Published online Oct 21, 2024. doi: 10.3748/wjg.v30.i39.4267
Revised: August 31, 2024
Accepted: September 19, 2024
Published online: October 21, 2024
Processing time: 166 Days and 13.1 Hours
Esophageal squamous cell carcinoma (ESCC) is the most common histological type of esophageal cancer with a poor prognosis. Early diagnosis and prognosis assessment are crucial for improving the survival rate of ESCC patients. With the advancement of artificial intelligence (AI) technology and the proliferation of medical digital information, AI has demonstrated promising sensitivity and accuracy in assisting precise detection, treatment decision-making, and prognosis assessment of ESCC. It has become a unique opportunity to enhance comprehen
Core Tip: This article provides an overview of the current status of esophageal squamous cell carcinoma (ESCC) diagnosis and treatment, emphasizing the pivotal role of artificial intelligence (AI)-based predictive models in enhancing the precision of ESCC management. It outlines the existing challenges and opportunities associated with the integration of AI technologies in the diagnosis and treatment of ESCC. Furthermore, the article discusses the future research directions necessary to advance the practical application of AI in clinical settings, aiming to improve the accuracy and efficacy of ESCC care.
- Citation: Zhang WY, Chang YJ, Shi RH. Artificial intelligence enhances the management of esophageal squamous cell carcinoma in the precision oncology era. World J Gastroenterol 2024; 30(39): 4267-4280
- URL: https://www.wjgnet.com/1007-9327/full/v30/i39/4267.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i39.4267
Esophageal squamous cell carcinoma (ESCC) is the predominant histological type of esophageal cancer (EC), which accounts for 85% of all EC cases[1]. Despite rapid advancements in medical technology over the past decade, approximately 70% of patients cannot undergo surgery due to a high tumor burden in the advanced stages[2].
The improvement of endoscopic techniques provides excellent opportunities for early screening and cancer cure[3]. Nevertheless, the misdiagnosis rate in ESCC screening is approximately 40% in conventional white light endoscopy (WLI), posing obstacles to subsequent treatment[4]. Assessing tumor depth of infiltration and the presence of metastasis holds crucial guidance for selecting appropriate treatment modalities[5,6]. However, commonly used clinical methods in evaluating lymph node metastasis, such as endoscopic ultrasonography (EUS), positron emission tomography (PET), and computed tomography (CT) exhibit accuracies of only 66%, 68%, and 63%, respectively[7,8]. Furthermore, the individual variations in late-stage patients regarding responses to radiotherapy, chemotherapy, immunotherapy, and targeted therapy have become significant challenges in treatment[9,10]. Therefore, a method that comprehensively considers patients’ specific conditions, is urgently needed to achieve early detection, prognosis assessment, and precise prediction of treatment outcomes for ESCC.
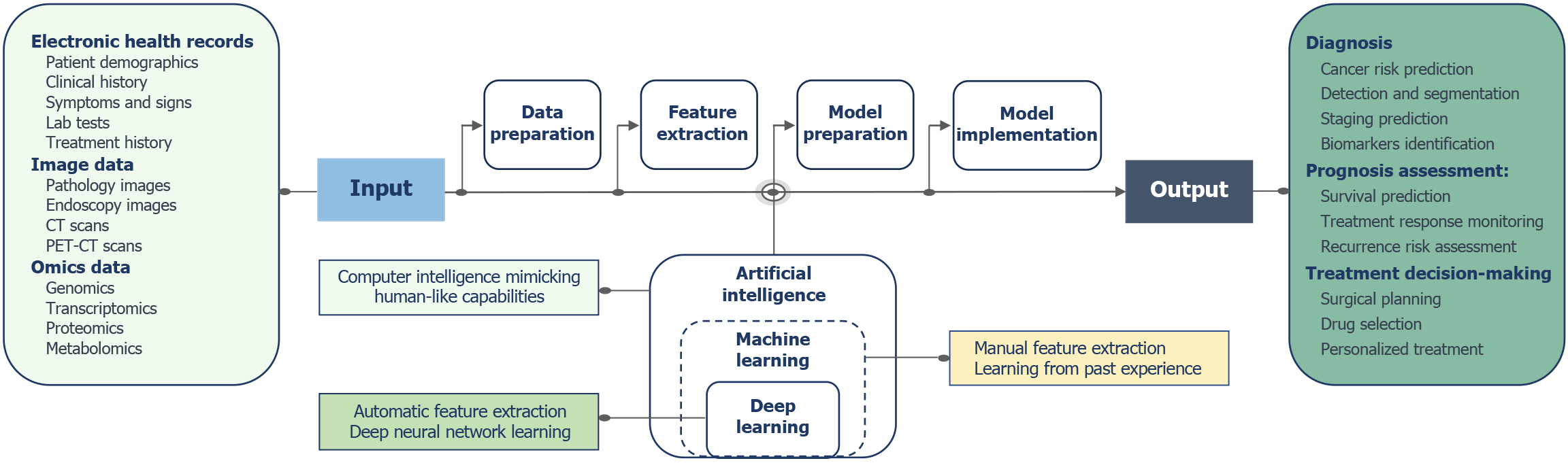
With the progress of artificial intelligence (AI) algorithms and the growth of medical digital information, AI technology has been widely applied in various aspects of cancer research[11]. AI has demonstrated expert-level diagnostic and predictive capabilities in the precise detection and prognosis assessment of ESCC, showing significant potential for expanding and enhancing the efficiency of clinical work (Figure 1)[12]. As a crucial component of AI, machine learning (ML) has exhibited excellent results in extracting “semantic” information[13]. ML can extract the value from a large number of electronic health records, learning logical patterns in data and making predictions about disease development trends. Deep learning (DL), an emerging technology in the AI field, is currently primarily applied in the processing and analysis of imaging data[14]. Computer vision plays a vital role in this process, enabling computers to gain advanced understanding from visual inputs and providing information supporting medical imaging experts. Convolutional neural networks (CNN) are the most widely used tools in computer vision applications in the medical field[15]. Computer vision models can significantly enhance image gradation, identifying features the human brain cannot recognize. Conversely, the model’s input layer quantitatively processes image information, aiding in objective judgment and ensuring repeatability. Recently, DL architectures based on transformer have proven to be more efficient in natural language processing and computer vision tasks, holding the potential for efficient fusion of multimodal clinical information[16,17]. With the development of genomic sequencing technologies, the establishment of databases has also propelled the application of ML and DL techniques in identifying molecular features, such as gene mutations[18,19]. Through learning large-scale genomic data, the AI models can automatically identify gene expression patterns, mutation features, and gene interactions that are useful for disease diagnosis, enabling the inference of potential genetic variations and effectively facilitating the diagnosis and prediction of diseases.
This study used the keywords “artificial intelligence”, “deep learning”, “machine learning”, and “esophageal squamous cell carcinoma” to search relevant literature published up to April 2024 in databases such as PubMed, Embase, and Web of Science. Moreover, it provides a comprehensive overview of the applications of AI in the detection, diagnosis, prognosis assessment, and treatment of ESCC. Additionally, we analyzed the difficulties and challenges encountered in the clinical translation of AI and proposed potential solutions. We hope that through the review and analysis of previous research, we can contribute to advancing the exploration of AI in the field of cancer, particularly in ESCC.
In light of the lower invasiveness and higher cost-effectiveness of non-endoscopic cell sampling methods, innovative cytological devices such as expandable balloons and sponges have been developed for large-scale community screening[20]. However, cytological diagnosis relies on subjective judgment of physicians, and the shortage of cytology experts during large-scale screening significantly impacts the accuracy and reliability of diagnostic outcomes[21]. In 2021, Chinese scholars introduced a novel esophageal cell collector utilizing sponge-like polymer materials for esophageal specimen collection, integrating AI technology to detect of abnormal cells on glass slides[22]. Results from studies conducted across multiple centers indicate that AI-assisted sponge cytology examinations exhibit high sensitivity and specificity in screening for abnormal proliferation of esophageal squamous epithelium[22,23]. Thus, Gao et al[24] integrating cytological and epidemiological features, utilized a CNN model to identify the risk of developing ESCC. The validated model achieved an area under the curve (AUC) value of 0.96, which was comparable to the diagnostic proficiency of cytologists. Importantly, the application of an AI-based fully automated screening method can additionally avert 92.8% of unnecessary endoscopic examinations, marking a milestone in EC screening[25].
Tumor biomarkers (TMs) are substances released by tumor cells or induced by host cells during tumor initiation and development[26]. Studies have shown a significant potential of blood and exhaled breath samples in identifying high-risk populations for ESCC[27]. However, the sensitivity and specificity of these markers, such as volatile organic compounds, circulating miRNA, and circular RNA, that can be used for ESCC screening are not ideal[28-31], and clinically applicable ESCC-TMs remain limited.
Utilizing AI methods to mine large-scale omics data can efficiently identify TMs with high specificity. Furthermore, constructing tumor classifier models using AI technology based on known or unknown biomarkers can prospectively assess the diagnostic value of these features in ESCC, enabling the possibility of an ESCC screening strategy based on TMs. Yuan et al[32] combined ML algorithms with serum lipidomics to identify a set of lipid TMs for screening ESCC, with a sensitivity of 90.7% in an independent validation cohort. Zhu et al[33] utilized a random forest algorithm for metabolomic analysis of plasma exosomes, using the selected four potential markers to construct an ML model. The results showed an AUC of 0.98 in predicting recurrence risk in patients with ESCC. Wang et al[34] developed a novel method for encoding untargeted metabolomic data obtained through liquid chromatography-mass spectrometry, the MetImage. Using multichannel images encoded by MetImage, a CNN-based DL model that accurately identified ESCC patients, exhibiting sensitivities, specificities, and AUC values of 85%, 92%, and 0.95, respectively.
The primary method for screening ESCC and detecting precancerous lesions is endoscopic examination[3]. Several studies have shown that regular endoscopic screening for high-risk patients with ESCC, with the option for endoscopic or surgical intervention as needed, can greatly improve prognosis[35,36]. Nonetheless, inexperienced physicians may overlook subtle changes in lesions during the examination, leading to missed diagnoses. Due to workload or fatigue, even experienced doctors may misdiagnose or overlook diseases.
DL models automatically extract complex features from endoscopic images through a multilayer network of structures. These models can detect subtle lesions or abnormal patterns and model intricate relationships through nonlinear transformations. This efficient feature learning and data processing ability allows AI models to excel in the diagnosis, differentiation assessment, and prediction of invasion depth of ESCC under endoscopy, significantly enhancing the diagnostic efficiency of clinicians (Table 1). Additionally, Yuan et al[37] developed an AI system based on deep CNN (DCNN) to identify various ESCC imaging patterns during endoscopy. The AI system can make a judgment in 17 microseconds, reducing diagnostic time by 300 times. Regarding diagnostic performance, the AI system’s judgments in all endoscopic imaging patterns were satisfactory, with sensitivity and specificity ranging from 92.5% to 99.7% and 78.5% to 89.0%, respectively. Zhao et al[38] reported that the DL model showed an accuracy approximately 15% higher than that of junior physicians in lesion classification. Li et al[39] integrated a fully automatic AI system into narrow-band imaging (NBI) and WLI. Following AI-guided results, the endoscopists’ specificity and accuracy in diagnosing ESCC lesions improved to 96.7% and 94.9%, respectively. These research findings indicate that AI serves as a diagnostic auxiliary tool and potentially functions as an effective training system, providing expert-level guidance for junior physicians to make accurate judgments.
| Ref. | Data sets | Endoscopic modality | Main study aim | AI algorithms | Best result |
| Yuan et al[37], 2022 | Image data set: 53933; images from 2621 patients; video data set: 142; videos from 19 patients | WLI, NM-NBI, ME -NBI | Detection of superficial ESCC | DCNN | Image data set: Sensitivity: 92.5%-99.7%; specificity: 78.5%-89.0%; AUC: 0.906-0.989; video data set: Sensitivity: 89.5%-100%; specificity: 73.7%-89.5% |
| Zhao et al[38], 2019 | Image data set: 1383; images from 219 patients | ME-NBI | Classification of IPCLs | FCN | Mean accuracy of senior IPCL: 92.0%; mid-level IPCL: 82.0%; junior IPCL: 73.3% |
| Li et al[39], 2021 | Image data set: 5367 images | NM-NBI | Detection of early ESCC | FCN | AUC: 0.9761; sensitivity: 91.0%; specificity: 96.7%; accuracy: 94.3% |
| Fukuda et al[41], 2020 | Image data set: 28333 images | ME-NBI | Detection and characterization of early ESCC | CNN | Detection: Sensitivity: 91.0%; specificity: 51.0%; accuracy: 63.0%; characterization: Sensitivity: 86.0% specificity: 89.0%; accuracy: 88.0% |
| Liu et al[42], 2022 | Image data set: 13083; WLI images from 1239 patients | WLI | Detection and delineation of ESCC margins | DCNN | Detection: Accuracy: 85.7% (internal validation) and 84.5% (external validation); delineation: Accuracy: 93.4% (internal validation) and 95.7% (external validation) |
| Ikenoyama et al[44], 2021 | Image data set: 7301; images from 667 patients | LCE | Predict multiple Lugol-voiding lesions | GoogLeNet deep neural network | Sensitivity: 84.4%; specificity: 70.0%; accuracy: 76.4% |
| Yuan et al[47], 2023 | Image data set: 10047; images from 1112 patients; video data set: 140; videos from 1183 lesions | ME-NBI | Detection and delineation of ESCC margins | DCNN | Detection: Accuracy: 92.4% (internal validation) and 89.9% (external validation); delineation: Accuracy: 88.9% (internal validation) and 87.0% (external validation) |
| Shimamoto et al[48], 2020 | Image data set: 23977; images from 909 patients; video data set: 102 videos | NM-NBI | Assessment of tumor infiltration depth | CNN | Sensitivity: 50.0%; specificity: 99.0%; accuracy: 87.0% |
| Wang et al[49], 2023 | ME-BLI data set: 2887; images from 246 patients; ME-NBI data set: 493; images from 81 patients | ME-BLI, ME-NBI | Identification of IPCL | R-CNN | Recall: 79.25%; precision: 75.54%; F1-score: 0.764; mean average precision: 74.95% |
| Zhang et al[53], 2023 | Image data set: 5119; images from 581 patients; video data set: 33 videos | ME-NBI | Infiltration depth prediction | AI-IDPS | For differentiating SM2-3; lesions: Image data set: Sensitivity: 85.7%; specificity: 86.3%; accuracy: 86.2%; video data set: Sensitivity: 87.5%; specificity: 84.0%; accuracy: 84.9% |
| Yuan et al[54], 2022 | Image data set: 7094; images from 685 patients | ME-NBI | Classification of IPCLs | DCNN | Accuracy (internal validation): 91.3%; accuracy (external validation): 89.8% |
WLI: Common methods for EC screening are observation of suspicious lesions and collection of biopsy samples under WLI. However, early EC lesions may manifest as subtle mucosal red changes that can be easily overlooked during WLI examinations[4]. However, AI algorithms are adept at recognizing subtle changes in endoscopic images. Utilizing techniques, such as data augmentation, multiscale and multiview analyses, and transfer learning, these algorithms conduct in-depth image analysis and make judgments that are more sensitive than those of endoscopy experts (sensitivity: 90.8% compared with 82.5%)[37]. To further reduce the risk of oversight, AI algorithms can also analyze video streams directly, automatically detecting and annotating potential lesion areas and providing real-time feedback. In a randomized controlled trial by Yuan et al[40], the diagnostic performance of the AI system was validated in real clinical settings across 12 centers. The researchers found that applying a video-based lesion detection AI system reduced the overall lesion miss rate and miss rate per patient by 5.0% and 3.2%, respectively[41].
The accurate delineation of the margins of early ESCC is crucial in evaluating endoscopic surgical margins. Liu et al[42] constructed a DCNN model that can outline lesion edges under WLI with an accuracy of 98%, surpassing the 95% accuracy of endoscopy experts. By combining endoscopic and pathological features, the AI model can predict the risk of lymph node metastasis, providing a valuable reference for tumor staging and subsequent treatment plan selection.
Lugol chromoendoscopy: Lugol chromoendoscopy (LCE) is performed based on the principle that abnormal esophageal mucosal cells, lacking glycogen, do not take up iodine stain. By assessing the staining pattern of the esophageal mucosa and performing targeted biopsies, LCE can further reduce the incidence of EC by approximately 70%[43]. When LCE videos are input into the DL model by Yuan et al[37], the diagnostic accuracy, sensitivity, and specificity for ESCC can reach 94.7%, 100%, and 89.5%, respectively. However, iodine staining may induce allergies and exacerbate hyper
NBI endoscopy: Although it may not be considered the optimal technique, NBI has emerged as a new endoscopic technology for EC screening. Meta-analysis shows that NBI has a sensitivity of 88% in identifying tumor patients, with the remaining 12% missed detection rate[45]. Inexperienced endoscopists who used NBI technology to identify ESCC may have a sensitivity as low as 53%[46]. Yuan et al[47] integrated an AI system into endoscopic devices for real-time detection of ESCC and precancerous lesions, as well as delineating tumor boundaries. The system demonstrated satisfactory performance, with lesion detection and boundary delineation accuracies of 91.4% and 85.9%, respectively, which was comparable to experienced endoscopists.
A crucial indication for esophagectomy is tumors with a risk of SM2-3 invasion > 25%. Shimamoto et al[48] developed a CNN-based tumor infiltration depth assessment system, which can analyze continuous image data from NBI videos in real-time, identify tumor lesions, and differentiate between SM1 and SM2-3 tumors. The system’s accuracy, sensitivity, and specificity were 87.3%, 50.0%, and 98.7%, respectively, surpassing the diagnostic proficiency of medical experts (84.7%, 44.9%, and 96.9%, respectively).
Magnifying endoscopy with NBI: The installation of a high-magnification lens on an endoscope, combined with NBI technology, allows a clearer observation of the fine structures of the esophageal mucosa epithelium and the morphological features of intrapapillary capillary loops (IPCLs), facilitating the assessment of tumor differentiation and infiltration depth. Wang et al[49] designed the PSA-HRNetV2p model based on region CNN, enabling automatic localization and identification of IPCL in magnifying endoscopy with NBI (ME-NBI). The Japan Esophageal Society proposed the JES classification based on different IPCL morphologies, categorizing IPCL into types A (normal or inflammatory tissue) and B (tumor tissue with varying degrees of infiltration)[50]. Zhao et al[38] introduced a dual-label fully convolutional network model that can identify IPCL types, with an accuracy of 87% in lesion classification.
Moreover, ME-NBI is an effective method for predicting the infiltration depth of ESCC, with an accuracy of 76% in distinguishing between tumor mucosal/submucosal infiltration[51]. The accuracy of assessing the invasion depth of ESCC can increase to 79% when used in conjunction with EUS[52]. Zhang et al[53] incorporated potential endoscopic visual features and developed the AI-based ESCC infiltration depth assessment system AI-IDPS. The accuracy of endoscopists in assessing the infiltration depth of ESCC can be further increased by 5% with the assistance of AI-IDPS. When evaluating tumor infiltration depth using the JES classification, the DCNN model by Yuan et al[54] achieved a comprehensive accuracy of approximately 90% in diagnosing IPCL subtypes and infiltration depth.
The gold standard for diagnosing ESCC is histopathological examination[3]. However, pathological diagnosis heavily relies on the individual experience of pathologists, and variations among different observers may lead to different diagnostic results or even misdiagnosis[55]. By using AI for the in-depth analysis of complex pathological data enables detailed quantitative analysis of pathological slide images, such as cell counting and morphological feature extraction. Subsequently, this approach provides an objective basis for tumor diagnosis, staging, and prognostic evaluation[56-58]. The CNN model by Yang et al[56] quantifies cell nucleus morphology in pathological images, accurately classifying the five stages of ESCC tissue transformation, with an accuracy of up to 99%. Kouzu et al[57] used a DL classifier to classify the proliferative response of connective tissue in pathological images, which achieved precise individualized prognosis prediction.
With the development of digital pathology, the advent of whole slide imaging (WSI) further promotes the application of AI in pathology[55]. In histological-based diagnosis, every cell in a sample can potentially be a tumor cell. Relying on expert identification and annotation of pixel regions that contain tumor cells for building large-scale datasets for model training is time-consuming and impractical, leading to the application of self-supervised methods[59]. Self-supervised learning models can autonomously learn from WSI information, which concludes by extracting potential features and patterns in the WSI without relying on pathologists’ annotations[58,60]. This fully automated architecture is suitable for large-scale image analysis, providing objective and reproducible results, and holding great potential in tumor pathology.
AI has been widely applied in medical imaging, demonstrating significant advantages in improving image quality and providing automated diagnoses. Using AI for CT image reconstruction helps identify and suppress noise from acquired projection signals, optimizing image clarity and contrast, which allows for more accurate observation of the morphological features of tumors[61]. Conversely, AI algorithms enable the quantitative analysis of medical imaging, providing objective diagnostic information[62]. CT scans are a routine radiological method to assess lymph node status preoperatively in patients with ESCC[8]. However, in clinical practice, > 20% of patients with ESCC may have occult lymph node metastasis, where the size and morphology of these lymph nodes do not show obvious changes in CT imaging. Neglecting these in preoperative staging examinations can directly lead to rapid tumor recurrence immediately postoperatively[63,64]. Zhang et al[65] found that ML models have significant advantages in detecting smaller occult lymph node metastases. This AI-based non-invasive predictive method effectively reduces the risk of postoperative recurrence and avoids unnecessary lymph node dissection in low-risk patients. Xie et al[66] input CT image radiomics features into a support vector machine (SVM) model to construct a single lymph node metastasis classification model, with an accuracy of 94.7%. These AI tools aid clinicians in making decisions regarding surgical approaches and the extent of lymph node dissection.
Compared with traditional surgical approaches, endoscopic resection and ablation techniques have advantages, such as minimal postoperative trauma, rapid recovery, and fewer complications, making them the first-line treatment for early-stage ESCC[67]. However, when the tumor infiltrates more than one-third of the submucosal layer with a depth > 200 μm, the risk of lymph node metastasis can be as high as 50%, necessitating surgical intervention[68]. As mentioned earlier, computer vision can identify tiny details and patterns in images from endoscopy, histopathology, and radiology, which can effectively assess tumor infiltration depth, extent of involvement, and lymph node metastasis. Accurate preoperative staging aids in selecting appropriate surgical approaches. However, clinical practice indicates that patients at the same tumor node metastasis (TNM) stage may have different risks of early postoperative recurrence, which may be attributed to the heterogeneity of ESCC, influencing biological behaviors, such as tumor cell migration and invasion[69]. Yang et al[70] integrated TNM staging, clinicopathological parameters, and tumor molecular markers to construct an SVM-based nomogram model, which predicts the risk of postoperative recurrence in patients and facilitates personalized and precise treatment strategies.
In 2003, robot-assisted minimally invasive esophagectomy (RAMIE) was initially introduced as a minimally invasive approach for the treatment of EC using robotic surgical systems. The use of high-definition 3D imaging technology provides an enlarged surgical view, which enhances the precision of the procedure. Additionally, the flexibility of robotic arms and tremor filtration technology further improved the stability and accuracy of the surgery. Several studies have validated its feasibility and safety. Van der Sluis et al[71] conducted a randomized controlled trial that compared RAMIE with open thoracoscopic esophagectomy (OTE) in terms of postoperative complications and other factors. They found that RAMIE and OTE performed equally well in achieving radical tumor resection, meeting the highest standards of contemporary practice. Compared with OTE, patients treated with RAMIE had lower rates of surgery-related complications and cardiopulmonary complications, along with better postoperative recovery. Yang et al[72] conducted a prospective comparative study of the postoperative outcomes of patients with ESCC treated with RAMIE and conventional minimally invasive esophagectomy. They demonstrated the safety of RAMIE in ESCC tumor treatment and also highlighted the significant advantages of RAMIE in identifying and dissecting metastatic lymph nodes. However, the effectiveness of RAMIE is highly influenced by the learning curve, and the postoperative outcomes also depend on the surgeon’s experience and expertise accumulation[73].
The standard treatment approaches for locally advanced resectable ESCC are neoadjuvant chemotherapy (NAC) and neoadjuvant chemoradiotherapy (NACRT)[7,67,74]. However, predicting patient response to NAC and NACRT is challenging due to tumor heterogeneity. Demographic characteristics, medical history, clinical staging, and other factors may contribute to treatment-related side effects and tumor recurrence after treatment[10]. Thus, establishing a non-invasive and accurate method for predicting patient responses to treatment before initiating therapy is crucial for guiding treatment decisions.
Kawahara et al[75] reported that DL models can predict pathological complete response (pCR) after NACRT by extracting and analyzing treatment-related features from endoscopic images. Hu et al[76] used CT radiomic features for training ML models, with an accuracy of 77.1%. Combining CT radiomic with clinical features, Wang et al[77] used ML algorithms to predict the pCR of primary tumor lesions and lymph node metastases. Subsequently, the combination of features from different dimensions significantly improved the predictive performance of the model. Interpreting the impact of genetic variations is the key to understanding an individual’s sensitivity to treatment regimens[78]. Sasagawa et al[79] developed an RF prediction model using immunogenomics and clinical information, which provided individualized predictions of patient responses to NAC and identified molecular markers closely associated with NAC response through feature analysis, opening new avenues for early assessment of patient treatment sensitivity.
Concurrent chemoradiotherapy: For patients with unresectable ESCC or ESCC who cannot tolerate surgery, the 2017 Japanese EC Practice Guidelines recommend curative concurrent chemoradiotherapy[67]. Huang et al[80] integrated pre-treatment CT imaging and EHR using ML methods to predict the objective response rates and progression-free survival (PFS) of patients with unresectable ESCC undergoing different chemoradiotherapy regimens. Moreover, this helps in formulating personalized treatment strategies. Li et al[81] designed a 3D-DLRM model based on ResNet34, which, by exploring high-dimensional imaging features in CT images, predicts the response of patients receiving radiation therapy to different radiotherapy plans, radiation doses, and radiation fields. Compared with less commonly used biomarkers and expensive PET-CT scans, CT-based DL methods contribute to achieving more accurate and convenient personalized treatment[82,83]. Clinical and radiomic features are high-risk factors associated with the prognosis of patients with ESCC. Cui et al[84] extracted clinical variables and contrast-enhanced CT images from patients, using a combined model to assess PFS and overall survival in patients with advanced ESCC receiving chemoradiotherapy. This achieved better performance than single-modal models.
Targeted therapy and immunotherapy: Due to individual differences in traditional chemoradiotherapy regimens, the treatment most effective in each patient is difficult to provide. Intratumoral genetic heterogeneity and clonal evolution also contribute to the development of chemotherapy resistance[69]. The local recurrence rate after treatment remains at 40%-60%, with a < 30% 5-year survival rate[64]. Moreover, traditional chemoradiotherapy lacks specificity and may damage not only tumor tissues but also normal tissue cells during treatment[85]. The development of targeted therapy and immunotherapy has brought survival benefits to patients[86]. However, only a few drugs are available for patients with ESCC patients[87-90]. New treatment targets, new targeted drugs, and screen biomarkers for immunotherapy should be identified to improve the prognosis of patients with ESCC[86]. Intratumoral heterogeneity (ITH) is a potential target for the development of combination treatment strategies[91]. Moreover, DL technology is recommended for integrating and analyzing multi-omics data, providing a powerful tool for dissecting ITH[69]. In addition, considering the intertumoral heterogeneity of tumors, building personalized molecular targeted drugs for specific patients also helps optimize treatment strategies[92]. Chen et al[93] combined metabolomics with ML algorithms to identify three potential therapeutic targets, SLC1A5, SLC7A5, and SLC16A10, in ESCC samples. Overall, AI models are expected to provide more precise and efficient tools for tumor immunotherapy and targeted therapy, which will contribute to the advancement of precision medicine in cancer.
Personalized prognostic evaluations contribute to understanding patient survival and treatment outcomes, enabling more informed treatment decisions. By analyzing extensive data, including genomics, pathology, and laboratory examinations, AI can identify key factors associated with patient prognosis, offering novel insights and directions for disease management. The tertiary lymphoid structure (TLS) is an ectopic lymphoid organ formed by the aggregation of various immune cells[94]. Ling et al[95] demonstrated that patients with ESCC with TLS-mature structures exhibit a 5-year disease-free survival rate approximately 20% higher than TLS-negative patients. A classification model based on inception-resnet-v2 has an accuracy of 95.3% in evaluating TLS maturity. Characterizing genomic information is crucial for understanding the functionality of each genomic component in the organism, which plays an important role in individualized clinical outcome assessments[96]. Li et al[97] utilized five ML algorithms and identified critical molecules that intersect in the prediction of ESCC prognosis, highlighting Stratifin protein as a key prognostic marker. Additionally, log-rank test results demonstrated the accuracy of Stratifin-based risk stratification in accurately characterizing the median survival time of patients.
Building AI predictive models based on potential factors can quantify patient prognosis, facilitating more precise and personalized treatment strategies. Table 2 highlights that AI models are currently extensively used for feature extraction and analysis in clinical indicators, CT images, and histopathological images, achieving outstanding results in predicting the survival of patients with ESCC[98-103].
| Ref. | Sample size | Data sources | AI algorithms | Best result | Highlight |
| Zhang et al[98], 2023 | Training: 1954 cases; testing: 487 cases | Clinicopathological and laboratory information | Rpart, Elastic Net, RF, GBM, GLMboost, and CoxPH | AUC (1-year overall survival): 0.725; AUC (3-year overall survival): 0.0.720; AUC (5-year overall survival): 0.752 | Prognosis evaluation after ESCC resection. First study comparing different AI algorithms’ performance in predicting the survival prognosis of ESCC |
| Zhang et al[99], 2023 | Training: 3612 cases; validation: 1204 cases testing: 1204 cases | Demographic, clinicopathological, and treatment information | DeepSurv | C-index: 0.732; AUC (3-year overall survival): 0.805; AUC (5-year overall survival): 0.825 | Survival prediction of ESCC. Creating a web-based tool for individual survival outcomes prediction |
| Ling et al[100], 2023 | Training: 4062 cases; testing: 1015 cases | Demographic, clinicopathological, and treatment information | WOA-XGBoost | AUC: 0.918; accuracy: 88.1% | Prognosis evaluation of stage IB-IVA ESCC after clinical treatment. The integration of IDPC and WOA-XGBoost realizes a more accurate prediction |
| Wang et al[101], 2021 | Training: 122 cases; testing: 31 cases | CT and histopathological image | Regularized Cox, Weibull Cox, RSF, and DeepSurv | C-index: 0.694 | Prognosis evaluation after ESCC resection. Successful survival analysis through combining multiple modal images together |
| Liu et al[102], 2020 | Training: 79 cases; testing: 71 cases | Transcriptome profile | RF, SVM | Specificity: 98.1%; sensitivity: 79.0% | Prognosis evaluation of patients with ESCC. Identifying novel genetic markers reflecting the prognosis of ESCC |
| Yi et al[103], 2020 | Training: 182 cases; testing: 179 cases | Tumor stemness index provided by one-class logistic regression | Combination of Lasso and Cox regression | AUC (1-year survival time): 0.947; AUC (3-year survival time): 0.760; AUC (5-year survival time): 0.770 | Survival prediction of ESCC. First study predicting the prognosis using the tumor intrinsic stemness pattern |
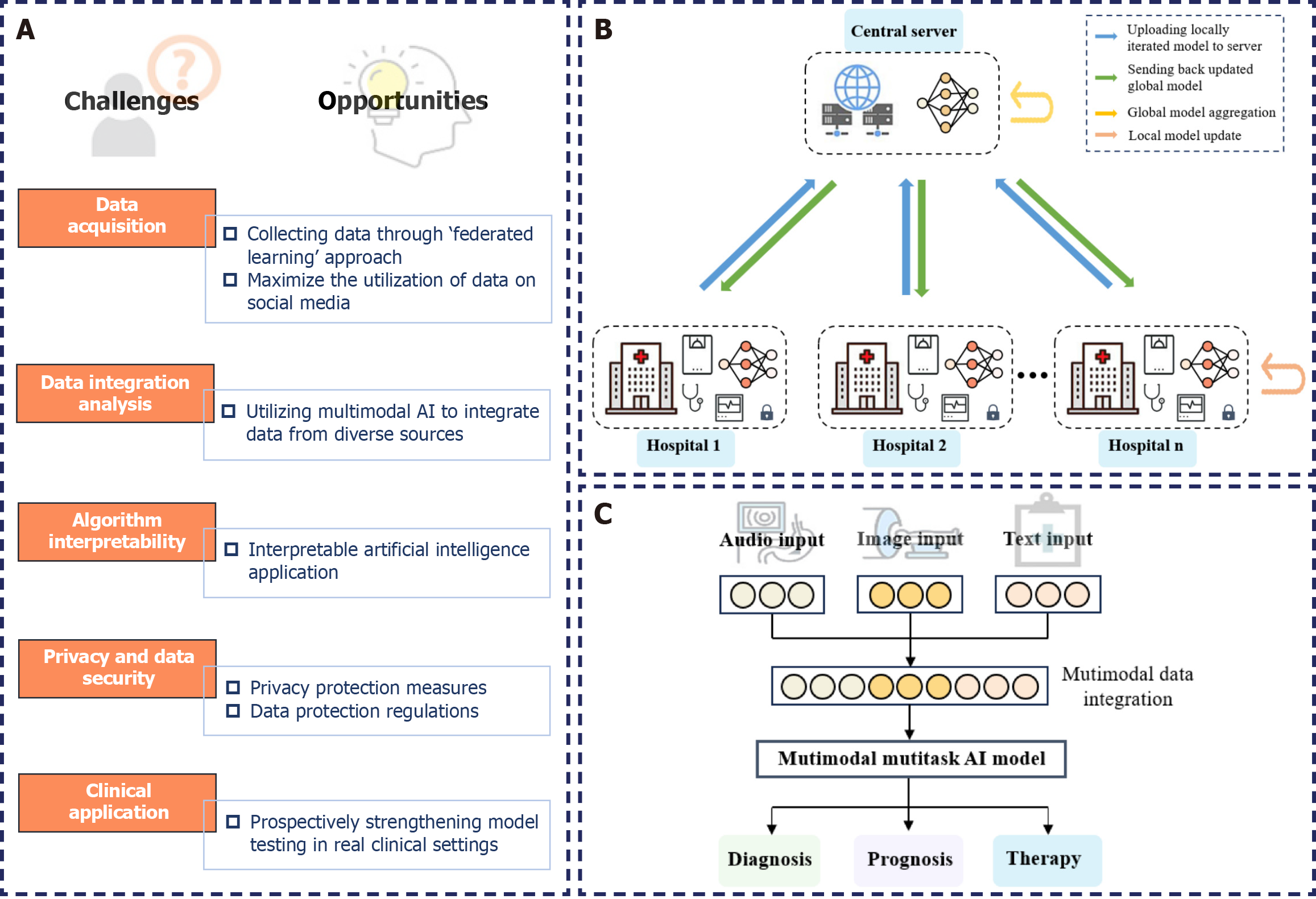
AI models, with their exceptional feature extraction and learning capabilities, have demonstrated high sensitivity and specificity in the diagnosis and treatment of ESCC. However, successfully integrating AI into clinical applications for ESCC remains challenging (Figure 2A).
We must consider issues, such as data bias caused by implicit or explicit biases existing in healthcare systems. Biases related to specific patient groups, sex, and ethnicity can influence the training and deployment of AI models[104]. Particularly in tumor prediction, AI models constructed based on minority populations lead to insufficient predictive performance and difficulties in generalization. Therefore, digitalization of the healthcare sector should be further enhanced, collaboration and communication should be facilitated among different medical institutions, and the application of healthcare AI should be expanded through large-scale data training and model optimization. In 2016, Google proposed the “federated learning” approach, enabling the aggregation of data from distributed health data clients, providing a feasible way to connect electronic medical record data across healthcare institutions (Figure 2B)[105]. Recently, Huang et al[106] curated a large dataset called OpenPath that used pathology images shared by clinical professionals on public forums, such as Twitter. This dataset was designed for training multimodal models with image and text understanding capabilities. They proposed that the development of social media platforms, such as Twitter, significantly facilitates the open sharing of medical information. Embracing this valuable yet underutilized opportunity is crucial for fostering the continued development of medical AI.
Clinical decisions are intricate. Relying on either single-image features or multimodal features alone may not adequately reflect the patient’s prognosis. A more comprehensive and accurate patient profile can be obtained by merging data from different modalities, enabling the formulation of more precise treatment plans[107]. However, current predictive models for ESCC often rely on single-modal data, overlooking crucial interactive information between modalities. In 2021, OpenAI initiated AI multimodal learning, which aimed to handle and correlate information from various modalities (Figure 2C)[108]. Integrating histopathological, radiological images, and even genomic information into models can provide supplementary evidence for tumor diagnosis and prediction, resulting in performance superior to unimodal models[109-111]. However, selecting effective fusion strategies to address challenges, such as the varying strengths of different modalities and noise due to misalignment, remains a major challenge. Thus, this will be a key focus of future AI research in the medical field.
The inherent “black box” nature of AI algorithms poses challenges in terms of interpretability, which limits the applicability of models[112]. In medicine, the reliability of model decision processes and the accuracy of prediction outcomes are equally crucial. Thus, improving decision transparency increases patient trust and enhances healthcare quality. In 2015, the United States Defense Advanced Research Projects Agency introduced the Explainable AI (XAI) program[113]. In 2016, the European Union enacted the General Data Protection Regulation (GDPR), incorporating provisions for the “right to explanation” concerning automated decisions by AI algorithms[114]. Addressing ethical concerns in the design and implementation of AI models, XAI provided interpretable insights into the decision-making process[115]. Lee et al[116] developed an XAI algorithm based on class activation mapping for localizing acute intracranial hemorrhage. Ming et al[117] introduced a visualization method named RuleMatrix based on rule extraction, aiding users in understanding how ML models classify tumor cells. By offering accessible explanations of AI system inferences, XAI contributes to building fair, transparent, and accountable AI decision systems.
The privacy and data security concerns associated with AI model training and validation must also be considered. Insufficient data encryption measures can expose stored and transmitted data to unauthorized access risks. Additionally, a lack of effective access control can lead to information leakage and data misuse[118]. Therefore, a range of supplementary measures should be implemented to ensure effective data utilization while protecting individual privacy. To ensure the legality and security of patient health data, healthcare institutions must comply with data protection regulations, such as the GDPR[119] and the Health Insurance Portability and Accountability Act[120].
Furthermore, most current research evaluates the effectiveness of models in retrospective datasets, overlooking the characteristic variability of the clinical environment, thereby limiting their clinical application. Thus, subsequent efforts should involve extensive testing of AI models in clinical settings to better comprehend and interpret the fluctuations and changes in data. Utilizing AI-assisted diagnostic tools for prospective assessments in real clinical environments will more convincingly demonstrate the model’s genuine clinical effectiveness in the face of dynamic conditions.
The era of AI has arrived, and the application of AI technologies is ushering in unprecedented transformations and opportunities in medicine. The potential of AI should be further explored in the diagnosis and personalized treatment of ESCC. While ensuring the safety and reliability of the technology, the utilization of AI should also be enhanced, assisting clinicians in decision-making, thereby achieving more efficient clinical diagnosis and treatment.
| 1. | Morgan E, Soerjomataram I, Rumgay H, Coleman HG, Thrift AP, Vignat J, Laversanne M, Ferlay J, Arnold M. The Global Landscape of Esophageal Squamous Cell Carcinoma and Esophageal Adenocarcinoma Incidence and Mortality in 2020 and Projections to 2040: New Estimates From GLOBOCAN 2020. Gastroenterology. 2022;163:649-658.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 583] [Cited by in RCA: 556] [Article Influence: 185.3] [Reference Citation Analysis (0)] |
| 2. | Zhou M, Wang H, Zeng X, Yin P, Zhu J, Chen W, Li X, Wang L, Wang L, Liu Y, Liu J, Zhang M, Qi J, Yu S, Afshin A, Gakidou E, Glenn S, Krish VS, Miller-Petrie MK, Mountjoy-Venning WC, Mullany EC, Redford SB, Liu H, Naghavi M, Hay SI, Wang L, Murray CJL, Liang X. Mortality, morbidity, and risk factors in China and its provinces, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2019;394:1145-1158. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2594] [Cited by in RCA: 2404] [Article Influence: 400.7] [Reference Citation Analysis (1)] |
| 3. | di Pietro M, Canto MI, Fitzgerald RC. Endoscopic Management of Early Adenocarcinoma and Squamous Cell Carcinoma of the Esophagus: Screening, Diagnosis, and Therapy. Gastroenterology. 2018;154:421-436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 173] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 4. | Lee CT, Chang CY, Lee YC, Tai CM, Wang WL, Tseng PH, Hwang JC, Hwang TZ, Wang CC, Lin JT. Narrow-band imaging with magnifying endoscopy for the screening of esophageal cancer in patients with primary head and neck cancers. Endoscopy. 2010;42:613-619. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 101] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 5. | Tian D, Li HX, Yang YS, Yan HJ, Jiang KY, Zheng YB, Zong ZD, Zhang HL, Guo XG, Wen HY, Chen LQ. The minimum number of examined lymph nodes for accurate nodal staging and optimal survival of stage T1-2 esophageal squamous cell carcinoma: A retrospective multicenter cohort with SEER database validation. Int J Surg. 2022;104:106764. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 6. | Wang Z, Mao Y, Gao S, Li Y, Tan L, Daiko H, Liu S, Chen C, Koyanagi K, He J. Lymph node dissection and recurrent laryngeal nerve protection in minimally invasive esophagectomy. Ann N Y Acad Sci. 2020;1481:20-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 7. | Kitagawa Y, Uno T, Oyama T, Kato K, Kato H, Kawakubo H, Kawamura O, Kusano M, Kuwano H, Takeuchi H, Toh Y, Doki Y, Naomoto Y, Nemoto K, Booka E, Matsubara H, Miyazaki T, Muto M, Yanagisawa A, Yoshida M. Esophageal cancer practice guidelines 2017 edited by the Japan Esophageal Society: part 1. Esophagus. 2019;16:1-24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 377] [Cited by in RCA: 390] [Article Influence: 65.0] [Reference Citation Analysis (0)] |
| 8. | Choi J, Kim SG, Kim JS, Jung HC, Song IS. Comparison of endoscopic ultrasonography (EUS), positron emission tomography (PET), and computed tomography (CT) in the preoperative locoregional staging of resectable esophageal cancer. Surg Endosc. 2010;24:1380-1386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 118] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 9. | Ilson DH, van Hillegersberg R. Management of Patients With Adenocarcinoma or Squamous Cancer of the Esophagus. Gastroenterology. 2018;154:437-451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 81] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 10. | Tang H, Wang H, Fang Y, Zhu JY, Yin J, Shen YX, Zeng ZC, Jiang DX, Hou YY, Du M, Lian CH, Zhao Q, Jiang HJ, Gong L, Li ZG, Liu J, Xie DY, Li WF, Chen C, Zheng B, Chen KN, Dai L, Liao YD, Li K, Li HC, Zhao NQ, Tan LJ. Neoadjuvant chemoradiotherapy versus neoadjuvant chemotherapy followed by minimally invasive esophagectomy for locally advanced esophageal squamous cell carcinoma: a prospective multicenter randomized clinical trial. Ann Oncol. 2023;34:163-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 100] [Article Influence: 50.0] [Reference Citation Analysis (0)] |
| 11. | Dankwa-Mullan I, Weeraratne D. Artificial Intelligence and Machine Learning Technologies in Cancer Care: Addressing Disparities, Bias, and Data Diversity. Cancer Discov. 2022;12:1423-1427. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 30] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 12. | Chen ZH, Lin L, Wu CF, Li CF, Xu RH, Sun Y. Artificial intelligence for assisting cancer diagnosis and treatment in the era of precision medicine. Cancer Commun (Lond). 2021;41:1100-1115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 107] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 13. | Jordan MI, Mitchell TM. Machine learning: Trends, perspectives, and prospects. Science. 2015;349:255-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2833] [Cited by in RCA: 1994] [Article Influence: 199.4] [Reference Citation Analysis (0)] |
| 14. | Esteva A, Robicquet A, Ramsundar B, Kuleshov V, DePristo M, Chou K, Cui C, Corrado G, Thrun S, Dean J. A guide to deep learning in healthcare. Nat Med. 2019;25:24-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1123] [Cited by in RCA: 1509] [Article Influence: 251.5] [Reference Citation Analysis (0)] |
| 15. | Gu J, Wang Z, Kuen J, Ma L, Shahroudy A, Shuai B, Liu T, Wang X, Wang G, Cai J, Chen T. Recent advances in convolutional neural networks. Pattern Recogni. 2018;77:354-377. [DOI] [Full Text] |
| 16. | Zhou HY, Yu Y, Wang C, Zhang S, Gao Y, Pan J, Shao J, Lu G, Zhang K, Li W. A transformer-based representation-learning model with unified processing of multimodal input for clinical diagnostics. Nat Biomed Eng. 2023;7:743-755. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 93] [Reference Citation Analysis (0)] |
| 17. | Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser L, Polosukhin I. Attention is all you need. 2017 Preprint. Available from: arXiv:1706.03762v1. [DOI] [Full Text] |
| 18. | Wong AK, Sealfon RSG, Theesfeld CL, Troyanskaya OG. Decoding disease: from genomes to networks to phenotypes. Nat Rev Genet. 2021;22:774-790. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 49] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 19. | Shao J, Ma J, Zhang Q, Li W, Wang C. Predicting gene mutation status via artificial intelligence technologies based on multimodal integration (MMI) to advance precision oncology. Semin Cancer Biol. 2023;91:1-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 33] [Reference Citation Analysis (0)] |
| 20. | Lao-Sirieix P, Fitzgerald RC. Screening for oesophageal cancer. Nat Rev Clin Oncol. 2012;9:278-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 125] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 21. | Pan QJ, Roth MJ, Guo HQ, Kochman ML, Wang GQ, Henry M, Wei WQ, Giffen CA, Lu N, Abnet CC, Hao CQ, Taylor PR, Qiao YL, Dawsey SM. Cytologic detection of esophageal squamous cell carcinoma and its precursor lesions using balloon samplers and liquid-based cytology in asymptomatic adults in Llinxian, China. Acta Cytol. 2008;52:14-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 54] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 22. | Gao Y, Xin L, Feng YD, Yao B, Lin H, Sun C, An W, Li ZS, Shi RH, Wang LW. Feasibility and Accuracy of Artificial Intelligence-Assisted Sponge Cytology for Community-Based Esophageal Squamous Cell Carcinoma Screening in China. Am J Gastroenterol. 2021;116:2207-2215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 23. | Feng Y, Wang B, Pan L, Yao B, Deng B, Liang Y, Sun Y, Zang J, Xu X, Song J, Li M, Xu G, Zhao K, Cheng CE, Shi R. Study protocol for artificial intelligence-assisted sponge cytology as pre-endoscopy screening for early esophegeal squmaous epithelial lesions in China. BMC Cancer. 2022;22:1105. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Reference Citation Analysis (0)] |
| 24. | Gao Y, Xin L, Lin H, Yao B, Zhang T, Zhou AJ, Huang S, Wang JH, Feng YD, Yao SH, Guo Y, Dang T, Meng XM, Yang ZZ, Jia WQ, Pang HF, Tian XJ, Deng B, Wang JP, Fan WC, Wang J, Shi LH, Yang GY, Sun C, Wang W, Zang JC, Li SY, Shi RH, Li ZS, Wang LW. Machine learning-based automated sponge cytology for screening of oesophageal squamous cell carcinoma and adenocarcinoma of the oesophagogastric junction: a nationwide, multicohort, prospective study. Lancet Gastroenterol Hepatol. 2023;8:432-445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 29] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 25. | Dawsey SM, Duits LC. A substantial advance for screening of oesophageal cancer. Lancet Gastroenterol Hepatol. 2023;8:393-395. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 26. | Crosby D, Bhatia S, Brindle KM, Coussens LM, Dive C, Emberton M, Esener S, Fitzgerald RC, Gambhir SS, Kuhn P, Rebbeck TR, Balasubramanian S. Early detection of cancer. Science. 2022;375:eaay9040. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 257] [Cited by in RCA: 516] [Article Influence: 172.0] [Reference Citation Analysis (0)] |
| 27. | Chu LY, Peng YH, Weng XF, Xie JJ, Xu YW. Blood-based biomarkers for early detection of esophageal squamous cell carcinoma. World J Gastroenterol. 2020;26:1708-1725. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 63] [Cited by in RCA: 61] [Article Influence: 12.2] [Reference Citation Analysis (2)] |
| 28. | Zou X, Zhou W, Lu Y, Shen C, Hu Z, Wang H, Jiang H, Chu Y. Exhaled gases online measurements for esophageal cancer patients and healthy people by proton transfer reaction mass spectrometry. J Gastroenterol Hepatol. 2016;31:1837-1843. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 39] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 29. | Liu F, Tian T, Xia LL, Ding Y, Cormier RT, He Y. Circulating miRNAs as novel potential biomarkers for esophageal squamous cell carcinoma diagnosis: a meta-analysis update. Dis Esophagus. 2017;30:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 30. | Qin Y, Wu CW, Taylor WR, Sawas T, Burger KN, Mahoney DW, Sun Z, Yab TC, Lidgard GP, Allawi HT, Buttar NS, Smyrk TC, Iyer PG, Katzka DA, Ahlquist DA, Kisiel JB. Discovery, Validation, and Application of Novel Methylated DNA Markers for Detection of Esophageal Cancer in Plasma. Clin Cancer Res. 2019;25:7396-7404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 46] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 31. | Ju C, He J, Wang C, Sheng J, Jia J, Du D, Li H, Zhou M, He F. Current advances and future perspectives on the functional roles and clinical implications of circular RNAs in esophageal squamous cell carcinoma: more influential than expected. Biomark Res. 2022;10:41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 17] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 32. | Yuan Y, Zhao Z, Xue L, Wang G, Song H, Pang R, Zhou J, Luo J, Song Y, Yin Y. Identification of diagnostic markers and lipid dysregulation in oesophageal squamous cell carcinoma through lipidomic analysis and machine learning. Br J Cancer. 2021;125:351-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 33. | Zhu Q, Huang L, Yang Q, Ao Z, Yang R, Krzesniak J, Lou D, Hu L, Dai X, Guo F, Liu F. Metabolomic analysis of exosomal-markers in esophageal squamous cell carcinoma. Nanoscale. 2021;13:16457-16464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 39] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 34. | Wang H, Yin Y, Zhu ZJ. Encoding LC-MS-Based Untargeted Metabolomics Data into Images toward AI-Based Clinical Diagnosis. Anal Chem. 2023;95:6533-6541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 35. | Altorki N, Mynard N, Nasar A, Spinelli C, Villena-Vargas J, Chow O, Lee B, Harrison S, Port J. Ten-Year Survival and Recurrence Patterns After Three-Field Lymph Node Dissection for Squamous Cell and Adenocarcinoma of the Esophagus. Ann Surg. 2023;278:e43-e50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 36. | Han J, Guo X, Zhao L, Zhang H, Ma S, Li Y, Zhao D, Wang J, Xue F. Development and Validation of Esophageal Squamous Cell Carcinoma Risk Prediction Models Based on an Endoscopic Screening Program. JAMA Netw Open. 2023;6:e2253148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 37. | Yuan XL, Guo LJ, Liu W, Zeng XH, Mou Y, Bai S, Pan ZG, Zhang T, Pu WF, Wen C, Wang J, Zhou ZD, Feng J, Hu B. Artificial intelligence for detecting superficial esophageal squamous cell carcinoma under multiple endoscopic imaging modalities: A multicenter study. J Gastroenterol Hepatol. 2022;37:169-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 38. | Zhao YY, Xue DX, Wang YL, Zhang R, Sun B, Cai YP, Feng H, Cai Y, Xu JM. Computer-assisted diagnosis of early esophageal squamous cell carcinoma using narrow-band imaging magnifying endoscopy. Endoscopy. 2019;51:333-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 86] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 39. | Li B, Cai SL, Tan WM, Li JC, Yalikong A, Feng XS, Yu HH, Lu PX, Feng Z, Yao LQ, Zhou PH, Yan B, Zhong YS. Comparative study on artificial intelligence systems for detecting early esophageal squamous cell carcinoma between narrow-band and white-light imaging. World J Gastroenterol. 2021;27:281-293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 17] [Cited by in RCA: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 40. | Yuan XL, Liu W, Lin YX, Deng QY, Gao YP, Wan L, Zhang B, Zhang T, Zhang WH, Bi XG, Yang GD, Zhu BH, Zhang F, Qin XB, Pan F, Zeng XH, Chaudhry H, Pang MY, Yang J, Zhang JY, Hu B. Effect of an artificial intelligence-assisted system on endoscopic diagnosis of superficial oesophageal squamous cell carcinoma and precancerous lesions: a multicentre, tandem, double-blind, randomised controlled trial. Lancet Gastroenterol Hepatol. 2024;9:34-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 24] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 41. | Fukuda H, Ishihara R, Kato Y, Matsunaga T, Nishida T, Yamada T, Ogiyama H, Horie M, Kinoshita K, Tada T. Comparison of performances of artificial intelligence versus expert endoscopists for real-time assisted diagnosis of esophageal squamous cell carcinoma (with video). Gastrointest Endosc. 2020;92:848-855. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 66] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 42. | Liu W, Yuan X, Guo L, Pan F, Wu C, Sun Z, Tian F, Yuan C, Zhang W, Bai S, Feng J, Hu Y, Hu B. Artificial Intelligence for Detecting and Delineating Margins of Early ESCC Under WLI Endoscopy. Clin Transl Gastroenterol. 2022;13:e00433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 20] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 43. | He Z, Liu Z, Liu M, Guo C, Xu R, Li F, Liu A, Yang H, Shen L, Wu Q, Duan L, Li X, Zhang C, Pan Y, Cai H, Ke Y. Efficacy of endoscopic screening for esophageal cancer in China (ESECC): design and preliminary results of a population-based randomised controlled trial. Gut. 2019;68:198-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 105] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 44. | Ikenoyama Y, Yoshio T, Tokura J, Naito S, Namikawa K, Tokai Y, Yoshimizu S, Horiuchi Y, Ishiyama A, Hirasawa T, Tsuchida T, Katayama N, Tada T, Fujisaki J. Artificial intelligence diagnostic system predicts multiple Lugol-voiding lesions in the esophagus and patients at high risk for esophageal squamous cell carcinoma. Endoscopy. 2021;53:1105-1113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 45. | Morita FH, Bernardo WM, Ide E, Rocha RS, Aquino JC, Minata MK, Yamazaki K, Marques SB, Sakai P, de Moura EG. Narrow band imaging versus lugol chromoendoscopy to diagnose squamous cell carcinoma of the esophagus: a systematic review and meta-analysis. BMC Cancer. 2017;17:54. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 117] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 46. | Ishihara R, Takeuchi Y, Chatani R, Kidu T, Inoue T, Hanaoka N, Yamamoto S, Higashino K, Uedo N, Iishi H, Tatsuta M, Tomita Y, Ishiguro S. Prospective evaluation of narrow-band imaging endoscopy for screening of esophageal squamous mucosal high-grade neoplasia in experienced and less experienced endoscopists. Dis Esophagus. 2010;23:480-486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 98] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 47. | Yuan XL, Zeng XH, Liu W, Mou Y, Zhang WH, Zhou ZD, Chen X, Hu YX, Hu B. Artificial intelligence for detecting and delineating the extent of superficial esophageal squamous cell carcinoma and precancerous lesions under narrow-band imaging (with video). Gastrointest Endosc. 2023;97:664-672.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 48. | Shimamoto Y, Ishihara R, Kato Y, Shoji A, Inoue T, Matsueda K, Miyake M, Waki K, Kono M, Fukuda H, Matsuura N, Nagaike K, Aoi K, Yamamoto K, Inoue T, Nakahara M, Nishihara A, Tada T. Real-time assessment of video images for esophageal squamous cell carcinoma invasion depth using artificial intelligence. J Gastroenterol. 2020;55:1037-1045. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 49. | Wang J, Long Q, Liang Y, Song J, Feng Y, Li P, Sun W, Zhao L. AI-assisted identification of intrapapillary capillary loops in magnification endoscopy for diagnosing early-stage esophageal squamous cell carcinoma: a preliminary study. Med Biol Eng Comput. 2023;61:1631-1648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 50. | Oyama T, Inoue H, Arima M, Momma K, Omori T, Ishihara R, Hirasawa D, Takeuchi M, Tomori A, Goda K. Prediction of the invasion depth of superficial squamous cell carcinoma based on microvessel morphology: magnifying endoscopic classification of the Japan Esophageal Society. Esophagus. 2017;14:105-112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 159] [Cited by in RCA: 224] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 51. | Lee MW, Kim GH, I H, Park DY, Baek DH, Lee BE, Song GA. Predicting the invasion depth of esophageal squamous cell carcinoma: comparison of endoscopic ultrasonography and magnifying endoscopy. Scand J Gastroenterol. 2014;49:853-861. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 29] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 52. | Su F, Zhu M, Feng R, Li Y. ME-NBI combined with endoscopic ultrasonography for diagnosing and staging the invasion depth of early esophageal cancer: a diagnostic meta-analysis. World J Surg Oncol. 2022;20:343. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 53. | Zhang L, Luo R, Tang D, Zhang J, Su Y, Mao X, Ye L, Yao L, Zhou W, Zhou J, Lu Z, Zhang M, Xu Y, Deng Y, Huang X, He C, Xiao Y, Wang J, Wu L, Li J, Zou X, Yu H. Human-Like Artificial Intelligent System for Predicting Invasion Depth of Esophageal Squamous Cell Carcinoma Using Magnifying Narrow-Band Imaging Endoscopy: A Retrospective Multicenter Study. Clin Transl Gastroenterol. 2023;14:e00606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 54. | Yuan XL, Liu W, Liu Y, Zeng XH, Mou Y, Wu CC, Ye LS, Zhang YH, He L, Feng J, Zhang WH, Wang J, Chen X, Hu YX, Zhang KH, Hu B. Artificial intelligence for diagnosing microvessels of precancerous lesions and superficial esophageal squamous cell carcinomas: a multicenter study. Surg Endosc. 2022;36:8651-8662. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 18] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 55. | Shmatko A, Ghaffari Laleh N, Gerstung M, Kather JN. Artificial intelligence in histopathology: enhancing cancer research and clinical oncology. Nat Cancer. 2022;3:1026-1038. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 205] [Article Influence: 68.3] [Reference Citation Analysis (0)] |
| 56. | Yang H, Li X, Zhang S, Li Y, Zhu Z, Shen J, Dai N, Zhou F. A one-dimensional convolutional neural network based deep learning for high accuracy classification of transformation stages in esophageal squamous cell carcinoma tissue using micro-FTIR. Spectrochim Acta A Mol Biomol Spectrosc. 2023;289:122210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 57. | Kouzu K, Nearchou IP, Kajiwara Y, Tsujimoto H, Lillard K, Kishi Y, Ueno H. Deep-learning-based classification of desmoplastic reaction on H&E predicts poor prognosis in oesophageal squamous cell carcinoma. Histopathology. 2022;81:255-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 58. | Wu S, Hong G, Xu A, Zeng H, Chen X, Wang Y, Luo Y, Wu P, Liu C, Jiang N, Dang Q, Yang C, Liu B, Shen R, Chen Z, Liao C, Lin Z, Wang J, Lin T. Artificial intelligence-based model for lymph node metastases detection on whole slide images in bladder cancer: a retrospective, multicentre, diagnostic study. Lancet Oncol. 2023;24:360-370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 46] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 59. | Krishnan R, Rajpurkar P, Topol EJ. Self-supervised learning in medicine and healthcare. Nat Biomed Eng. 2022;6:1346-1352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 183] [Article Influence: 61.0] [Reference Citation Analysis (0)] |
| 60. | Ehteshami Bejnordi B, Balkenhol M, Litjens G, Holland R, Bult P, Karssemeijer N, van der Laak JA. Automated Detection of DCIS in Whole-Slide H&E Stained Breast Histopathology Images. IEEE Trans Med Imaging. 2016;35:2141-2150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 39] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 61. | Koetzier LR, Mastrodicasa D, Szczykutowicz TP, van der Werf NR, Wang AS, Sandfort V, van der Molen AJ, Fleischmann D, Willemink MJ. Deep Learning Image Reconstruction for CT: Technical Principles and Clinical Prospects. Radiology. 2023;306:e221257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 149] [Article Influence: 74.5] [Reference Citation Analysis (0)] |
| 62. | Avanzo M, Wei L, Stancanello J, Vallières M, Rao A, Morin O, Mattonen SA, El Naqa I. Machine and deep learning methods for radiomics. Med Phys. 2020;47:e185-e202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 321] [Article Influence: 80.3] [Reference Citation Analysis (0)] |
| 63. | Yun JK, Kim HR, Park SI, Kim YH. Risk prediction of occult lymph node metastasis in patients with clinical T1 through T2 N0 esophageal squamous cell carcinoma. J Thorac Cardiovasc Surg. 2022;164:265-275.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 64. | Smyth EC, Lagergren J, Fitzgerald RC, Lordick F, Shah MA, Lagergren P, Cunningham D. Oesophageal cancer. Nat Rev Dis Primers. 2017;3:17048. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 587] [Cited by in RCA: 737] [Article Influence: 92.1] [Reference Citation Analysis (2)] |
| 65. | Zhang Y, Zhang L, Li B, Ye T, Zhang Y, Yu Y, Ma Y, Sun Y, Xiang J, Li Y, Chen H. Machine learning to predict occult metastatic lymph nodes along the recurrent laryngeal nerves in thoracic esophageal squamous cell carcinoma. BMC Cancer. 2023;23:197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 66. | Xie C, Hu Y, Han L, Fu J, Vardhanabhuti V, Yang H. Prediction of Individual Lymph Node Metastatic Status in Esophageal Squamous Cell Carcinoma Using Routine Computed Tomography Imaging: Comparison of Size-Based Measurements and Radiomics-Based Models. Ann Surg Oncol. 2022;29:8117-8126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 19] [Article Influence: 6.3] [Reference Citation Analysis (1)] |
| 67. | Kitagawa Y, Uno T, Oyama T, Kato K, Kato H, Kawakubo H, Kawamura O, Kusano M, Kuwano H, Takeuchi H, Toh Y, Doki Y, Naomoto Y, Nemoto K, Booka E, Matsubara H, Miyazaki T, Muto M, Yanagisawa A, Yoshida M. Esophageal cancer practice guidelines 2017 edited by the Japan esophageal society: part 2. Esophagus. 2019;16:25-43. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 233] [Cited by in RCA: 328] [Article Influence: 54.7] [Reference Citation Analysis (0)] |
| 68. | Markar SR, Gronnier C, Pasquer A, Duhamel A, Behal H, Théreaux J, Gagnière J, Lebreton G, Brigand C, Renaud F, Piessen G, Meunier B, Collet D, Mariette C; FREGAT Working Group—FRENCH—AFC. Discrepancy Between Clinical and Pathologic Nodal Status of Esophageal Cancer and Impact on Prognosis and Therapeutic Strategy. Ann Surg Oncol. 2017;24:3911-3920. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 69. | Li J, Li L, You P, Wei Y, Xu B. Towards artificial intelligence to multi-omics characterization of tumor heterogeneity in esophageal cancer. Semin Cancer Biol. 2023;91:35-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 26] [Reference Citation Analysis (0)] |
| 70. | Yang HX, Feng W, Wei JC, Zeng TS, Li ZD, Zhang LJ, Lin P, Luo RZ, He JH, Fu JH. Support vector machine-based nomogram predicts postoperative distant metastasis for patients with oesophageal squamous cell carcinoma. Br J Cancer. 2013;109:1109-1116. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 71. | van der Sluis PC, van der Horst S, May AM, Schippers C, Brosens LAA, Joore HCA, Kroese CC, Haj Mohammad N, Mook S, Vleggaar FP, Borel Rinkes IHM, Ruurda JP, van Hillegersberg R. Robot-assisted Minimally Invasive Thoracolaparoscopic Esophagectomy Versus Open Transthoracic Esophagectomy for Resectable Esophageal Cancer: A Randomized Controlled Trial. Ann Surg. 2019;269:621-630. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 247] [Cited by in RCA: 422] [Article Influence: 84.4] [Reference Citation Analysis (0)] |
| 72. | Yang Y, Li B, Yi J, Hua R, Chen H, Tan L, Li H, He Y, Guo X, Sun Y, Yu B, Li Z. Robot-assisted Versus Conventional Minimally Invasive Esophagectomy for Resectable Esophageal Squamous Cell Carcinoma: Early Results of a Multicenter Randomized Controlled Trial: the RAMIE Trial. Ann Surg. 2022;275:646-653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 101] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 73. | Esagian SM, Ziogas IA, Skarentzos K, Katsaros I, Tsoulfas G, Molena D, Karamouzis MV, Rouvelas I, Nilsson M, Schizas D. Robot-Assisted Minimally Invasive Esophagectomy versus Open Esophagectomy for Esophageal Cancer: A Systematic Review and Meta-Analysis. Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 74. | van Hagen P, Hulshof MC, van Lanschot JJ, Steyerberg EW, van Berge Henegouwen MI, Wijnhoven BP, Richel DJ, Nieuwenhuijzen GA, Hospers GA, Bonenkamp JJ, Cuesta MA, Blaisse RJ, Busch OR, ten Kate FJ, Creemers GJ, Punt CJ, Plukker JT, Verheul HM, Spillenaar Bilgen EJ, van Dekken H, van der Sangen MJ, Rozema T, Biermann K, Beukema JC, Piet AH, van Rij CM, Reinders JG, Tilanus HW, van der Gaast A; CROSS Group. Preoperative chemoradiotherapy for esophageal or junctional cancer. N Engl J Med. 2012;366:2074-2084. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3288] [Cited by in RCA: 4080] [Article Influence: 313.8] [Reference Citation Analysis (0)] |
| 75. | Kawahara D, Murakami Y, Tani S, Nagata Y. A prediction model for pathological findings after neoadjuvant chemoradiotherapy for resectable locally advanced esophageal squamous cell carcinoma based on endoscopic images using deep learning. Br J Radiol. 2022;95:20210934. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Reference Citation Analysis (0)] |
| 76. | Hu Y, Xie C, Yang H, Ho JWK, Wen J, Han L, Lam KO, Wong IYH, Law SYK, Chiu KWH, Vardhanabhuti V, Fu J. Computed tomography-based deep-learning prediction of neoadjuvant chemoradiotherapy treatment response in esophageal squamous cell carcinoma. Radiother Oncol. 2021;154:6-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 98] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 77. | Wang J, Zhu X, Zeng J, Liu C, Shen W, Sun X, Lin Q, Fang J, Chen Q, Ji Y. Using clinical and radiomic feature-based machine learning models to predict pathological complete response in patients with esophageal squamous cell carcinoma receiving neoadjuvant chemoradiation. Eur Radiol. 2023;33:8554-8563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 78. | Bruni D, Angell HK, Galon J. The immune contexture and Immunoscore in cancer prognosis and therapeutic efficacy. Nat Rev Cancer. 2020;20:662-680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 529] [Cited by in RCA: 1054] [Article Influence: 210.8] [Reference Citation Analysis (0)] |
| 79. | Sasagawa S, Kato H, Nagaoka K, Sun C, Imano M, Sato T, Johnson TA, Fujita M, Maejima K, Okawa Y, Kakimi K, Yasuda T, Nakagawa H. Immuno-genomic profiling of biopsy specimens predicts neoadjuvant chemotherapy response in esophageal squamous cell carcinoma. Cell Rep Med. 2022;3:100705. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 80. | Huang Y, Huang X, Wang A, Chen Q, Chen G, Ye J, Wang Y, Qin Z, Xu K. Individualized treatment decision model for inoperable elderly esophageal squamous cell carcinoma based on multi-modal data fusion. BMC Med Inform Decis Mak. 2023;23:237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 81. | Li X, Gao H, Zhu J, Huang Y, Zhu Y, Huang W, Li Z, Sun K, Liu Z, Tian J, Li B. 3D Deep Learning Model for the Pretreatment Evaluation of Treatment Response in Esophageal Carcinoma: A Prospective Study (ChiCTR2000039279). Int J Radiat Oncol Biol Phys. 2021;111:926-935. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 82. | Cao Q, Li Y, Li Z, An D, Li B, Lin Q. Development and validation of a radiomics signature on differentially expressed features of (18)F-FDG PET to predict treatment response of concurrent chemoradiotherapy in thoracic esophagus squamous cell carcinoma. Radiother Oncol. 2020;146:9-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 83. | Azad TD, Chaudhuri AA, Fang P, Qiao Y, Esfahani MS, Chabon JJ, Hamilton EG, Yang YD, Lovejoy A, Newman AM, Kurtz DM, Jin M, Schroers-Martin J, Stehr H, Liu CL, Hui AB, Patel V, Maru D, Lin SH, Alizadeh AA, Diehn M. Circulating Tumor DNA Analysis for Detection of Minimal Residual Disease After Chemoradiotherapy for Localized Esophageal Cancer. Gastroenterology. 2020;158:494-505.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 181] [Article Influence: 36.2] [Reference Citation Analysis (0)] |
| 84. | Cui Y, Li Z, Xiang M, Han D, Yin Y, Ma C. Machine learning models predict overall survival and progression free survival of non-surgical esophageal cancer patients with chemoradiotherapy based on CT image radiomics signatures. Radiat Oncol. 2022;17:212. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 31] [Reference Citation Analysis (0)] |
| 85. | Salapa J, Bushman A, Lowe K, Irudayaraj J. Nano drug delivery systems in upper gastrointestinal cancer therapy. Nano Converg. 2020;7:38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 86. | Yang YM, Hong P, Xu WW, He QY, Li B. Advances in targeted therapy for esophageal cancer. Signal Transduct Target Ther. 2020;5:229. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 209] [Cited by in RCA: 314] [Article Influence: 62.8] [Reference Citation Analysis (0)] |
| 87. | Moehler M, Maderer A, Thuss-Patience PC, Brenner B, Meiler J, Ettrich TJ, Hofheinz RD, Al-Batran SE, Vogel A, Mueller L, Lutz MP, Lordick F, Alsina M, Borchert K, Greil R, Eisterer W, Schad A, Slotta-Huspenina J, Van Cutsem E, Lorenzen S. Cisplatin and 5-fluorouracil with or without epidermal growth factor receptor inhibition panitumumab for patients with non-resectable, advanced or metastatic oesophageal squamous cell cancer: a prospective, open-label, randomised phase III AIO/EORTC trial (POWER). Ann Oncol. 2020;31:228-235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 73] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 88. | Dutton SJ, Ferry DR, Blazeby JM, Abbas H, Dahle-Smith A, Mansoor W, Thompson J, Harrison M, Chatterjee A, Falk S, Garcia-Alonso A, Fyfe DW, Hubner RA, Gamble T, Peachey L, Davoudianfar M, Pearson SR, Julier P, Jankowski J, Kerr R, Petty RD. Gefitinib for oesophageal cancer progressing after chemotherapy (COG): a phase 3, multicentre, double-blind, placebo-controlled randomised trial. Lancet Oncol. 2014;15:894-904. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 259] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 89. | Wang ZQ, Wang DS, Wang FH, Ren C, Tan Q, Li YH. Recombinant human endostatin plus paclitaxel/nedaplatin for recurrent or metastatic advanced esophageal squamous cell carcinoma: a prospective, single-arm, open-label, phase II study. Invest New Drugs. 2021;39:516-523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 90. | Kojima T, Shah MA, Muro K, Francois E, Adenis A, Hsu CH, Doi T, Moriwaki T, Kim SB, Lee SH, Bennouna J, Kato K, Shen L, Enzinger P, Qin SK, Ferreira P, Chen J, Girotto G, de la Fouchardiere C, Senellart H, Al-Rajabi R, Lordick F, Wang R, Suryawanshi S, Bhagia P, Kang SP, Metges JP; KEYNOTE-181 Investigators. Randomized Phase III KEYNOTE-181 Study of Pembrolizumab Versus Chemotherapy in Advanced Esophageal Cancer. J Clin Oncol. 2020;38:4138-4148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 299] [Cited by in RCA: 690] [Article Influence: 138.0] [Reference Citation Analysis (0)] |
| 91. | Vitale I, Shema E, Loi S, Galluzzi L. Intratumoral heterogeneity in cancer progression and response to immunotherapy. Nat Med. 2021;27:212-224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 478] [Article Influence: 119.5] [Reference Citation Analysis (0)] |
| 92. | Yang H, Li X, Yang W. Advances in targeted therapy and immunotherapy for esophageal cancer. Chin Med J (Engl). 2023;136:1910-1922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 93. | Chen Z, Gao Y, Huang X, Yao Y, Chen K, Zeng S, Mao W. Tissue-based metabolomics reveals metabolic biomarkers and potential therapeutic targets for esophageal squamous cell carcinoma. J Pharm Biomed Anal. 2021;197:113937. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 94. | Fridman WH, Meylan M, Petitprez F, Sun CM, Italiano A, Sautès-Fridman C. B cells and tertiary lymphoid structures as determinants of tumour immune contexture and clinical outcome. Nat Rev Clin Oncol. 2022;19:441-457. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 367] [Article Influence: 122.3] [Reference Citation Analysis (0)] |
| 95. | Ling Y, Zhong J, Weng Z, Lin G, Liu C, Pan C, Yang H, Wei X, Xie X, Wei X, Zhang H, Wang G, Fu J, Wen J. The prognostic value and molecular properties of tertiary lymphoid structures in oesophageal squamous cell carcinoma. Clin Transl Med. 2022;12:e1074. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 54] [Reference Citation Analysis (0)] |
| 96. | Eraslan G, Avsec Ž, Gagneur J, Theis FJ. Deep learning: new computational modelling techniques for genomics. Nat Rev Genet. 2019;20:389-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 423] [Cited by in RCA: 599] [Article Influence: 99.8] [Reference Citation Analysis (0)] |
| 97. | Li MX, Sun XM, Cheng WG, Ruan HJ, Liu K, Chen P, Xu HJ, Gao SG, Feng XS, Qi YJ. Using a machine learning approach to identify key prognostic molecules for esophageal squamous cell carcinoma. BMC Cancer. 2021;21:906. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 98. | Zhang K, Ye B, Wu L, Ni S, Li Y, Wang Q, Zhang P, Wang D. Machine learningbased prediction of survival prognosis in esophageal squamous cell carcinoma. Sci Rep. 2023;13:13532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 23] [Reference Citation Analysis (0)] |
| 99. | Zhang H, Jiang X, Yu Q, Yu H, Xu C. A novel staging system based on deep learning for overall survival in patients with esophageal squamous cell carcinoma. J Cancer Res Clin Oncol. 2023;149:8935-8944. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Reference Citation Analysis (0)] |
| 100. | Ling D, Liu A, Sun J, Wang Y, Wang L, Song X, Zhao X. Integration of IDPC Clustering Analysis and Interpretable Machine Learning for Survival Risk Prediction of Patients with ESCC. Interdiscip Sci. 2023;15:480-498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 101. | Wang J, Wu LL, Zhang Y, Ma G, Lu Y. Establishing a survival prediction model for esophageal squamous cell carcinoma based on CT and histopathological images. Phys Med Biol. 2021;66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 102. | Liu T, Fang P, Han C, Ma Z, Xu W, Xia W, Hu J, Xu Y, Xu L, Yin R, Wang S, Zhang Q. Four transcription profile-based models identify novel prognostic signatures in oesophageal cancer. J Cell Mol Med. 2020;24:711-721. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 103. | Yi L, Huang P, Zou X, Guo L, Gu Y, Wen C, Wu G. Integrative stemness characteristics associated with prognosis and the immune microenvironment in esophageal cancer. Pharmacol Res. 2020;161:105144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 104. | Seyyed-Kalantari L, Zhang H, McDermott MBA, Chen IY, Ghassemi M. Underdiagnosis bias of artificial intelligence algorithms applied to chest radiographs in under-served patient populations. Nat Med. 2021;27:2176-2182. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 312] [Article Influence: 78.0] [Reference Citation Analysis (0)] |
| 105. | McMahan HB, Moore E, Ramage D, Hampson S, Arcas BA. Communication-Efficient Learning of Deep Networks from Decentralized Data. 2016 Preprint. Available from: arXiv:1602.05629. [DOI] [Full Text] |
| 106. | Huang Z, Bianchi F, Yuksekgonul M, Montine TJ, Zou J. A visual-language foundation model for pathology image analysis using medical Twitter. Nat Med. 2023;29:2307-2316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 154] [Article Influence: 77.0] [Reference Citation Analysis (0)] |
| 107. | Acosta JN, Falcone GJ, Rajpurkar P, Topol EJ. Multimodal biomedical AI. Nat Med. 2022;28:1773-1784. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 384] [Article Influence: 128.0] [Reference Citation Analysis (0)] |
| 108. | Radford A, Kim J W, Hallacy C, Ramesh A, Goh G, Agarwal S, Sastry G, Askell A, Mishkin P, Clark J, Krueger G, Sutskever I. Learning transferable visual models from natural language supervision. Proceedings of the 38th International Conference on Machine Learning, PMLR 139:8748-8763. 2021. [cited 13 September 2024]. Available from: https://proceedings.mlr.press/v139/radford21a/radford21a.pdf. |
| 109. | Boehm KM, Khosravi P, Vanguri R, Gao J, Shah SP. Harnessing multimodal data integration to advance precision oncology. Nat Rev Cancer. 2022;22:114-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 250] [Article Influence: 83.3] [Reference Citation Analysis (0)] |
| 110. | Nguyen HH, Blaschko MB, Saarakkala S, Tiulpin A. Clinically-Inspired Multi-Agent Transformers for Disease Trajectory Forecasting From Multimodal Data. IEEE Trans Med Imaging. 2024;43:529-541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 111. | Li Z, Jiang Y, Lu M, Li R, Xia Y. Survival Prediction via Hierarchical Multimodal Co-Attention Transformer: A Computational Histology-Radiology Solution. IEEE Trans Med Imaging. 2023;42:2678-2689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 112. | Loh HW, Ooi CP, Seoni S, Barua PD, Molinari F, Acharya UR. Application of explainable artificial intelligence for healthcare: A systematic review of the last decade (2011-2022). Comput Methods Programs Biomed. 2022;226:107161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 188] [Article Influence: 62.7] [Reference Citation Analysis (0)] |
| 113. | Guidotti R, Monreale A, Ruggieri S, Turini F, Giannotti F, Pedreschi D. A Survey of Methods for Explaining Black Box Models. ACM Comput Surv. 2019;51:1-42. [DOI] [Full Text] |
| 114. | Voigt P, von dem Bussche A. Introduction and ‘Checklist’. In: The EU General Data Protection Regulation (GDPR). Berlin: Springer, 2017. [DOI] [Full Text] |
| 115. | Yang G, Ye Q, Xia J. Unbox the black-box for the medical explainable AI via multi-modal and multi-centre data fusion: A mini-review, two showcases and beyond. Inf Fusion. 2022;77:29-52. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 267] [Cited by in RCA: 204] [Article Influence: 68.0] [Reference Citation Analysis (0)] |
| 116. | Lee H, Yune S, Mansouri M, Kim M, Tajmir SH, Guerrier CE, Ebert SA, Pomerantz SR, Romero JM, Kamalian S, Gonzalez RG, Lev MH, Do S. An explainable deep-learning algorithm for the detection of acute intracranial haemorrhage from small datasets. Nat Biomed Eng. 2019;3:173-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 216] [Article Influence: 30.9] [Reference Citation Analysis (0)] |
| 117. | Ming Y, Qu H, Bertini E. RuleMatrix: Visualizing and Understanding Classifiers with Rules. IEEE Trans Vis Comput Graph. 2018;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 55] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 118. | Day S, Zweig M. Beyond wellness for the healthy: Digital health consumer adoption 2018. Feb 11, 2019. [cited 13 September 2024]. Available from: https://rockhealth.com/insights/beyond-wellness-for-the-healthy-digital-health-consumer-adoption-2018/. |
| 119. | Hoofnagle CJ, van der Sloot B, Borgesius FZ. The European Union general data protection regulation: what it is and what it means. Inf Commun. 2019;28:65-98. [DOI] [Full Text] |
| 120. | Act A. Health insurance portability and accountability act of 1996. Aug 20, 1996. [cited 13 September 2024]. Available from: https://aspe.hhs.gov/reports/health-insurance-portability-accountability-act-1996. |