu C. Radiomic analysis of Gd-EOB-DTPA-enhanced MRI predicts Ki-67 expression in hepatocellular carcinoma. BMC Med Imaging. 2021;21:100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 33] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 98. | Gao F, Qiao K, Yan B, Wu M, Wang L, Chen J, Shi D. Hybrid network with difference degree and attention mechanism combined with radiomics (H-DARnet) for MVI prediction in HCC. Magn Reson Imaging. 2021;83:27-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 99. | Li X, Cheng L, Li C, Hu X, Tan L, Li Q, Liu C, Wang J. Associating Preoperative MRI Features and Gene Expression Signatures of Early-stage Hepatocellular Carcinoma Patients using Machine Learning. J Clin Transl Hepatol. 2022;10:63-71. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 100. | Shi Z, Cai W, Feng X, Cai J, Liang Y, Xu J, Zhen J, Liang X. Radiomics Analysis of Gd-EOB-DTPA Enhanced Hepatic MRI for Assessment of Functional Liver Reserve. Acad Radiol. 2022;29:213-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 101. | Dai H, Lu M, Huang B, Tang M, Pang T, Liao B, Cai H, Huang M, Zhou Y, Chen X, Ding H, Feng ST. Considerable effects of imaging sequences, feature extraction, feature selection, and classifiers on radiomics-based prediction of microvascular invasion in hepatocellular carcinoma using magnetic resonance imaging. Quant Imaging Med Surg. 2021;11:1836-1853. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 37] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 102. | Fan Y, Yu Y, Wang X, Hu M, Du M, Guo L, Sun S, Hu C. Texture Analysis Based on Gd-EOB-DTPA-Enhanced MRI for Identifying Vessels Encapsulating Tumor Clusters (VETC)-Positive Hepatocellular Carcinoma. J Hepatocell Carcinoma. 2021;8:349-359. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 103. | Yang X, Yuan C, Zhang Y, Wang Z. Magnetic resonance radiomics signatures for predicting poorly differentiated hepatocellular carcinoma: A SQUIRE-compliant study. Medicine (Baltimore). 2021;100:e25838. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 104. | Chen Y, Xia Y, Tolat PP, Long L, Jiang Z, Huang Z, Tang Q. Comparison of Conventional Gadoxetate Disodium-Enhanced MRI Features and Radiomics Signatures With Machine Learning for Diagnosing Microvascular Invasion. AJR Am J Roentgenol. 2021;216:1510-1520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 105. | Zhao J, Gao S, Sun W, Grimm R, Fu C, Han J, Sheng R, Zeng M. Magnetic resonance imaging and diffusion-weighted imaging-based histogram analyses in predicting glypican 3-positive hepatocellular carcinoma. Eur J Radiol. 2021;139:109732. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 106. | Song D, Wang Y, Wang W, Cai J, Zhu K, Lv M, Gao Q, Zhou J, Fan J, Rao S, Wang M, Wang X. Using deep learning to predict microvascular invasion in hepatocellular carcinoma based on dynamic contrast-enhanced MRI combined with clinical parameters. J Cancer Res Clin Oncol. 2021;147:3757-3767. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 60] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 107. | Zhong X, Guan T, Tang D, Li J, Lu B, Cui S, Tang H. Differentiation of small (≤ 3 cm) hepatocellular carcinomas from benign nodules in cirrhotic liver: the added additive value of MRI-based radiomics analysis to LI-RADS version 2018 algorithm. BMC Gastroenterol. 2021;21:155. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 108. | Chen Y, Liu Z, Mo Y, Li B, Zhou Q, Peng S, Li S, Kuang M. Prediction of Post-hepatectomy Liver Failure in Patients With Hepatocellular Carcinoma Based on Radiomics Using Gd-EOB-DTPA-Enhanced MRI: The Liver Failure Model. Front Oncol. 2021;11:605296. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 109. | Liang H, Hu C, Lu J, Zhang T, Jiang J, Ding D, Du S, Duan S. Correlation of radiomic features on dynamic contrast-enhanced magnetic resonance with microvessel density in hepatocellular carcinoma based on different models. J Int Med Res. 2021;49:300060521997586. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 110. | Zhang L, Hu J, Hou J, Jiang X, Guo L, Tian L. Radiomics-based model using gadoxetic acid disodium-enhanced MR images: associations with recurrence-free survival of patients with hepatocellular carcinoma treated by surgical resection. Abdom Radiol (NY). 2021;46:3845-3854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 111. | Zhang L, Cai P, Hou J, Luo M, Li Y, Jiang X. Radiomics Model Based on Gadoxetic Acid Disodium-Enhanced MR Imaging to Predict Hepatocellular Carcinoma Recurrence After Curative Ablation. Cancer Manag Res. 2021;13:2785-2796. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 112. | Zhang Y, Shu Z, Ye Q, Chen J, Zhong J, Jiang H, Wu C, Yu T, Pang P, Ma T, Lin C. Preoperative Prediction of Microvascular Invasion in Hepatocellular Carcinoma via Multi-Parametric MRI Radiomics. Front Oncol. 2021;11:633596. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 113. | Zhao Y, Wang N, Wu J, Zhang Q, Lin T, Yao Y, Chen Z, Wang M, Sheng L, Liu J, Song Q, Wang F, An X, Guo Y, Li X, Wu T, Liu AL. Radiomics Analysis Based on Contrast-Enhanced MRI for Prediction of Therapeutic Response to Transarterial Chemoembolization in Hepatocellular Carcinoma. Front Oncol. 2021;11:582788. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 114. | Kuang Y, Li R, Jia P, Ye W, Zhou R, Zhu R, Wang J, Lin S, Pang P, Ji W. MRI-Based Radiomics: Nomograms predicting the short-term response after transcatheter arterial chemoembolization (TACE) in hepatocellular carcinoma patients with diameter less than 5 cm. Abdom Radiol (NY). 2021;46:3772-3789. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 33] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 115. | Meng XP, Wang YC, Zhou JY, Yu Q, Lu CQ, Xia C, Tang TY, Xu J, Sun K, Xiao W, Ju S. Comparison of MRI and CT for the Prediction of Microvascular Invasion in Solitary Hepatocellular Carcinoma Based on a Non-Radiomics and Radiomics Method: Which Imaging Modality Is Better? J Magn Reson Imaging. 2021;54:526-536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 37] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 116. | Zhu Y, Weng S, Li Y, Yan C, Ye R, Wen L, Zhou L, Gao L. A radiomics nomogram based on contrast-enhanced MRI for preoperative prediction of macrotrabecular-massive hepatocellular carcinoma. Abdom Radiol (NY). 2021;46:3139-3148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 117. | Liu J, Pei Y, Zhang Y, Wu Y, Liu F, Gu S. Predicting the prognosis of hepatocellular carcinoma with the treatment of transcatheter arterial chemoembolization combined with microwave ablation using pretreatment MR imaging texture features. Abdom Radiol (NY). 2021;46:3748-3757. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 118. | Chong HH, Yang L, Sheng RF, Yu YL, Wu DJ, Rao SX, Yang C, Zeng MS. Multi-scale and multi-parametric radiomics of gadoxetate disodium-enhanced MRI predicts microvascular invasion and outcome in patients with solitary hepatocellular carcinoma ≤ 5 cm. Eur Radiol. 2021;31:4824-4838. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 149] [Article Influence: 37.3] [Reference Citation Analysis (1)] |
| 119. | Gu D, Xie Y, Wei J, Li W, Ye Z, Zhu Z, Tian J, Li X. MRI-Based Radiomics Signature: A Potential Biomarker for Identifying Glypican 3-Positive Hepatocellular Carcinoma. J Magn Reson Imaging. 2020;52:1679-1687. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 43] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 120. | Zhao Y, Wu J, Zhang Q, Hua Z, Qi W, Wang N, Lin T, Sheng L, Cui D, Liu J, Song Q, Li X, Wu T, Guo Y, Cui J, Liu A. Radiomics Analysis Based on Multiparametric MRI for Predicting Early Recurrence in Hepatocellular Carcinoma After Partial Hepatectomy. J Magn Reson Imaging. 2021;53:1066-1079. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 55] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 121. | Ai Z, Han Q, Huang Z, Wu J, Xiang Z. The value of multiparametric histogram features based on intravoxel incoherent motion diffusion-weighted imaging (IVIM-DWI) for the differential diagnosis of liver lesions. Ann Transl Med. 2020;8:1128. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 122. | Shaghaghi M, Aliyari Ghasabeh M, Ameli S, Ghadimi M, Hazhirkarzar B, Rezvani Habibabadi R, Khoshpouri P, Pandey A, Pandey P, Kamel IR. Post-TACE changes in ADC histogram predict overall and transplant-free survival in patients with well-defined HCC: a retrospective cohort with up to 10 years follow-up. Eur Radiol. 2021;31:1378-1390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 123. | Li J, Xue F, Xu X, Wang Q, Zhang X. Dynamic contrast-enhanced MRI differentiates hepatocellular carcinoma from hepatic metastasis of rectal cancer by extracting pharmacokinetic parameters and radiomic features. Exp Ther Med. 2020;20:3643-3652. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 124. | Geng Z, Zhang Y, Wang S, Li H, Zhang C, Yin S, Xie C, Dai Y. Radiomics Analysis of Susceptibility Weighted Imaging for Hepatocellular Carcinoma: Exploring the Correlation between Histopathology and Radiomics Features. Magn Reson Med Sci. 2021;20:253-263. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 125. | Zhang J, Wang X, Zhang L, Yao L, Xue X, Zhang S, Li X, Chen Y, Pang P, Sun D, Xu J, Shi Y, Chen F. Radiomics predict postoperative survival of patients with primary liver cancer with different pathological types. Ann Transl Med. 2020;8:820. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 126. | Zhang Z, Chen J, Jiang H, Wei Y, Zhang X, Cao L, Duan T, Ye Z, Yao S, Pan X, Song B. Gadoxetic acid-enhanced MRI radiomics signature: prediction of clinical outcome in hepatocellular carcinoma after surgical resection. Ann Transl Med. 2020;8:870. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 127. | Hectors SJ, Lewis S, Kennedy P, Bane O, Said D, Segall M, Schwartz M, Kim E, Taouli B. Assessment of Hepatocellular Carcinoma Response to (90)Y Radioembolization Using Dynamic Contrast Material-enhanced MRI and Intravoxel Incoherent Motion Diffusion-weighted Imaging. Radiol Imaging Cancer. 2020;2:e190094. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 128. | Shi G, Han X, Wang Q, Ding Y, Liu H, Zhang Y, Dai Y. Evaluation of Multiple Prognostic Factors of Hepatocellular Carcinoma with Intra-Voxel Incoherent Motions Imaging by Extracting the Histogram Metrics. Cancer Manag Res. 2020;12:6019-6031. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 129. | Feng M, Zhang M, Liu Y, Jiang N, Meng Q, Wang J, Yao Z, Gan W, Dai H. Texture analysis of MR images to identify the differentiated degree in hepatocellular carcinoma: a retrospective study. BMC Cancer. 2020;20:611. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (1)] |
| 130. | Nebbia G, Zhang Q, Arefan D, Zhao X, Wu S. Pre-operative Microvascular Invasion Prediction Using Multi-parametric Liver MRI Radiomics. J Digit Imaging. 2020;33:1376-1386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 55] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 131. | Schobert IT, Savic LJ, Chapiro J, Bousabarah K, Chen E, Laage-Gaupp F, Tefera J, Nezami N, Lin M, Pollak J, Schlachter T. Neutrophil-to-lymphocyte and platelet-to-lymphocyte ratios as predictors of tumor response in hepatocellular carcinoma after DEB-TACE. Eur Radiol. 2020;30:5663-5673. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 84] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 132. | Sun Y, Bai H, Xia W, Wang D, Zhou B, Zhao X, Yang G, Xu L, Zhang W, Liu P, Xu J, Meng S, Liu R, Gao X. Predicting the Outcome of Transcatheter Arterial Embolization Therapy for Unresectable Hepatocellular Carcinoma Based on Radiomics of Preoperative Multiparameter MRI. J Magn Reson Imaging. 2020;52:1083-1090. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 44] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 133. | Wilson GC, Cannella R, Fiorentini G, Shen C, Borhani A, Furlan A, Tsung A. Texture analysis on preoperative contrast-enhanced magnetic resonance imaging identifies microvascular invasion in hepatocellular carcinoma. HPB (Oxford). 2020;22:1622-1630. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 134. | Hectors SJ, Lewis S, Besa C, King MJ, Said D, Putra J, Ward S, Higashi T, Thung S, Yao S, Laface I, Schwartz M, Gnjatic S, Merad M, Hoshida Y, Taouli B. MRI radiomics features predict immuno-oncological characteristics of hepatocellular carcinoma. Eur Radiol. 2020;30:3759-3769. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 114] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 135. | Wang W, Gu D, Wei J, Ding Y, Yang L, Zhu K, Luo R, Rao SX, Tian J, Zeng M. A radiomics-based biomarker for cytokeratin 19 status of hepatocellular carcinoma with gadoxetic acid-enhanced MRI. Eur Radiol. 2020;30:3004-3014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 64] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 136. | Wang XH, Long LH, Cui Y, Jia AY, Zhu XG, Wang HZ, Wang Z, Zhan CM, Wang ZH, Wang WH. MRI-based radiomics model for preoperative prediction of 5-year survival in patients with hepatocellular carcinoma. Br J Cancer. 2020;122:978-985. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 91] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 137. | Song W, Yu X, Guo D, Liu H, Tang Z, Liu X, Zhou J, Zhang H, Liu Y. MRI-Based Radiomics: Associations With the Recurrence-Free Survival of Patients With Hepatocellular Carcinoma Treated With Conventional Transcatheter Arterial Chemoembolization. J Magn Reson Imaging. 2020;52:461-473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 65] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 138. | Zhang J, Liu X, Zhang H, He X, Liu Y, Zhou J, Guo D. Texture Analysis Based on Preoperative Magnetic Resonance Imaging (MRI) and Conventional MRI Features for Predicting the Early Recurrence of Single Hepatocellular Carcinoma after Hepatectomy. Acad Radiol. 2019;26:1164-1173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 139. | Huang X, Long L, Wei J, Li Y, Xia Y, Zuo P, Chai X. Radiomics for diagnosis of dual-phenotype hepatocellular carcinoma using Gd-EOB-DTPA-enhanced MRI and patient prognosis. J Cancer Res Clin Oncol. 2019;145:2995-3003. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 140. | Ye Z, Jiang H, Chen J, Liu X, Wei Y, Xia C, Duan T, Cao L, Zhang Z, Song B. Texture analysis on gadoxetic acid enhanced-MRI for predicting Ki-67 status in hepatocellular carcinoma: A prospective study. Chin J Cancer Res. 2019;31:806-817. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 141. | Zhang R, Xu L, Wen X, Zhang J, Yang P, Zhang L, Xue X, Wang X, Huang Q, Guo C, Shi Y, Niu T, Chen F. A nomogram based on bi-regional radiomics features from multimodal magnetic resonance imaging for preoperative prediction of microvascular invasion in hepatocellular carcinoma. Quant Imaging Med Surg. 2019;9:1503-1515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 70] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 142. | Chen J, Wu Z, Xia C, Jiang H, Liu X, Duan T, Cao L, Ye Z, Zhang Z, Ma L, Song B, Shi Y. Noninvasive prediction of HCC with progenitor phenotype based on gadoxetic acid-enhanced MRI. Eur Radiol. 2020;30:1232-1242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 143. | Xu YS, Liu HF, Xi DL, Li JK, Liu Z, Yan RF, Lei JQ. Whole-lesion histogram analysis metrics of the apparent diffusion coefficient: a correlation study with histological grade of hepatocellular carcinoma. Abdom Radiol (NY). 2019;44:3089-3098. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 144. | Li Y, Yan C, Weng S, Shi Z, Sun H, Chen J, Xu X, Ye R, Hong J. Texture analysis of multi-phase MRI images to detect expression of Ki67 in hepatocellular carcinoma. Clin Radiol. 2019;74:813.e19-813.e27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 145. | Oyama A, Hiraoka Y, Obayashi I, Saikawa Y, Furui S, Shiraishi K, Kumagai S, Hayashi T, Kotoku J. Hepatic tumor classification using texture and topology analysis of non-contrast-enhanced three-dimensional T1-weighted MR images with a radiomics approach. Sci Rep. 2019;9:8764. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 56] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 146. | Wang HQ, Yang C, Zeng MS, Rao SX, Ji Y, Weng X, Wang JY, Sheng RF. Magnetic resonance texture analysis for the identification of cytokeratin 19-positive hepatocellular carcinoma. Eur J Radiol. 2019;117:164-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 147. | Zhu YJ, Feng B, Wang S, Wang LM, Wu JF, Ma XH, Zhao XM. Model-based three-dimensional texture analysis of contrast-enhanced magnetic resonance imaging as a potential tool for preoperative prediction of microvascular invasion in hepatocellular carcinoma. Oncol Lett. 2019;18:720-732. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 148. | Zhang Z, Jiang H, Chen J, Wei Y, Cao L, Ye Z, Li X, Ma L, Song B. Hepatocellular carcinoma: radiomics nomogram on gadoxetic acid-enhanced MR imaging for early postoperative recurrence prediction. Cancer Imaging. 2019;19:22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 98] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 149. | Gordic S, Wagner M, Zanato R, Hectors S, Besa C, Kihira S, Kim E, Taouli B. Prediction of hepatocellular carcinoma response to (90)Yttrium radioembolization using volumetric ADC histogram quantification: preliminary results. Cancer Imaging. 2019;19:29. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 150. | Jansen MJA, Kuijf HJ, Veldhuis WB, Wessels FJ, Viergever MA, Pluim JPW. Automatic classification of focal liver lesions based on MRI and risk factors. PLoS One. 2019;14:e0217053. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 151. | Ma X, Ouyang H, Wang S, Wang M, Zhou C, Zhao X. Histogram analysis of apparent diffusion coefficient predicts response to radiofrequency ablation in hepatocellular carcinoma. Chin J Cancer Res. 2019;31:366-374. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 152. | Wu J, Liu A, Cui J, Chen A, Song Q, Xie L. Radiomics-based classification of hepatocellular carcinoma and hepatic haemangioma on precontrast magnetic resonance images. BMC Med Imaging. 2019;19:23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 70] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 153. | Kim S, Shin J, Kim DY, Choi GH, Kim MJ, Choi JY. Radiomics on Gadoxetic Acid-Enhanced Magnetic Resonance Imaging for Prediction of Postoperative Early and Late Recurrence of Single Hepatocellular Carcinoma. Clin Cancer Res. 2019;25:3847-3855. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 151] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 154. | Lewis S, Peti S, Hectors SJ, King M, Rosen A, Kamath A, Putra J, Thung S, Taouli B. Volumetric quantitative histogram analysis using diffusion-weighted magnetic resonance imaging to differentiate HCC from other primary liver cancers. Abdom Radiol (NY). 2019;44:912-922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 155. | Chen S, Feng S, Wei J, Liu F, Li B, Li X, Hou Y, Gu D, Tang M, Xiao H, Jia Y, Peng S, Tian J, Kuang M. Pretreatment prediction of immunoscore in hepatocellular cancer: a radiomics-based clinical model based on Gd-EOB-DTPA-enhanced MRI imaging. Eur Radiol. 2019;29:4177-4187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 125] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 156. | Feng ST, Jia Y, Liao B, Huang B, Zhou Q, Li X, Wei K, Chen L, Li B, Wang W, Chen S, He X, Wang H, Peng S, Chen ZB, Tang M, Chen Z, Hou Y, Peng Z, Kuang M. Preoperative prediction of microvascular invasion in hepatocellular cancer: a radiomics model using Gd-EOB-DTPA-enhanced MRI. Eur Radiol. 2019;29:4648-4659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 148] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 157. | Wu M, Tan H, Gao F, Hai J, Ning P, Chen J, Zhu S, Wang M, Dou S, Shi D. Predicting the grade of hepatocellular carcinoma based on non-contrast-enhanced MRI radiomics signature. Eur Radiol. 2019;29:2802-2811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 134] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 158. | Yang L, Gu D, Wei J, Yang C, Rao S, Wang W, Chen C, Ding Y, Tian J, Zeng M. A Radiomics Nomogram for Preoperative Prediction of Microvascular Invasion in Hepatocellular Carcinoma. Liver Cancer. 2019;8:373-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 238] [Article Influence: 39.7] [Reference Citation Analysis (0)] |
| 159. | Stocker D, Marquez HP, Wagner MW, Raptis DA, Clavien PA, Boss A, Fischer MA, Wurnig MC. MRI texture analysis for differentiation of malignant and benign hepatocellular tumors in the non-cirrhotic liver. Heliyon. 2018;4:e00987. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 160. | Ahn SJ, Kim JH, Park SJ, Kim ST, Han JK. Hepatocellular carcinoma: preoperative gadoxetic acid-enhanced MR imaging can predict early recurrence after curative resection using image features and texture analysis. Abdom Radiol (NY). 2019;44:539-548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 74] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 161. | Hui TCH, Chuah TK, Low HM, Tan CH. Predicting early recurrence of hepatocellular carcinoma with texture analysis of preoperative MRI: a radiomics study. Clin Radiol. 2018;73:1056.e11-1056.e16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 53] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 162. | Zou X, Luo Y, Li Z, Hu Y, Li H, Tang H, Shen Y, Hu D, Kamel IR. Volumetric Apparent Diffusion Coefficient Histogram Analysis in Differentiating Intrahepatic Mass-Forming Cholangiocarcinoma From Hepatocellular Carcinoma. J Magn Reson Imaging. 2019;49:975-983. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 163. | Li H, Zhang J, Zheng Z, Guo Y, Chen M, Xie C, Zhang Z, Mei Y, Feng Y, Xu Y. Preoperative histogram analysis of intravoxel incoherent motion (IVIM) for predicting microvascular invasion in patients with single hepatocellular carcinoma. Eur J Radiol. 2018;105:65-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 46] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 164. | Wu LF, Rao SX, Xu PJ, Yang L, Chen CZ, Liu H, Huang JF, Fu CX, Halim A, Zeng MS. Pre-TACE kurtosis of ADC(total) derived from histogram analysis for diffusion-weighted imaging is the best independent predictor of prognosis in hepatocellular carcinoma. Eur Radiol. 2019;29:213-223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 165. | Li Z, Mao Y, Huang W, Li H, Zhu J, Li W, Li B. Texture-based classification of different single liver lesion based on SPAIR T2W MRI images. BMC Med Imaging. 2017;17:42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 77] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 166. | Moriya T, Saito K, Tajima Y, Harada TL, Araki Y, Sugimoto K, Tokuuye K. 3D analysis of apparent diffusion coefficient histograms in hepatocellular carcinoma: correlation with histological grade. Cancer Imaging. 2017;17:1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |