Published online Sep 14, 2024. doi: 10.3748/wjg.v30.i34.3894
Revised: August 6, 2024
Accepted: August 16, 2024
Published online: September 14, 2024
Processing time: 96 Days and 20.5 Hours
Immunotherapy presents both promises and challenges in treating hepatocellular carcinoma (HCC) due to its complex immunological microenvironment. The role of B cells, a key part of the immune system, remains uncertain in HCC.
To identify B-cell-specific signatures and reveal novel immunophenotyping and therapeutic targets for HCC.
Using the Tumor Immune Single-cell Hub 2 database, we identified B-cell-related genes (BRGs) in HCC. Gene enrichment analysis was performed to explore the possible collaboration between B cells and T cells in HCC. We conducted univa
The risk score derived from the prognostic model emerged as an independent prognostic factor for HCC. Analysis of the immune microenvironment and cell infiltration revealed the immune status of various risk groups, supporting the cooperation of B and T cells in suppressing HCC. The BRGs model identified new molecular subtypes of HCC, each with distinct immune characteristics. Drug sensitivity analysis identified targeted drugs effective for each HCC subtype, enabling precision therapy and guiding clinical decisions.
We clarified the role of B cells in HCC and propose that the BRGs model offers promising targets for personalized immunotherapy.
Core tip: We have established a reliable B-cell-related genes (BRGs) prognostic model and novel molecular subtypes in hepatocellular carcinoma (HCC). The BRGs model revealed the immune status and personalized treatment options of HCC molecular subtypes. B cells may play a role in tumor cytotoxicity by activating CD4+ and CD8+ T cells in HCC.
- Citation: Xu KQ, Gong Z, Yang JL, Xia CQ, Zhao JY, Chen X. B-cell-specific signatures reveal novel immunophenotyping and therapeutic targets for hepatocellular carcinoma. World J Gastroenterol 2024; 30(34): 3894-3925
- URL: https://www.wjgnet.com/1007-9327/full/v30/i34/3894.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i34.3894
Primary liver cancer is the eighth most common cause of cancer-related mortality, ranking third among all cancer-related deaths[1]. Hepatocellular carcinoma (HCC) constitutes 80% of all primary liver cancer cases. In 2019, approximately 747000 cases of HCC were reported globally, marking a 70% increase since 1990, with 480000 deaths attributed to HCC[1]. Given the malignancy of HCC, compounded by its central location within the vascular system, early systemic dissemination and metastasis are common, resulting in high recurrence rates even with curative surgery and combined radiochemotherapy[1,2]. Immunotherapy, purportedly capable of activating T cells or rejuvenating immunosurveillance against cancer[3], is regarded as the most promising approach for curing HCC[4,5]. Several multikinase inhibitors and antivascular endothelial growth factor receptor 2 antibodies have gained approval for advanced HCC treatment[6]. Nonetheless, immunotherapy exhibits only a modest response rate of 15%–30% among HCC patients[7]. This could be attributed to the current research predominantly focusing on T cells, leaving our understanding of the overall tumor immune microenvironment in HCC limited, thereby lacking specificity in identifying immunotherapeutic targets tailored to individual HCC patients. Consequently, there is an urgent need to elucidate the roles of other crucial immune cells such as B cells in HCC, facilitating improved immune stratification of patients for precision therapy, thus elevating the cure rate of HCC.
Currently, immunotherapy for tumors predominantly focuses on T-cell-mediated adaptive immunity. While B cells consistently represent a rich cellular component within tumors, little is known about their activation status and biological functions in human tumors, thus leading to a significant oversight of B cells in tumor immunotherapy[8]. Traditionally associated with humoral immune responses against viral and bacterial infections, the role of B cells in tumor immunity has long been debated and may exhibit cancer-specific characteristics[9]. The role of B cells in HCC remains obscure, yet mounting evidence from both preclinical and clinical studies suggests that B cells can induce potent anticancer immunity through humoral and cellular immune responses[8]; a hypothesis awaiting formal validation through analysis of HCC molecular features. B cells infiltrating tumors can be identified at various stages of HCC development, with their presence varying according to stage and histological subtype[10]. Given their influence on both humoral and cellular immunity, a deeper understanding of B-cell biology can offer significant opportunities for HCC immunotherapy[11]. Therefore, a thorough analysis of the specific role of B cells in HCC, along with their interplay with other crucial immune cells in HCC, is crucial for reshaping the immune landscape of the tumor microenvironment and treating HCC effectively.
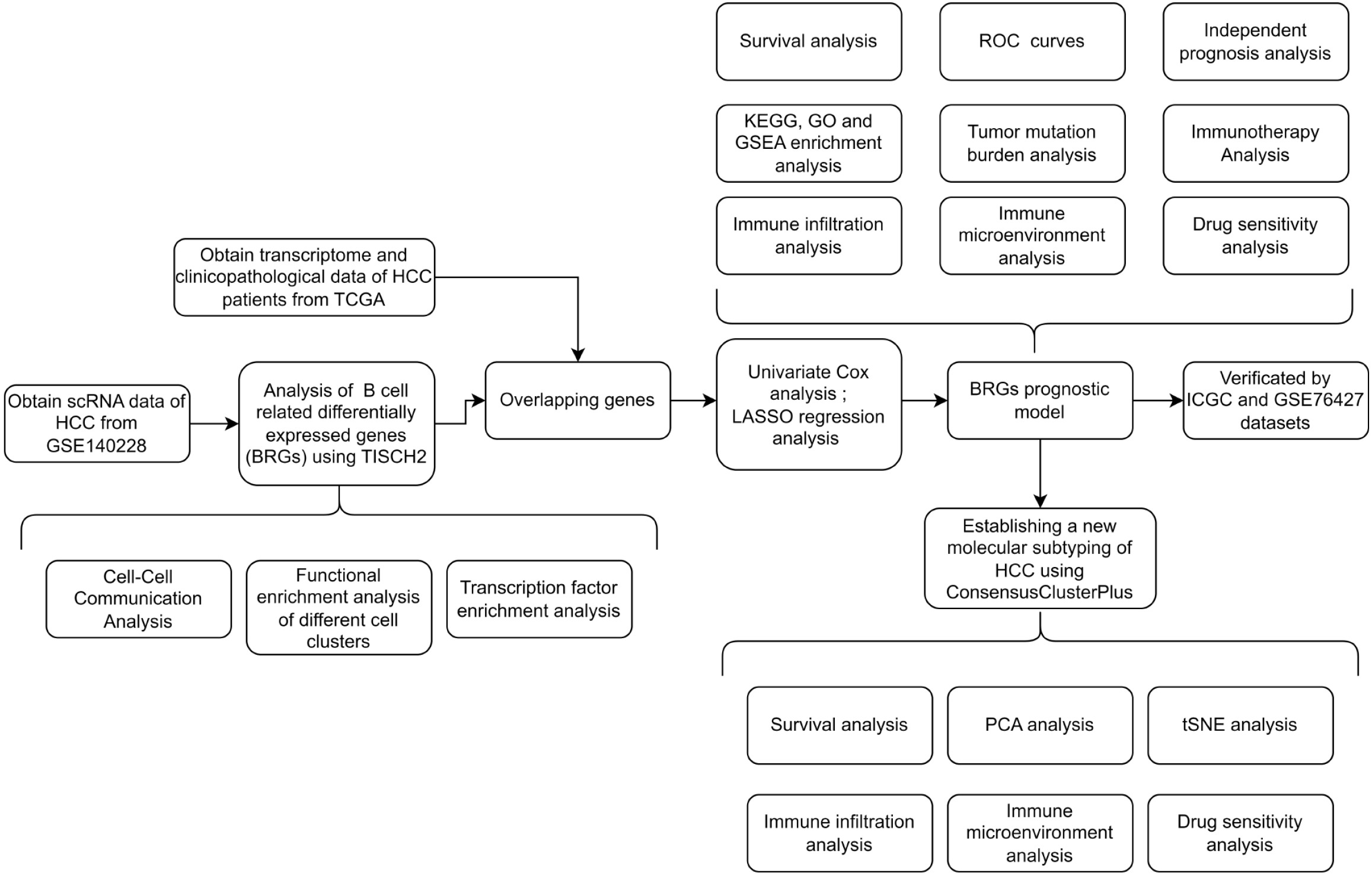
In this study, we identified differentially expressed B-cell-related genes (BRGs) in HCC using the Tumor Immune Single-Cell Hub 2 (TISCH2) database. Gene enrichment analysis suggested potential synergy between B cells and T cells in HCC. Subsequently, using The Cancer Genome Atlas Liver Hepatocellular Carcinoma Collection (TCGA-LIHC) dataset, we conducted single-factor Cox regression analysis to screen BRGs associated with HCC prognosis. We used least absolute shrinkage and selection operator (LASSO) regression analysis to construct a prognostic evaluation model composed of 11 BRGs. The risk score derived from the BRG prognostic model was validated as an independent prognostic factor for HCC, confirmed by the International Cancer Genome Consortium (ICGC) dataset and GSE76427. Extensive clinical correlation analysis demonstrated the association between the prognostic model and clinicopathological factors. Immune microenvironment and immune cell infiltration analyses revealed the immune status of different risk score groups, supporting the notion of B cells and T cells synergistically inhibiting HCC. Finally, through the BRGs model, we characterized three novel molecular subtypes of HCC, delineating their immune characteristics and identifying targeted drugs effective for each subtype, thus facilitating precision therapy for HCC and aiding clinical decision-making.
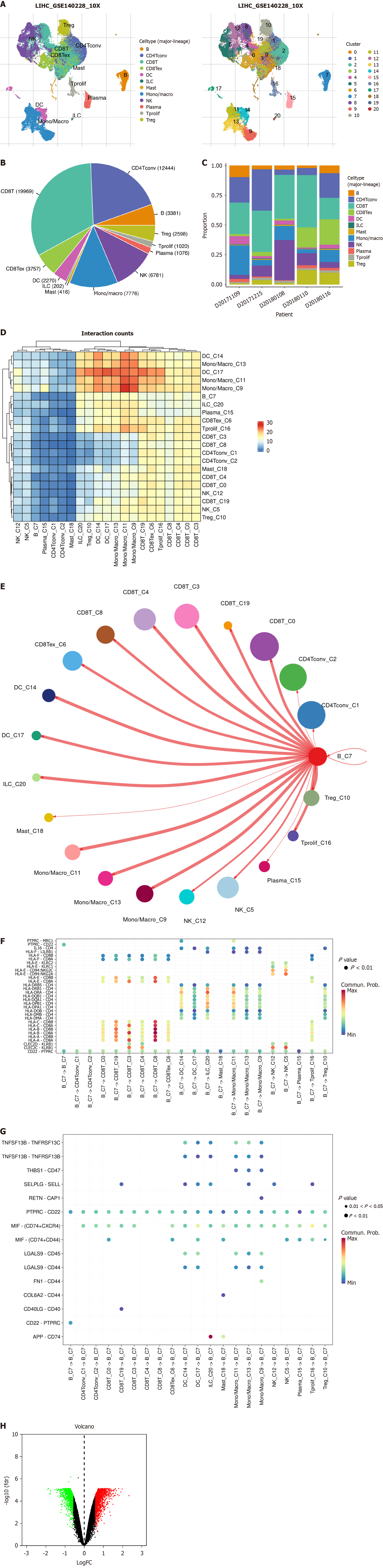
We downloaded single-cell RNA data of HCC, comprising 62 530 cells, from the Gene Expression Omnibus website GSE140228 dataset. We analyzed the single-cell data using the Transcriptomic-Immunohistochemical Single-Cell Hybridization (TISH2) website[12]. We used principal component analysis (PCA) for dimensionality reduction, K-nearest neighbors, and Louvain algorithms for cluster identification, and annotated cell types using marker genes[13,14]. By applying the Wilcoxon test, we identified differentially expressed genes in B-cell clusters compared to all other cells based on the logarithmic fold change (|fold change| ≥ 1.5) and false discovery rate (FDR) < 0.05[15].
We conducted Cell Chat analysis[16] using the TISH2 website[12] to assess cell–cell communication based on the expression of known ligand–receptor (L–R) pairs across different clusters. We utilized the netVisual_circle function from the pheatmap R package and the CellChat R package to compute and display the quantity of significant L–R interaction pairs and the communication probability between two clusters. Significant L–R interaction pairs were identified for each cluster, designating them as either source or target cells, with a P value threshold set at 0.05[12].
To characterize the functionality of distinct cell type populations, we conducted gene set enrichment analysis (GSEA) using the TISH2 website, ranking genes based on fold changes derived from differential analysis[12]. We used the Kyoto Encyclopedia of Genes and Genomes (KEGG), Gene Ontology (GO), and GSEA analyses to identify and visualize significantly upregulated and downregulated pathways within each cell cluster (FDR ≤ 0.05), facilitating functional enrichment analysis across different clusters[12].
We obtained transcriptomic data from the TCGA-LIHC dataset, comprising 375 HCC samples and 50 paired adjacent normal tissues. Among these, 365 patients had complete survival data, clinicopathological information, and somatic mutation data. Additionally, we collected transcriptomic and survival data from 232 HCC patients from the ICGC database. Transcriptomic and survival data from 115 HCC patients were retrieved from the GSE76427 dataset.
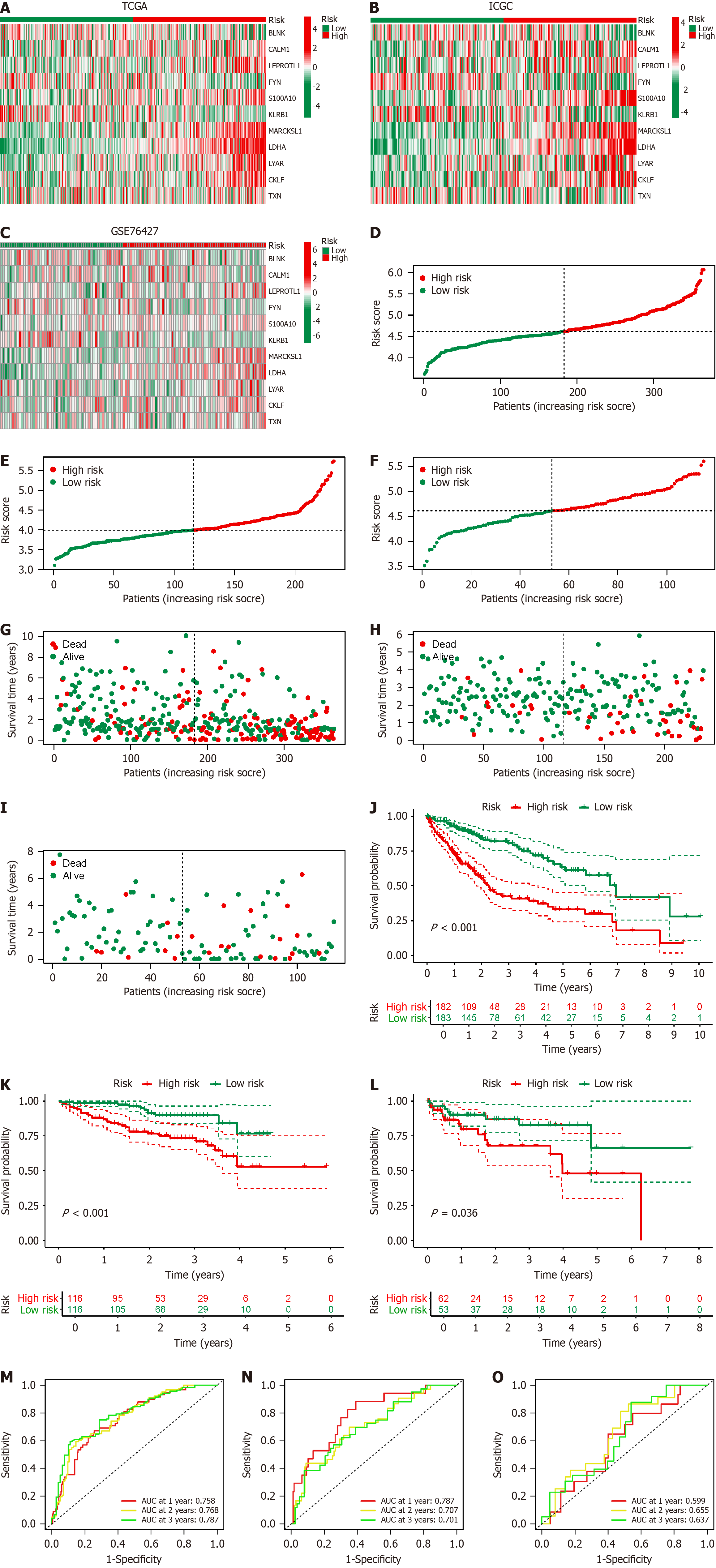
Using the TCGA-LIHC database as the training set, we conducted univariate Cox regression analysis (FDR < 0.05) to identify BRGs associated with HCC prognosis. LASSO regression analysis was performed using the R glm Sparse Net package[17] to mitigate model overfitting, resulting in the derivation of an HCC prognosis model composed of 11 BRGs. The BRGs incorporated into the prognosis model and their corresponding gene coefficients are presented in Supplementary Table 1. Subsequently, we computed the risk scores for each HCC sample in TCGA-LIHC. Based on a 1:1 cutoff value, TCGA-LIHC samples were split into low- and high-risk groups. We used Kaplan–Meier survival and receiver operating characteristic (ROC)[17] analysis in R survival and R time ROC to evaluate the predictive accuracy of the model for HCC prognosis. The reliability of the BRGs model was validated using the ICGC dataset and GSE76427 as validation datasets.
Differential gene analysis between high-risk and low-risk groups in TCGA-LIHC was conducted using the limma package in R (|log fold change| > 1, FDR < 0.05). We used cluster Profiler[17] in R for KEGG and GO analysis to explore molecular mechanisms among HCC risk score groups. GSEA assessed biological functional changes between low- and high-risk subgroups (|normalized enrichment score| > 1, FDR < 0.05).
To demonstrate that the risk score was an independent prognostic factor for HCC, we conducted univariate Cox regression analysis (FDR < 0.05) and multivariate Cox regression analysis (FDR < 0.05) between the risk score and other clinicopathological factors of HCC using the survival package in R. We utilized the survminer[17] and time ROC[17] packages in R to generate ROC curves, illustrating the comparative efficacy of the risk score and other clinicopathological factors for HCC prognosis. Visualization of the correlation between the risk score and other clinicopathological factors of HCC was achieved using the Complex Heatmap and reshape2 packages in R.
Various algorithms, including tumor immune estimation resource, single-sample GSEA (ssGSEA), microenvironment cell population counter, QUANTitative immune single-cell expression, estimation, cell-type identification by estimating relative subsets of RNA transcripts (CIBERSORT), and evaluating the proportion of immune cells[18], were used to calculate the relative levels of tumor-infiltrating immune cells in TCGA-LIHC samples. ssGSEA and CIBERSORT algorithms were utilized to compute the abundance of cell subpopulations in TCGA-LIHC samples. The ssGSEA algorithm was applied to assess the immune functional status in TCGA-LIHC samples.
We used the R packages limma and reshape 2[18] to identify differential expression of immune checkpoint genes across various risk score groups. Elevated expression levels of these genes may correlate with improved efficacy of immune checkpoint inhibitors. For the evaluation of immunotherapy effectiveness, a prognostic model based on Biomarker Response to Immunotherapy Group was applied to 348 malignant tumor patients undergoing immunotherapy within the IMvigor210 dataset.
The R package consensus cluster plus[18] was used for unsupervised consensus clustering to identify novel molecular subtypes of HCC. This package generated several key outputs: the consensus matrix (CM), cumulative distribution function (CDF) plot, and consensus heatmap were essential tools for identifying the ideal number of clusters for classifying HCC subtypes. The CDF plot provided a glimpse into the stability of clustering outcomes across various cluster numbers. The CM, displayed as a matrix, indicated the frequency with which sample pairs were grouped together across iterations, providing a quantitative measure of clustering robustness. The consensus heatmap visually represented the CM, facilitating a clearer interpretation of the clustering results.
All statistical analyses were performed using R version 4.3.1. The Kruskal–Wallis test was utilized to examine variances in immune scores, immune checkpoint gene expression, and drug sensitivity across different clusters. To compare the differences in patient survival rates between two risk groups, a log-rank test from the R survival package was used for Kaplan–Meier analysis. The statistical tests were two-sided, where P < 0.05 was considered significant. Statistical significance levels are denoted with asterisks: aP < 0.05, bP < 0.01, and cP < 0.001.
The entire study followed the screening and analysis process outlined in Figure 1. We analyzed scRNA data from GSE140228 using TISCH2, and Figure 2A clearly shows the distinct segregation of B-cell subgroups. Both the pie chart and bar graph depict the number and proportion of B cells, demonstrating that they fell within the normal range (Figure 2B and C). In the tumor microenvironment, cellular interactions regulated cell function and influenced the surrounding immune environment and tumor progression. By using the cell chat algorithm in TISCH2, we predicted interactions between different cell types. The analysis showed strong interactions among B cells, most T cells, macrophages, and dendritic cells (Figure 2D and E), suggesting that B cells play a key role in the crosstalk within the HCC immune microenvironment. We examined the gene pairs involved in the interactions of B cells as donors (Figure 2F) or recipients (Figure 2G) with other cells. This revealed the probability of interactions among specific gene pairs, demonstrating that B cells significantly regulate various crucial genes of CD8 T cells such as CD8A and CD8D. To identify BRGs compared with genes of other cells, we utilized Wilcoxon tests available in TISCH2. We applied a log transformation fold change (|fold change|≥ 1.5) and FDR < 0.05 to recognize BRGs. The analysis identified 74 upregulated and 171 downregulated BRGs.
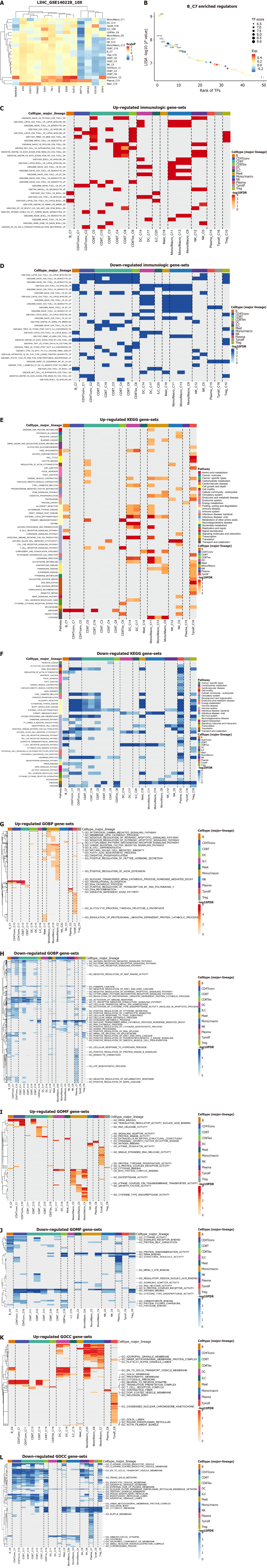
Various transcription factors play critical roles in the development and progression of HCC. We used the local indicators of spatial association algorithm within TISCH2 to infer the transcription factors regulating gene expression in each cell cluster. Using heatmaps, we demonstrated the expression patterns of key transcription factors across all cell clusters in the dataset. The expression patterns of transcription factors in B cells and T cells were almost identical (Figure 3A), supporting the potential synergistic immune role of B cells and T cells in HCC. Subsequently, we sought to identify cell-type-specific transcription factors. In B cells, transcription factors such as myelocytomatosis proto-oncogene, signal transducer and activator of transcription 5B, plant homeodomain finger protein 8, core-binding factor subunit, and E26 transformation-specific-related genes, NRF1, FLI1, EGR3, TFAP4, and RUNX1 showed specific expression (Figure 3B). This specificity highlighted the unique regulatory mechanisms at play within B cells in the HCC immune environment.
To elucidate the specific functions that B cells may regulate in HCC and their synergistic interaction with T cells, we utilized TISCH2 GSEA along with a collection of 16 626 gene sets from the molecular signatures database to characterize the unique functions of B cells. These gene sets encompassed various domains including KEGG pathways, hallmark gene sets, GO terms, immunological features, oncogenic signatures, and transcription factor targets. We observed a strong correlation between B cells and multiple T-cell-related immunological gene sets (Figure 3C and D). In terms of KEGG enrichment analysis, B cells and T cells exhibited highly consistent regulation of pathways such as T-cell receptor signaling, natural-killer-cell-mediated cytotoxicity, NOD-like receptor signaling, Toll-like receptor signaling, and ribosome pathways (Figure 3E and F). Regarding GO analysis, both B cells and T cells showed significant enrichment in pathways including nuclear transcribed mRNA catabolic processes involved in nonsense-mediated decay, translational elongation, RNA catabolic processes, presynaptic membranes, and cytosolic ribosomes (Figure 3G–L). These findings collectively suggest that B cells and T cells jointly regulate mRNA metabolism, mRNA translation, and subsequent immune responses in HCC.
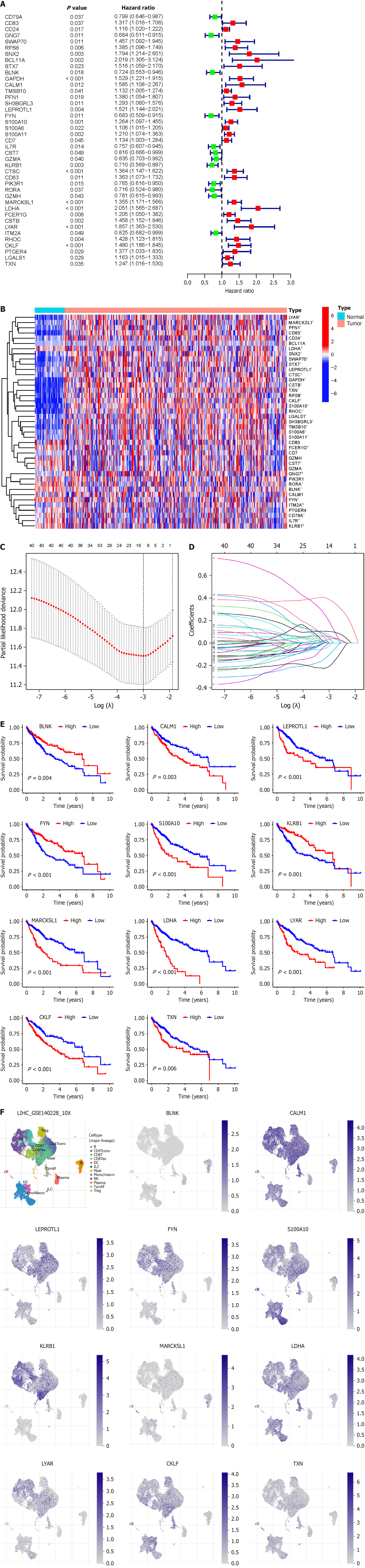
Based on the BRGs selected from Figure 1H, we extracted the expression levels of these BRGs from the TCGA-LIHC database, and merged them with clinical, pathological, and prognostic data, resulting in an integrated table containing complete information for 365 patients. Through single-factor Cox regression analysis, we identified 41 BRGs associated with prognosis of HCC patients (Figure 4A), displaying their expression levels in HCC compared to paired adjacent cancer samples (Figure 4B). We used LASSO regression analysis to mitigate model overfitting, and derived an HCC prognostic model composed of 11 BRGs (Figure 4C and D, Supplementary Table 1). Utilizing Kaplan–Meier survival estimates, we evaluated the relationship between these BRGs and HCC prognosis, revealing that high expression levels of BLNK, FYN, and KLRB1 were associated with favorable prognosis, whereas elevated expression of CALM1, LEPROTL1, S100A10, MARCKSL1, LDHA, LYAR, CKLF, and TXN correlated with poor prognosis (Figure 4E). Additionally, we presented the expression of these BRGs in B cells of HCC, noting high expression of BLNK and MARCKSL1 in B cells, while FYN, KLRB1, CALM1, LEPROTL1, S100A10, LDHA, LYAR, CKLF, and TXN exhibited low expression (Figure 4F).
To evaluate the prognostic model of BRGs, we utilized TCGA-LIHC data as the training set, with ICGC and GSE76427 serving as validation sets. The expression patterns of the 11 BRGs in the prognostic model were consistent between the training and validation sets (Figure 5A–C). The distribution of HCC patients across different risk score groups (Figure 5D–F) and their survival status (Figure 5G–I) were largely consistent across various risk score groups. We depicted the prognostic performance of patients in different risk score groups within the training and validation sets. Consistently, patients in the high-risk score group exhibited poorer prognosis compared with those in the low-risk score group (Figure 5J–L). We constructed ROC curves to validate the effectiveness of the model. The area under the ROC curve (AUC) for 1, 2, and 3 years in the TCGA-LIHC dataset was 0.758, 0.768, and 0.787 respectively (Figure 5M); 0.787, 0.707, and 0.701 in the ICGC database (Figure 5N); and 0.599, 0.655, and 0.637 in GSE76427 (Figure 5O). These results collectively affirm the efficacy of our BRG prognostic model in predicting the prognosis of HCC patients.
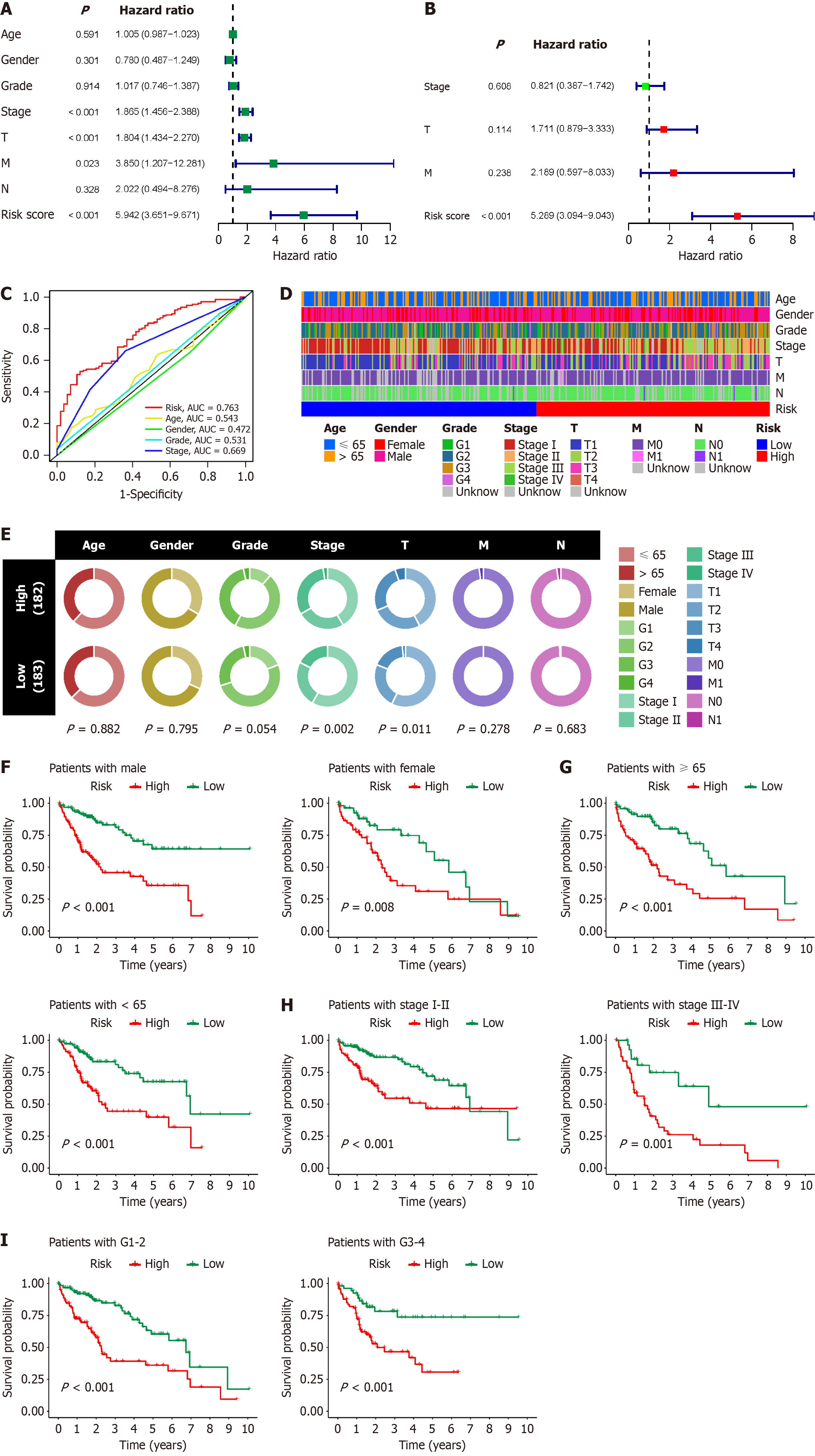
To assess the guiding value of the BRGs prognostic model in clinical practice, we conducted Cox regression analysis to compare its predictive superiority regarding HCC patient prognosis with other clinicopathological factors. Single-factor Cox regression analysis revealed that the stage, T stage, M stage, and risk score of HCC were significant factors associated with poor prognosis, with risk score exhibiting the highest hazard ratio (Figure 6A). Multifactor Cox regression analysis indicated that risk score was the sole independent prognostic factor for HCC, suggesting its potential for accurate stand-alone diagnosis of HCC (Figure 6B). Compared with clinically common factors such as age, gender, grade, and stage in assessing HCC prognosis, risk score demonstrated significantly higher AUC values, underscoring the substantial advantages and efficacy of the BRGs prognostic model in evaluating HCC prognosis (Figure 6C). We also assessed the correlation between risk score and clinicopathological factors, revealing that HCC with high risk score had higher stages and T stages, while risk score showed no significant correlation with age, gender, grade, M stage, or N stage of HCC patients (Figure 6D and E). We further evaluated the value of the BRGs prognostic model in different clinical subgroups by stratifying HCC patients into different clinical subsets based on age, gender, grade, and stage. The results demonstrated that patients in the high-risk score group exhibited poorer prognosis across all clinical subsets (Figure 6F–I). These findings underscore the significant value and potential of the BRGs prognostic model in clinical diagnosis and predicting the prognosis of HCC patients.
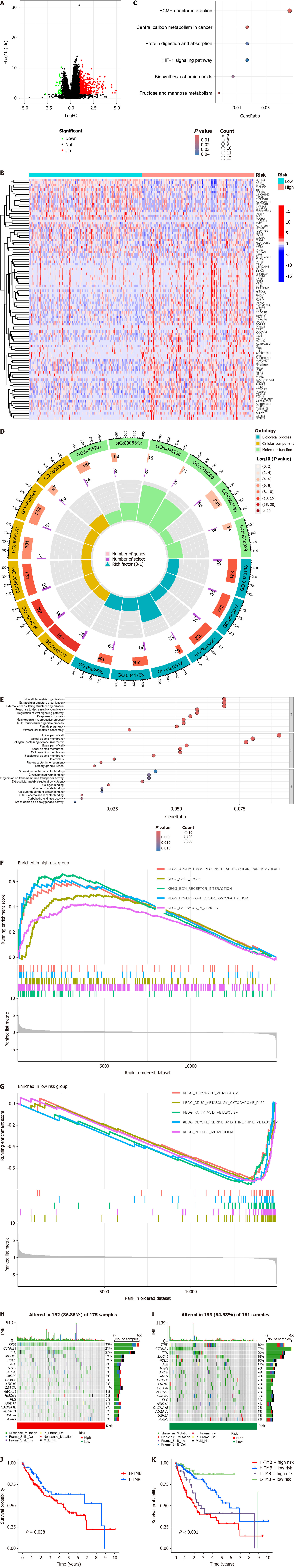
To delve into the distinct features of various risk score groups and elucidate the potential role of B cells in regulating HCC, we conducted differential gene expression analysis on different risk score groups within the TCGA-LIHC dataset. We identified 420 upregulated and 28 downregulated genes (Figure 7A), with the top 50 differentially expressed genes in TCGA-LIHC displayed (Figure 7B). Subsequently, KEGG enrichment analysis was performed to identify differential pathways within different risk score groups. Pathways such as extracellular matrix (ECM)–receptor interaction, central carbon metabolism in cancer, protein digestion and absorption, hypoxia-inducible factor (HIF)-1 signaling pathway, biosynthesis of amino acids, and fructose and mannose metabolism were significantly enriched with differential genes (Figure 7C). Further GO analysis suggested that B cells were involved in crucial cellular molecular functions and biological processes in HCC, including ECM organization, apical part of cells, and G-protein-coupled receptor binding (Figure 7D and E). Lastly, GSEA was conducted to validate the significance of the enriched pathways. Results indicated that in the high-risk score group, KEGG cell cycle and KEGG ECM–receptor interaction were potentially important enriched pathways (Figure 7F), whereas in the low-risk score group, it was KEGG butanoate metabolism and KEGG fatty acid metabolism (Figure 7G).
The mutation of key genes in tumor cells is closely associated with the progression and prognosis of HCC[19]. We sought to determine whether the functionality of B cells in HCC correlates with this. Patients with HCC in the high-risk score group exhibited significantly higher somatic mutation rates in key genes, such as TP53 (33% vs 19%) and TTN (27% vs 20%), compared with those in the low-risk score group (Figure 7H and I). HCC patients with a high tumor mutation burden (TMB) had significantly poorer prognosis than those with a low TMB (Figure 7J). When evaluating the prognosis of HCC patients using both TMB and risk score, those with high TMB and high risk scores had the worst prognosis. Conversely, those with low TMB and low risk scores had the best prognosis. Patients with either high TMB or high risk scores alone had prognoses that fell between these extremes (Figure 7K). These results suggest that the functionality of B cells in HCC is associated with somatic mutations and that the BRGs prognosis model, when combined with TMB, can enhance the prediction of prognosis in HCC patients.
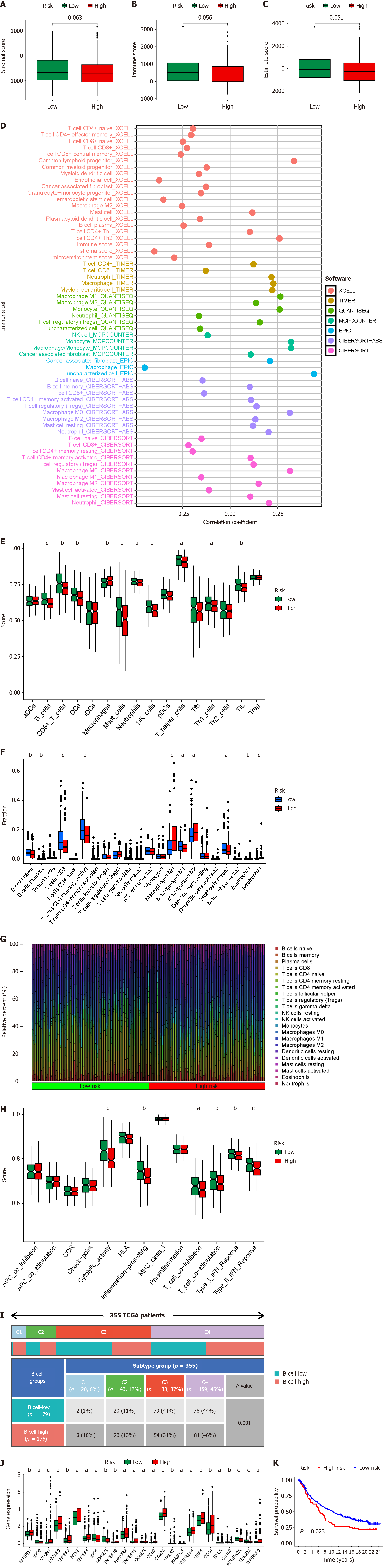
B cells, as crucial immune cells, are likely to play a significant role in the regulation of the immune microenvironment in HCC. Regarding immune microenvironment analysis, patients with HCC in the high-risk score group showed a trend towards decreased stromal score (P = 0.063), immune score (P = 0.056), and estimate score (P = 0.051), although the lack of significant differences may be attributed to the small sample size (Figure 8A–C). Concerning immune cell infiltration, there was a significant negative correlation between risk score and B-cell infiltration, with various types of T cells showing a similar trend to B cells (Figure 8D). ssGSEA algorithm or CIBERSORT algorithm both revealed significantly suppressed B-cell infiltration in patients with high risk scores, with T cells exhibiting a similar infiltration pattern (Figure 8E–G). These findings align with those shown in Figures 2 and 3, indicating a probable collaboration between B cells and T cells in HCC. Based on the ssGSEA algorithm, we also observed significant suppression of functions related to cytolytic activity, inflammation promotion, T-cell co-inhibition, and T-cell co-stimulation in high-risk patients (Figure 8H), further corroborating our standpoint.
The TCGA official team has classified HCC into four subtypes: (1) Wound Healing (Immune C1); (2) Interferon- dominant (Immune C2); (3) Inflammatory (Immune C3); and (4) Lymphocyte depleted (Immune C4), which has become a significant reference for classification[20]. Our findings indicate that the BRGs prognosis model can differentiate immune subtypes, particularly with a notable increase in the number of Immune C1 patients with high-risk scores and a marked decrease in the number of Immune C3 patients (Figure 8I).
Our findings indicate that the BRGs prognosis model can differentiate immune subtypes, particularly with a notable increase in the number of Immune C1 patients with high-risk scores and a marked decrease in the number of Immune C3 patients (Figure 8I). Immune checkpoints, crucial factors expressed on immune cells capable of modulating immune activation levels, play a pivotal role. These genes, under normal circumstances, can inhibit T-cell function to prevent excessive immune system activation. Simultaneously, in tumor tissues, they might be exploited by tumors to facilitate immune evasion. The effectiveness of immune checkpoint inhibitors in treating HCC has been demonstrated[21,22]. Our analysis suggests that in the high-risk score group, expression of most immune checkpoint genes (17 of 25) significantly increased (Figure 8J), indicating that patients in the high-risk score group may benefit from these immune checkpoint inhibitors. ENTPD1, NT5E, and HAVCR2 are also immune checkpoint genes coexpressed at high levels in both B cells and T cells[23], further indicating potential synergistic action between B cells and T cells in HCC. We evaluated the efficacy of immunotherapy in the IMvigor210 immune treatment cohort using the BRGs prognosis model and found that high-risk patients showed significantly poor response to immunotherapy (Figure 8K). This suggests that high-risk group patients may require alternative therapeutic agents to improve efficacy. Therefore, based on drug sensitivity analysis, we have identified potential targeted drugs such as osimertinib, gefitinib, GDC0810, and paclitaxel (Supplementary Figure 1) that may be beneficial for high-risk group patients. These drugs, in combination with immunotherapy, could serve as future references for further validation in clinical experiments.
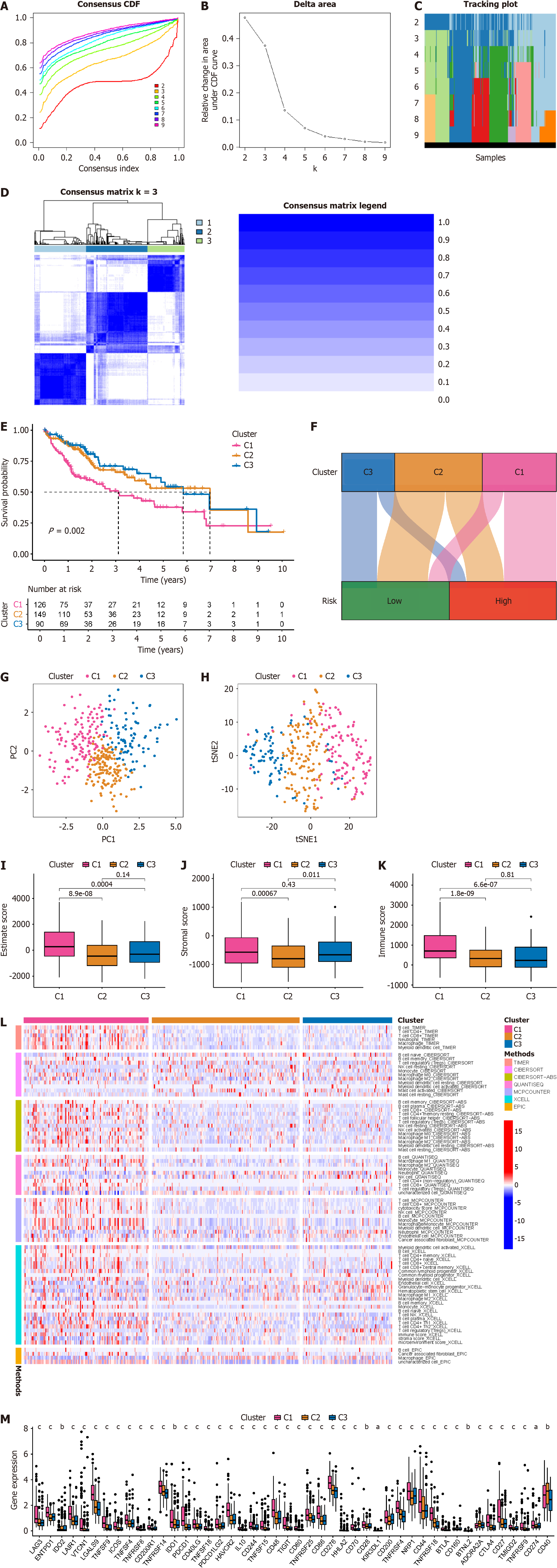
Current immunotherapy for HCC patients yields only a 15%–30% remission rate, possibly due to inadequate differentiation among patients with distinct immune microenvironments, thus failing to achieve personalized precision treatment. Therefore, identifying novel molecular subtyping for HCC is imperative[7]. Leveraging all 11 BRGs in the prognosis model, we classified HCC patients into molecular subtypes using consensus cluster plus. Based on delta area and CDF curves, we observed good stability in sample clustering when k = 3 (Figure 9A and B). We depicted sample distribution under different k values (Figure 9C). A heatmap of CM demonstrated consistent blue shading when k = 3 (Figure 9D). Subsequently, the C1 group had the poorest prognosis, while those in the C2 and C3 groups exhibited better prognosis based on survival analysis (Figure 9E). A Sankey diagram illustrated the correspondence between novel molecular subtyping and prognosis model grouping (Figure 9F). PCA and t-distributed stochastic neighbor embedding analyses both distinguished HCC patients based on the novel molecular subtyping (Figure 9G and H). We attempted to ascertain whether the novel molecular subtyping could differentiate HCC patients with distinct immune features. Analysis of the immune microenvironment indicated that patients in the C1 group had high estimate scores, stromal scores, and immune scores, whereas those in the C2 group had low scores, with C3 group patients falling between the two (Figure 9I–K). Regarding immune cell infiltration, C1 group patients exhibited markedly elevated levels across various immune cell types, possibly contributing to their poorer prognosis due to immune resource depletion, whereas C2 and C3 group patients showed low levels of immune cell infiltration (Figure 9L). Immune checkpoint analysis revealed significant upregulation of most immune checkpoint genes (42 of 46) in C1 group patients, indicating their potential benefit from immune checkpoint inhibitors. C2 patients might benefit from CD40 inhibitors, while C3 patients might benefit from IDO2, KIR3DL1, and CD160 inhibitors (Figure 9M). We analyzed potential targeted drugs suitable for HCC patients in each immune subtype. C1 group patients were most likely to benefit from staurosporine, pevonedistat, and midkine (MK)-8776, while C2 group patients might benefit from podophyllotoxin bromide and elephantin, and C3 group patients might benefit from epirubicin, extracellular regulated protein kinases (ERK)-2440, and Janus kinase (JAK)-8517 (Supplementary Figure 2).
Although the role of T cells in antitumor immunity has been extensively studied, the role of B cells, including in HCC, remains poorly understood, hindering efforts to utilize B-cell responses for cancer immunotherapy[24]. B cells, as a crucial subset of antigen-presenting cells, play a significant role, which is often overlooked in tumor immunotherapy, including HCC. Unlike dendritic cells and macrophages, which primarily present antigens through phagocytosis, B cells predominantly present antigens through the high-affinity binding of the B-cell receptor (BCR) to the antigen, thereby effectively recognizing low levels of antigens and enhancing antigen presentation efficiency. Upon binding of antigens to specific BCRs, the process of antigen internalization initiates, and the antigens are displayed on the surface of B cells via MHC class II molecules, activating cognate CD4+ helper T cells, thus inducing antibody production[25]. Based on gene enrichment analysis of BRGs, B cells and T cells (including CD4+ and CD8+ T cells) significantly enrich pathways associated with HCC occurrence and development, such as T-cell receptor signaling pathway and natural-killer-cell mediated cytotoxicity. B cells and T cells jointly regulate important biological processes such as GO nuclear transcribed mRNA catabolic process nonsense mediated decay and GO translational elongation. Our analysis reveals a potential synergistic interaction between B cells and T cells in HCC. Our findings suggest that B cells play a crucial role in activating CD4+ T cells and may act in conjunction with CD8+ T cells. This discovery represents a novel breakthrough in tumor therapy.
The role of B cells in most tumors, including HCC, remains a subject of considerable controversy[25]. Among the 11 key genes in our BRGs prognostic model, BLNK is associated with a favorable prognosis in HCC, exhibiting high expression in B cells within HCC. Conversely, CALM1, LEPROTL1, S100A10, LDHA, LYAR, CKLF, and TXN are linked to an adverse prognosis in HCC, displaying low expression in B cells within HCC. The alignment of these BRGs with the prognostic outcomes of HCC, as reflected in their expression within B cells, suggests that B-cell activation is a favorable prognostic factor for HCC. However, certain BRGs exhibit discrepancies between their overall expression in HCC and their expression specifically within B cells. For instance, FYN and KLRB1 are associated with a favorable prognosis in HCC but demonstrate low expression in B cells within HCC, while MARCKSL1 is linked to an adverse prognosis in HCC yet exhibits high expression in B cells within HCC. These findings suggest potential complexities in the roles of these BRGs, indicating that their functions may extend beyond B cells, warranting further investigation. The high risk score group identified by the BRGs prognostic model shows significant enrichment in pathways such as ECM–receptor interaction, central carbon metabolism in cancer, protein digestion and absorption, HIF-1 signaling pathway, biosynthesis of amino acids, and fructose and mannose metabolism. This implies that B cells may regulate the occurrence and progression of HCC through these pathways, necessitating further validation in future studies.
Similar to regulatory T cells, B cells can also exhibit regulatory phenotypes to suppress immune responses. They express inhibitory receptors on their surface, akin to immune checkpoint genes. These include a variety of immune checkpoint molecules like programmed cell death protein 1 (PD-1) and its ligands PD ligand 1 (PD-L1)/PD ligand 2, which play diverse regulatory roles in both humoral and cellular immunity[26]. Recent findings in HCC reveal that B cells with elevated PD-1 expression, when encountering PD-L1-expressing cells, induce T-cell suppression by releasing interleukin-10. Blocking PD-1 on B cells with checkpoint blockade antibodies has shown promise in enhancing cancer immunotherapy[27]. Anti-PD-1 therapy has become the preferred immunotherapy for treating HCC. However, anti-PD1 therapy targeting T cells has only achieved a 15% response rate in HCC patients[7]. In this context, anti-PD-1 therapy targeting B cells may potentially become an important approach to improving the efficacy of immunotherapy for HCC patients. Our results indicate significant upregulation in expression of most T-cell immune checkpoint genes in the high risk scoring group of HCC patients, with ENTPD1, NT5E, and HAVCR2 being among the immune checkpoint genes highly expressed in B cells[23]. Targeting inhibitors against these shared immune checkpoint genes in both B cells and T cells may effectively harness the synergistic action of these cells in HCC therapy; a hypothesis deserving validation in future multicenter large-scale clinical trials.
Our prognostic model for HCC, termed BRGs, offers guidance in diagnosis and prognosis prediction. Through univariate Cox regression and LASSO regression analysis, we constructed the BRGs prognostic model. Our analysis indicates a significant reduction in overall survival for patients in the high-risk group compared to those in the low-risk group. Furthermore, through visualizations such as risk curves, risk heatmaps, ROC curves, and Kaplan–Meier survival curves, we validated the robust predictive power of our risk model, which outperformed other significant risk factors in HCC such as age, grade, and stage. Importantly, similar robust results were replicated in two external validation sets for HCC (ICGC and GSE76427), bolstering the reliability and reproducibility of our research findings.
Beyond elucidating the role of B cells in HCC, identifying precise molecular subtypes is crucial for enhancing personalized treatment, given the widespread resistance of HCC to targeted therapies[28]. Leveraging 11 BRGs from our prognostic model, we classified HCC patients into three molecular subtypes. Our results indicate that C1 patients exhibited the most extensive immune cell infiltration, potentially indicating immune exhaustion and consequently the poorest prognosis. However, these patients may benefit from targeted drugs such as staurosporine, pevonedistat, and MK-8776. Conversely, C2 and C3 patients showed better prognosis. C2 patients may improve their prognosis with drugs such as podophyllotoxin bromide and elephantin, while C3 patients may benefit from drugs such as epirubicin, ERK-2440, and JAK-8517. Our proposed novel HCC molecular subtypes hold significant implications for precise HCC treatment.
Our research suggests that in HCC, B cells play a role in tumor cytotoxicity by activating CD4+ and CD8+ T cells. We have established a reliable BRGs prognostic model and novel molecular subtypes, offering pivotal and valuable references for the diagnosis, prognosis, and personalized treatment strategies of HCC.
| 1. | Toh MR, Wong EYT, Wong SH, Ng AWT, Loo LH, Chow PK, Ngeow J. Global Epidemiology and Genetics of Hepatocellular Carcinoma. Gastroenterology. 2023;164:766-782. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 266] [Reference Citation Analysis (0)] |
| 2. | Xu K, Wu T, Xia P, Chen X, Yuan Y. Alternative splicing: a bridge connecting NAFLD and HCC. Trends Mol Med. 2023;29:859-872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 19] [Reference Citation Analysis (0)] |
| 3. | Pfister D, Núñez NG, Pinyol R, Govaere O, Pinter M, Szydlowska M, Gupta R, Qiu M, Deczkowska A, Weiner A, Müller F, Sinha A, Friebel E, Engleitner T, Lenggenhager D, Moncsek A, Heide D, Stirm K, Kosla J, Kotsiliti E, Leone V, Dudek M, Yousuf S, Inverso D, Singh I, Teijeiro A, Castet F, Montironi C, Haber PK, Tiniakos D, Bedossa P, Cockell S, Younes R, Vacca M, Marra F, Schattenberg JM, Allison M, Bugianesi E, Ratziu V, Pressiani T, D'Alessio A, Personeni N, Rimassa L, Daly AK, Scheiner B, Pomej K, Kirstein MM, Vogel A, Peck-Radosavljevic M, Hucke F, Finkelmeier F, Waidmann O, Trojan J, Schulze K, Wege H, Koch S, Weinmann A, Bueter M, Rössler F, Siebenhüner A, De Dosso S, Mallm JP, Umansky V, Jugold M, Luedde T, Schietinger A, Schirmacher P, Emu B, Augustin HG, Billeter A, Müller-Stich B, Kikuchi H, Duda DG, Kütting F, Waldschmidt DT, Ebert MP, Rahbari N, Mei HE, Schulz AR, Ringelhan M, Malek N, Spahn S, Bitzer M, Ruiz de Galarreta M, Lujambio A, Dufour JF, Marron TU, Kaseb A, Kudo M, Huang YH, Djouder N, Wolter K, Zender L, Marche PN, Decaens T, Pinato DJ, Rad R, Mertens JC, Weber A, Unger K, Meissner F, Roth S, Jilkova ZM, Claassen M, Anstee QM, Amit I, Knolle P, Becher B, Llovet JM, Heikenwalder M. NASH limits anti-tumour surveillance in immunotherapy-treated HCC. Nature. 2021;592:450-456. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 259] [Cited by in RCA: 839] [Article Influence: 209.8] [Reference Citation Analysis (1)] |
| 4. | Li X, Ramadori P, Pfister D, Seehawer M, Zender L, Heikenwalder M. The immunological and metabolic landscape in primary and metastatic liver cancer. Nat Rev Cancer. 2021;21:541-557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 345] [Article Influence: 86.3] [Reference Citation Analysis (0)] |
| 5. | Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, Coussens LM, Gabrilovich DI, Ostrand-Rosenberg S, Hedrick CC, Vonderheide RH, Pittet MJ, Jain RK, Zou W, Howcroft TK, Woodhouse EC, Weinberg RA, Krummel MF. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24:541-550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2019] [Cited by in RCA: 3802] [Article Influence: 543.1] [Reference Citation Analysis (0)] |
| 6. | Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S, Lencioni R, Koike K, Zucman-Rossi J, Finn RS. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7:6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4432] [Cited by in RCA: 3852] [Article Influence: 963.0] [Reference Citation Analysis (3)] |
| 7. | Finn RS, Qin S, Ikeda M, Galle PR, Ducreux M, Kim TY, Kudo M, Breder V, Merle P, Kaseb AO, Li D, Verret W, Xu DZ, Hernandez S, Liu J, Huang C, Mulla S, Wang Y, Lim HY, Zhu AX, Cheng AL; IMbrave150 Investigators. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N Engl J Med. 2020;382:1894-1905. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2542] [Cited by in RCA: 4670] [Article Influence: 934.0] [Reference Citation Analysis (2)] |
| 8. | Shi Y. PLAN B for immunotherapy: Promoting and leveraging anti-tumor B cell immunity. J Control Release. 2021;339:156-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 9. | Qin M, Wang D, Fang Y, Zheng Z, Liu X, Wu F, Wang L, Li X, Hui B, Ma S, Tang W, Pan X. Current Perspectives on B Lymphocytes in the Immunobiology of Hepatocellular Carcinoma. Front Oncol. 2021;11:647854. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 10. | Saffo S, Taddei TH. Systemic Management for Advanced Hepatocellular Carcinoma: A Review of the Molecular Pathways of Carcinogenesis, Current and Emerging Therapies, and Novel Treatment Strategies. Dig Dis Sci. 2019;64:1016-1029. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 11. | Okusaka T, Ikeda M. Immunotherapy for hepatocellular carcinoma: current status and future perspectives. ESMO Open. 2018;3:e000455. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 70] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 12. | Han Y, Wang Y, Dong X, Sun D, Liu Z, Yue J, Wang H, Li T, Wang C. TISCH2: expanded datasets and new tools for single-cell transcriptome analyses of the tumor microenvironment. Nucleic Acids Res. 2023;51:D1425-D1431. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 278] [Article Influence: 92.7] [Reference Citation Analysis (0)] |
| 13. | Zhang L, Yu X, Zheng L, Zhang Y, Li Y, Fang Q, Gao R, Kang B, Zhang Q, Huang JY, Konno H, Guo X, Ye Y, Gao S, Wang S, Hu X, Ren X, Shen Z, Ouyang W, Zhang Z. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature. 2018;564:268-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 427] [Cited by in RCA: 886] [Article Influence: 126.6] [Reference Citation Analysis (0)] |
| 14. | Yost KE, Satpathy AT, Wells DK, Qi Y, Wang C, Kageyama R, McNamara KL, Granja JM, Sarin KY, Brown RA, Gupta RK, Curtis C, Bucktrout SL, Davis MM, Chang ALS, Chang HY. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat Med. 2019;25:1251-1259. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1020] [Cited by in RCA: 1037] [Article Influence: 172.8] [Reference Citation Analysis (0)] |
| 15. | Sun D, Wang J, Han Y, Dong X, Ge J, Zheng R, Shi X, Wang B, Li Z, Ren P, Sun L, Yan Y, Zhang P, Zhang F, Li T, Wang C. TISCH: a comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic Acids Res. 2021;49:D1420-D1430. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 373] [Cited by in RCA: 707] [Article Influence: 176.8] [Reference Citation Analysis (0)] |
| 16. | Vu R, Jin S, Sun P, Haensel D, Nguyen QH, Dragan M, Kessenbrock K, Nie Q, Dai X. Wound healing in aged skin exhibits systems-level alterations in cellular composition and cell-cell communication. Cell Rep. 2022;40:111155. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 66] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 17. | Xu K, Xia P, Liu P, Zhang X. A six lipid metabolism related gene signature for predicting the prognosis of hepatocellular carcinoma. Sci Rep. 2022;12:20781. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 18. | Xu K, Dai C, Yang J, Xu J, Xia C, Li J, Zhang C, Xu N, Wu T. Disulfidptosis-related lncRNA signatures assess immune microenvironment and drug sensitivity in hepatocellular carcinoma. Comput Biol Med. 2024;169:107930. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 19. | Zhang L, Zhang X, Liu H, Yang C, Yu J, Zhao W, Guo J, Zhou B, Jiang N. MTFR2-dependent mitochondrial fission promotes HCC progression. J Transl Med. 2024;22:73. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 20. | Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA, Ziv E, Culhane AC, Paull EO, Sivakumar IKA, Gentles AJ, Malhotra R, Farshidfar F, Colaprico A, Parker JS, Mose LE, Vo NS, Liu J, Liu Y, Rader J, Dhankani V, Reynolds SM, Bowlby R, Califano A, Cherniack AD, Anastassiou D, Bedognetti D, Mokrab Y, Newman AM, Rao A, Chen K, Krasnitz A, Hu H, Malta TM, Noushmehr H, Pedamallu CS, Bullman S, Ojesina AI, Lamb A, Zhou W, Shen H, Choueiri TK, Weinstein JN, Guinney J, Saltz J, Holt RA, Rabkin CS; Cancer Genome Atlas Research Network, Lazar AJ, Serody JS, Demicco EG, Disis ML, Vincent BG, Shmulevich I. The Immune Landscape of Cancer. Immunity. 2018;48:812-830.e14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4007] [Cited by in RCA: 3812] [Article Influence: 544.6] [Reference Citation Analysis (0)] |
| 21. | Llovet JM, Castet F, Heikenwalder M, Maini MK, Mazzaferro V, Pinato DJ, Pikarsky E, Zhu AX, Finn RS. Immunotherapies for hepatocellular carcinoma. Nat Rev Clin Oncol. 2022;19:151-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 1005] [Article Influence: 335.0] [Reference Citation Analysis (2)] |
| 22. | Postow MA, Sidlow R, Hellmann MD. Immune-Related Adverse Events Associated with Immune Checkpoint Blockade. N Engl J Med. 2018;378:158-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2308] [Cited by in RCA: 3141] [Article Influence: 448.7] [Reference Citation Analysis (0)] |
| 23. | Bod L, Kye YC, Shi J, Torlai Triglia E, Schnell A, Fessler J, Ostrowski SM, Von-Franque MY, Kuchroo JR, Barilla RM, Zaghouani S, Christian E, Delorey TM, Mohib K, Xiao S, Slingerland N, Giuliano CJ, Ashenberg O, Li Z, Rothstein DM, Fisher DE, Rozenblatt-Rosen O, Sharpe AH, Quintana FJ, Apetoh L, Regev A, Kuchroo VK. B-cell-specific checkpoint molecules that regulate anti-tumour immunity. Nature. 2023;619:348-356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 86] [Article Influence: 43.0] [Reference Citation Analysis (0)] |
| 24. | Zhang Q, Chen Y, Bai X, Liang T. Immune Checkpoint Blockade Therapy for Hepatocellular Carcinoma: Clinical Challenges and Considerations. Front Oncol. 2020;10:590058. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 25. | Cyster JG, Allen CDC. B Cell Responses: Cell Interaction Dynamics and Decisions. Cell. 2019;177:524-540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 594] [Cited by in RCA: 632] [Article Influence: 105.3] [Reference Citation Analysis (0)] |
| 26. | Veh J, Ludwig C, Schrezenmeier H, Jahrsdörfer B. Regulatory B Cells-Immunopathological and Prognostic Potential in Humans. Cells. 2024;13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 15] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 27. | Shalapour S, Lin XJ, Bastian IN, Brain J, Burt AD, Aksenov AA, Vrbanac AF, Li W, Perkins A, Matsutani T, Zhong Z, Dhar D, Navas-Molina JA, Xu J, Loomba R, Downes M, Yu RT, Evans RM, Dorrestein PC, Knight R, Benner C, Anstee QM, Karin M. Inflammation-induced IgA+ cells dismantle anti-liver cancer immunity. Nature. 2017;551:340-345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 259] [Cited by in RCA: 424] [Article Influence: 53.0] [Reference Citation Analysis (0)] |
| 28. | Ladd AD, Duarte S, Sahin I, Zarrinpar A. Mechanisms of drug resistance in HCC. Hepatology. 2024;79:926-940. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 92] [Article Influence: 92.0] [Reference Citation Analysis (0)] |