Published online Jun 28, 2024. doi: 10.3748/wjg.v30.i24.3036
Revised: April 30, 2024
Accepted: June 4, 2024
Published online: June 28, 2024
Processing time: 102 Days and 3 Hours
Autophagy, a conserved cellular degradation process, is crucial for various cellular processes such as immune responses, inflammation, metabolic and oxidative stress adaptation, cell proliferation, development, and tissue repair and remodeling. Dysregulation of autophagy is suspected in numerous diseases, including cancer, neurodegenerative diseases, digestive disorders, metabolic syndromes, and infectious and inflammatory diseases. If autophagy is disrupted, for example, this can have serious consequences and lead to chronic inflammation and tissue damage, as occurs in diseases such as Chron's disease and ulcerative colitis. On the other hand, the influence of autophagy on the development and progression of cancer is not clear. Autophagy can both suppress and promote the progression and metastasis of cancer at various stages. From inflammatory bowel diseases to gastrointestinal cancer, researchers are discovering the intricate role of autophagy in maintaining gut health and its potential as a therapeutic target. Researchers should carefully consider the nature and progression of diseases such as cancer when trying to determine whether inhibiting or stimulating autophagy is likely to be beneficial. Multidisciplinary approaches that combine cutting-edge research with clinical expertise are key to unlocking the full therapeutic potential of autophagy in digestive diseases.
Core Tip: There are still many unanswered questions regarding the role of autophagy in the onset and development of diseases. Before induction or inhibition of autophagy can be into therapeutic protocols, the mysteries surrounding autophagy need to be solved through detailed experimental studies.
- Citation: Esrefoglu M. Harnessing autophagy: A potential breakthrough in digestive disease treatment. World J Gastroenterol 2024; 30(24): 3036-3043
- URL: https://www.wjgnet.com/1007-9327/full/v30/i24/3036.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i24.3036
Autophagy, also known as autophagocytosis, is a conserved cellular degradation process that plays a role in the survival and maintenance of the cell by degrading cytoplasmic organelles, proteins, and macromolecules, and recycling the degradation products. Through a lysosome-dependent, regulated mechanism, autophagy removes unnecessary or dysfunctional components. Autophagy is crucial for maintaining cellular and organismal homeostasis[1,2], facilitating immune responses[3], preventing inflammation[4], adapting to metabolic, oxidative, and inflammatory stress conditions[5,6], supporting energy metabolism[1,7], contributing to various aspects of development[2,8-10], regulating cell proliferation[11], and tissue repair and remodeling[12]. Dysregulation of autophagy has been implicated in numerous diseases, including cancer, neurodegenerative disorders, digestive diseases, metabolic syndromes, and infectious and inflammatory diseases, highlighting its importance in overall health and disease prevention.
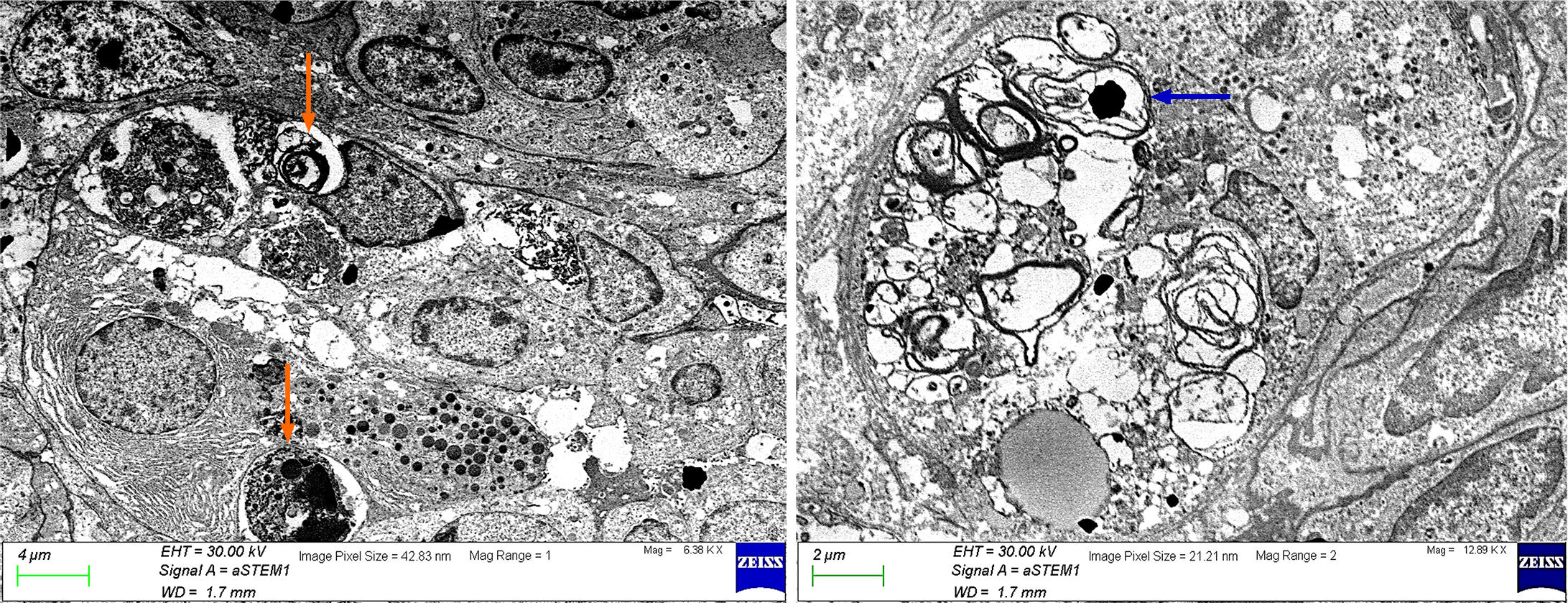
Autophagy is a generic term for all pathways by which cytoplasmic materials are released into the lysosomes in animal cells or into the vacuole in plant and yeast cells. The resultant degradation products can be reused in protein synthesis, energy production, and gluconeogenesis[12]. Autophagy refers to the formation of autophagosomes that surround the organelles and proteins that need to be degraded in the cells. The standard pathway of mammalian autophagy includes induction, formation of the phagophore, an isolation membrane, expansion of phagophore to form the autophagosome, and fusion of the autophagosome with the lysosome forming autolysosome where degradation occurs. Phagophores and autophagosomes are double-membraned structures[13,14]. Autophagosomes are formed on or near the endoplasmic reticulum (ER)[13,15]. However, some recent studies suggest that additional membrane sources such as the Golgi complex, mitochondria, and the plasma membrane also contribute to autophagosome formation[13,15-17]. Three-dimensional analysis of correlative light and electron microscopy by Takahashi et al[18] shows that ER is associated with all phagophores, 92% of autophagosomes, and 79% of autolysosomes suggesting that most autophagosomes remain on the ER after closure and even fuse with the lysosomes. They also reported that the ER is the organelle most frequently engulfed by phagophores and autophagosomes, with a percentage of 65%. Sometimes the content is undefinable although membranous, and sometimes it is non-membranous but only cytosol. Therefore, the electron microscopic features of the autophagosomes can vary in size and shape, reflecting the diversity of cellular cargo being degraded. Engulfed organelles, cytoplasmic components, cytosol (Figure 1), or pathogens can be observed in the autophagosomes.
Although autophagy was previously thought to be a non-selective degradation mechanism that indiscriminately degrades cytoplasmic components, more recent research has shown that it can be selective in some situations[19]. Depending on which organelle is removed, selective autophagy has different names. For example, if the target cargo is the mitochondria, the process is called mitophagy, if the target cargo is the nucleus, it is called nucleophagy[20]. With most of these names such as ER-phagy, lysophagy, and lipophagy, it is easy to recognize which cellular element is involved in the cargo. Some names such as xenophagy (target is cellular pathogens)[21], allophagy (target is paternal organelles)[22], aggrephagy (target is abnormal protein aggregates)[23] do not give a clear idea of what the content of the cargo is.
Autophagy is broadly divided into macroautophagy, chaperone-mediated autophagy (CMA), and microautophagy. The term autophagy often refers to macroautophagy[19]. The degradation of cargo by macroautophagy enables the recycling of the resultant macromolecules to maintain cellular homeostasis. The substrates of cytoplasmic macroautophagy are entrapped in the autophagosome, a temporary organelle with double membrane that eventually fuses with lysosomes and vacuoles[2,24]. CMA is a type of selective autophagy that focuses on proteins for lysosomal destruction. The supply of free amino acids generated after protein degradation is the main function of this pathway. In addition, CMA influences glucose and lipid metabolism and controls cell metabolism, which in turn influences the body's overall energy metabolism[25]. In microautophagy, autophagic tubing facilitates both the rupture of vesicles and access to the cytoplasmic lumen by directly engulfing the cytoplasmic cargo[26]. In this way, eukaryotic cells directly degrade a variety of autophagic cargoes through a lysosomal degradation process[27]. It contributes to the maintenance of cell survival and homeostasis[26].
A collection of proteins known as class III phosphatidylinositol3-kinase, also known as Vps34, and BH3 domain protein Beclin-1 (BECN1) interacting with B-cell lymphoma 2 (Bcl-2) make up the intracellular machinery of autophagy. Both proteins are required for the development of autophagosomes[28]. The mammalian target of the rapamycin signaling pathway, a serine/threonine kinase involved in various pathways of cell survival, can initiate signal transduction[29].
Autophagy has been described as a novel mechanism for cell death as well as a stress-adaptive mechanism to support cell survival. In stressful conditions, autophagy ensures that the cells receive the metabolic substrates required for energy for cell growth and survival[30,31]. If carbon sources such as amino acids and glucose are insufficient to maintain the rate of protein synthesis or to provide sufficient amounts of ATP needed to sustain metabolic reactions, cells turn on autophagy to rapidly degrade obsolete and depleted fractions and recycle the available biomolecule library. Through negative feedback, autophagy can be induced to provide energy and building blocks to restore homeostasis while eliminating oxidative damage. From this perspective, cells also need autophagy to overcome oxidative stress[5]. Autophagy delays cell death by removing mitochondria, ER components, peroxisomes, and proteins damaged by oxidative stress. There is a close relationship between autophagy and oxidative stress[32-34]. Accordingly, defects of autophagic genes lead to increased production of reactive oxygen species (ROS) and accumulation of damaged organelles and DNA which in turn promote metabolic reprogramming and induce tumorigenesis[35]. Cancer cells have an increased metabolism for proliferation, and they usually need to grow under hypoxic conditions until angiogenesis is adequately established. As a result, cancer cells, especially those with RAS mutations, which are one of the most common mutations in all malignancies, are highly dependent on and dependent on autophagy[36].
Because the baseline level of autophagy is important for maintaining normal cellular homeostasis which allows physiological turnover of damaged organelles, and cytoplasmic content, it should be tightly regulated. Large-scale degradation is important in autophagic function, but it also carries some risk because unregulated degradation of the cytoplasm is likely to be lethal[37]. The most important lesson is probably that autophagy can be harmful either in excess or insufficient amounts. This is complicated by the fact that autophagy plays a dual role in both cytoprotection and cell death. Since silencing of autophagy-related gene (ATG) results in cell death occurring more rapidly rather than gradually, autophagy is thought to play a cytoprotective role in response to most forms of cellular stress[38,39]. Conversely, if ATG1 is upregulated, the resulting upregulation of autophagy can lead to apoptotic cell death[40]. The mechanisms linking autophagy and apoptosis are not fully understood; however, it is known that many signals that induce activation of apoptosis also induce autophagy, while signals that inhibit apoptosis also inhibit autophagy[41-44]. In addition, several apoptotic proteins such as Bcl-2-associated X protein, NOXA, and NIX modulate autophagy, and several autophagic proteins are implicated in intrinsic and extrinsic apoptosis[44]. The coordinated regulation of ‘self-digestion’ by autophagy and ‘self-killing’ by apoptosis may underlie various aspects of development, tissue homeostasis, and disease pathogenesis[37].
It is well known that cells in the digestive system, like every part of the body, rely on autophagy to maintain their function and health. For example, basal autophagy metabolizes cytoplasmic components to prevent the accumulation of degenerated proteins and organelles in hepatocytes, thereby preventing hepatocellular degeneration[13]. Loss of ATG7 in mouse hepatocytes has been shown to lead to an accumulation of edematous and deformed mitochondria, an increase in the number of peroxisomes and lipid droplets, and the formation of protein aggregates[45]. The mutant mice suffering from severe hepatomegaly develop hepatitis[46]. In addition, spontaneous benign tumorigenesis is observed in the livers of mice with systemic mosaic deletion of ATG5 or with ATG7-specific disruption in hepatocytes[35].
Similar to the liver, basal autophagy is also important for the maintenance of beta cell volume and function in the endocrine pancreas[13]. Autophagy is necessary to maintain the structure, mass, and function of pancreatic beta cells. Impairment of autophagy leads to insulin deficiency and hyperglycemia as the turnover and function of cellular organelles are impaired[46].
The most important link between autophagy and the intestinal epithelium is centered on Paneth cells. In vivo studies have shown that autophagy or ATG genes play a role in maintaining the normal function of Paneth cells. ATG16L1 hypomorphic mice, which express a very low level of ATG16L1, exhibit structurally abnormal and disorganized Paneth cell granules[47]. ATG5- and ATG7-deficient mice have also shown morphological abnormalities of Paneth cells[48]. The lineage between autophagy and Paneth cells expresses several microbial pattern recognition receptors (PRRs), including nucleotide-binding oligomerization domain-containing protein 2, (NOD2) and Toll-like receptors[49,50], which can stimulate the release of granule contents of Paneth cells[51,52]. The antimicrobial proteins produced by Paneth cells can protect against pathogens and alter the composition of the commensal microbiota[53]. Increased ER stress in autophagy deficiency can lead to abnormalities in Paneth cell and goblet cell functions, and damage the barrier of normal intestinal antimicrobial and mucous proteins, thereby causing continuous inflammatory stimulation and promoting the occurrence of inflammatory bowel diseases (IBDs)[54].
PRRs induce autophagy, and via autophagic adaptors, autophagy provides a mechanism for the clearance of intracellular microorganisms. Autophagy controls inflammation by regulating interactions with innate immune signaling pathways, by removing endogenous inflammasome agonists, and by affecting the secretion of immune mediators[55]. In many cases, signaling associated with inflammation may also regulate autophagy. Recent studies suggest that in addition to excessive inflammation, alterations in immune response, and imbalance of gut flora, abnormal autophagy may be associated with IBDs. Single-nucleotide polymorphisms (SNPs) of various genes related to autophagy are associated with susceptibility to IBDs[54].
Autophagy has shown promise to understand and treat numerous diseases, including digestive diseases, infectious and inflammatory diseases, and cancers. From IBD to gastrointestinal cancer, researchers are discovering the complex role of autophagy in maintaining gut health and its potential as a therapeutic target. Chron’s disease (CD) and ulcerative colitis (UC) are the two major forms of IBD characterized by damage to the intestinal epithelium which forms a barrier between luminal contents and the mucosal immune system. Modulation of tight junctions, leading to increased permeability of the barrier, has been associated with IBDs[56]. The intestinal epithelium disrupted by altered tight junction expression allows more microbiota or luminal antigens to cross the barrier, leading to stimulation of local immune cells, production of chemokines, and subsequent infiltration of immune cells. One of the main mechanisms of chronic long-term inflammation in UC is the excessive inflammation caused by these events[57]. Autophagy and its regulatory mechanisms are involved in both homeostasis and repair of the intestine. They support intestinal barrier function in response to cellular stress by regulating tight junction proteins and protecting against cell death[58,59]. Previous studies have shown that autophagy can increase the expression of occluding via terminating its BECN1-mediated endocytosis[60], conversely, it can downregulate claudin-2 via lysosomal degradation or TNF-mediated inhibition[59,61,62]. Previous studies have demonstrated that occludin expression is reduced in the colonic mucosa of UC patients and is positively related to intestinal mucosal healing[63].
The association between the polymorphisms of specific autophagy genes including ATG16L1, immunity-related GTPase family M protein, and leucine-rich repeat kinase 2, and susceptibility to CD has been described extensively in the literature[58,64]. For example, the SNP of the ATG16L1 gene are important risk factors for the pathogenesis of CD and are closely associated with CD[65]. Genetic polymorphisms in NOD2 are also associated with the highest risk of developing CD and remain the greatest genetic risk factor[66]. ATG16L1 inhibits pro-inflammatory signaling downstream of NOD2[67]. Recent studies have shown that NOD2 acts as a sensor for intracellular bacteria by triggering autophagy through interaction with ATG16L1[68]. Interestingly, NOD2 and ATG16L1 have only been associated with CD, but not UC[69]. However, mutations in the X-box binding protein 1 gene, which is involved in the unfolded protein response, as risk factors for both UC and CD[70,71].
Although defects in autophagic genes lead to increased production of ROS and accumulation of damaged organelles and DNA which in turn promote metabolic reprogramming and induce tumorigenesis[35], research indicates that autophagy can either promote or suppress tumor growth, depending on the context. In some cases, autophagy acts as a tumor suppressor, facilitating the clearance of damaged organelles and protein aggregates that might otherwise promote oncogenesis. Conversely, in certain circumstances, autophagy can promote tumor survival and support cancer growth by providing nutrients and alleviating metabolic stress.
Autophagy can regulate the cell cycle thus influence cell proliferation[11,72]. Although the mechanisms underlying the interaction between autophagy and cell proliferation are not clear, various cell cycle proteins regulate autophagy, and autophagy in turn can modulate the expression of cell cycle proteins[11]. Basal autophagy in normal cells prevents carcinogenesis but basal autophagy in tumor cells promotes tumor growth and cancer progression[73]. Disruption of autophagic cell quality control can lead to tumor development. Tumor growth is also related to the supply of nutrients to the cells through the activation of autophagy. Therefore, both inhibition and activation of autophagy contribute to tumor development and growth through different pathways[20]. Recent data suggest that autophagy may promote cancer cell proliferation by degrading cyclin A2 to prevent G2/M cell cycle arrest. Autophagy also regulates cyclin D1 to promote proliferation. Cyclin D1 is frequently overexpressed in cancer cells[74,75]. Current evidence suggests that cyclin D1 is not degraded by autophagy. It is not known whether autophagy-induced promotion/maintenance of cyclin D1 and/or degradation of cyclin A2 is a universal mechanism to promote cancer cell proliferation by autophagy[73]. Reduction of cyclin D1 and suppression of proliferation in cancer cells can be induced by inhibition of autophagy via downregulation of autophagy genes such as RB1-inducible coiled-coil protein 1/focal adhesion kinase family interacting protein of 200 kDa[76], BECN[77], and ATG4B[78]. By contrast, one study reported that knockdown of ATG4B in colorectal cancer cells inhibited cyclin D1 expression, cell proliferation, and tumor growth but increased autophagy flux[79]. In addition, the autophagosome marker light chain 3-II protein is overexpressed in advanced colorectal cancer compared to normal surrounding tissues[80]. Furthermore, the expression of BECN1, a regulatory protein of autophagy, is increased in colorectal and gastric cancers without any significant association with invasion, metastasis, and stage[81].
Altered expression and/or mutations of autophagy genes including BECN, ATG10, and ATG5 have been reported in human gastric and colorectal cancers[82]. Downregulation of several autophagy genes including BECN1, ATG3, and ATG5 result in suppression of proliferation[83,84]. Increased autophagy activity has been found in human colorectal cancer[82,85]. Autophagy may play a role in the development and/or progression of colorectal cancer.
Taken together, autophagy can both suppress and promote cancer progression and metastasis at different stages. This complicates therapeutic intervention and makes it necessary to evaluate the type of tumor cell, its genetic background, the stage of tumor progression, and the tumor microenvironment to achieve the desired effect of autophagy modulation and avoid potential disease exacerbation[6].
In recent years, the complicated relationship between dysfunction of autophagy and digestive disorders has become increasingly evident. Autophagy, a self-degradation process, serves as a guardian of gut homeostasis, clearing damaged organelles, combating microbial invaders, and regulating inflammation. If autophagy is disrupted, for example, this can have serious consequences and lead to chronic inflammation and tissue damage, as occurs in diseases such as CD and UC. Targeting autophagy pathways holds immense potential for modulating immune responses, reducing inflammation, and restoring intestinal tissue integrity. Multidisciplinary approaches that combine cutting-edge research with clinical expertise are key to unlocking the full therapeutic potential of autophagy in digestive diseases. Researchers should carefully consider the nature and progression of diseases such as cancer when attempting to determine whether autophagy inhibition or stimulation is likely to be beneficial. For instance, autophagy acts as a tumor suppressor, facilitating the removal of damaged organelles and toxic protein aggregates that could otherwise promote oncogenesis. Conversely, in certain microenvironments or under specific genetic alterations, autophagy can promote tumor survival and fuel cancer growth by providing nutrients and mitigating metabolic stress. Nevertheless, translating autophagy research from the laboratory to the clinic presents its own set of challenges. While preclinical studies have yielded promising results, clinical trials must navigate the complexities of patient heterogeneity, treatment resistance, and off-target effects. Additionally, ensuring the safety and efficacy of autophagy-modulating therapies requires meticulous oversight and rigorous evaluation.
| 1. | Eskelinen EL. Autophagy: Supporting cellular and organismal homeostasis by self-eating. Int J Biochem Cell Biol. 2019;111:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 79] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 2. | Chang YF, Li JJ, Liu T, Wei CQ, Ma LW, Nikolenko VN, Chang WL. Morphological and biochemical characteristics associated with autophagy in gastrointestinal diseases. World J Gastroenterol. 2024;30:1524-1532. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 3] [Reference Citation Analysis (0)] |
| 3. | Gkikas I, Palikaras K, Tavernarakis N. The Role of Mitophagy in Innate Immunity. Front Immunol. 2018;9:1283. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 182] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 4. | Deretic V, Levine B. Autophagy balances inflammation in innate immunity. Autophagy. 2018;14:243-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 266] [Cited by in RCA: 415] [Article Influence: 59.3] [Reference Citation Analysis (0)] |
| 5. | Filomeni G, De Zio D, Cecconi F. Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell Death Differ. 2015;22:377-388. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1427] [Cited by in RCA: 1607] [Article Influence: 160.7] [Reference Citation Analysis (0)] |
| 6. | Dikic I, Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol. 2018;19:349-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1271] [Cited by in RCA: 2031] [Article Influence: 338.5] [Reference Citation Analysis (0)] |
| 7. | Yang J, Zhou R, Ma Z. Autophagy and Energy Metabolism. Adv Exp Med Biol. 2019;1206:329-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 75] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 8. | Tsukamoto S, Kuma A, Murakami M, Kishi C, Yamamoto A, Mizushima N. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008;321:117-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 413] [Cited by in RCA: 420] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 9. | Sato M, Sato K. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science. 2011;334:1141-1144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 384] [Cited by in RCA: 350] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 10. | Al Rawi S, Louvet-Vallée S, Djeddi A, Sachse M, Culetto E, Hajjar C, Boyd L, Legouis R, Galy V. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science. 2011;334:1144-1147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 347] [Cited by in RCA: 358] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 11. | Zheng K, He Z, Kitazato K, Wang Y. Selective Autophagy Regulates Cell Cycle in Cancer Therapy. Theranostics. 2019;9:104-125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 160] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 12. | Migneault F, Hébert MJ. Autophagy, tissue repair, and fibrosis: a delicate balance. Matrix Biol. 2021;100-101:182-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 46] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 13. | Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728-741. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3678] [Cited by in RCA: 4852] [Article Influence: 346.6] [Reference Citation Analysis (0)] |
| 14. | Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27-42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5765] [Cited by in RCA: 5742] [Article Influence: 337.8] [Reference Citation Analysis (1)] |
| 15. | Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol. 2010;12:831-835. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 405] [Cited by in RCA: 439] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 16. | Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656-667. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1137] [Cited by in RCA: 1061] [Article Influence: 70.7] [Reference Citation Analysis (0)] |
| 17. | Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010;12:747-757. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 723] [Cited by in RCA: 694] [Article Influence: 46.3] [Reference Citation Analysis (0)] |
| 18. | Takahashi S, Saito C, Koyama-Honda I, Mizushima N. Quantitative 3D correlative light and electron microscopy of organelle association during autophagy. Cell Struct Funct. 2022;47:89-99. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 19] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 19. | Zaffagnini G, Martens S. Mechanisms of Selective Autophagy. J Mol Biol. 2016;428:1714-1724. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 356] [Cited by in RCA: 450] [Article Influence: 50.0] [Reference Citation Analysis (0)] |
| 20. | Ichimiya T, Yamakawa T, Hirano T, Yokoyama Y, Hayashi Y, Hirayama D, Wagatsuma K, Itoi T, Nakase H. Autophagy and Autophagy-Related Diseases: A Review. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 216] [Article Influence: 43.2] [Reference Citation Analysis (0)] |
| 21. | Kwon DH, Song HK. A Structural View of Xenophagy, a Battle between Host and Microbes. Mol Cells. 2018;41:27-34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 34] [Reference Citation Analysis (0)] |
| 22. | Sato M, Sato K. Maternal inheritance of mitochondrial DNA: degradation of paternal mitochondria by allogeneic organelle autophagy, allophagy. Autophagy. 2012;8:424-425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 77] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 23. | Lamark T, Johansen T. Aggrephagy: selective disposal of protein aggregates by macroautophagy. Int J Cell Biol. 2012;2012:736905. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 278] [Cited by in RCA: 363] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 24. | Mizushima N, Ohsumi Y, Yoshimori T. Autophagosome formation in mammalian cells. Cell Struct Funct. 2002;27:421-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 674] [Cited by in RCA: 690] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 25. | Tasset I, Cuervo AM. Role of chaperone-mediated autophagy in metabolism. FEBS J. 2016;283:2403-2413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 99] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 26. | Li WW, Li J, Bao JK. Microautophagy: lesser-known self-eating. Cell Mol Life Sci. 2012;69:1125-1136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 568] [Cited by in RCA: 535] [Article Influence: 41.2] [Reference Citation Analysis (0)] |
| 27. | Wang L, Klionsky DJ, Shen HM. The emerging mechanisms and functions of microautophagy. Nat Rev Mol Cell Biol. 2023;24:186-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 247] [Article Influence: 123.5] [Reference Citation Analysis (0)] |
| 28. | Funderburk SF, Wang QJ, Yue Z. The Beclin 1-VPS34 complex--at the crossroads of autophagy and beyond. Trends Cell Biol. 2010;20:355-362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 549] [Cited by in RCA: 643] [Article Influence: 42.9] [Reference Citation Analysis (0)] |
| 29. | Nunes T, Bernardazzi C, de Souza HS. Cell death and inflammatory bowel diseases: apoptosis, necrosis, and autophagy in the intestinal epithelium. Biomed Res Int. 2014;2014:218493. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 100] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 30. | Bhutia SK, Kegelman TP, Das SK, Azab B, Su ZZ, Lee SG, Sarkar D, Fisher PB. Astrocyte elevated gene-1 induces protective autophagy. Proc Natl Acad Sci U S A. 2010;107:22243-22248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 98] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 31. | Ouyang L, Shi Z, Zhao S, Wang FT, Zhou TT, Liu B, Bao JK. Programmed cell death pathways in cancer: a review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 2012;45:487-498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 938] [Cited by in RCA: 1056] [Article Influence: 81.2] [Reference Citation Analysis (0)] |
| 32. | Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749-1760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1509] [Cited by in RCA: 1626] [Article Influence: 90.3] [Reference Citation Analysis (0)] |
| 33. | Wu D, Cederbaum AI. Inhibition of autophagy promotes CYP2E1-dependent toxicity in HepG2 cells via elevated oxidative stress, mitochondria dysfunction and activation of p38 and JNK MAPK. Redox Biol. 2013;1:552-565. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 69] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 34. | Wang T, Wang Q, Song R, Zhang Y, Zhang K, Yuan Y, Bian J, Liu X, Gu J, Liu Z. Autophagy Plays a Cytoprotective Role During Cadmium-Induced Oxidative Damage in Primary Neuronal Cultures. Biol Trace Elem Res. 2015;168:481-489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 35. | Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, Eishi Y, Hino O, Tanaka K, Mizushima N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795-800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 923] [Cited by in RCA: 1059] [Article Influence: 75.6] [Reference Citation Analysis (0)] |
| 36. | Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, Bause A, Li Y, Stommel JM, Dell'antonio G, Mautner J, Tonon G, Haigis M, Shirihai OS, Doglioni C, Bardeesy N, Kimmelman AC. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717-729. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1005] [Cited by in RCA: 1188] [Article Influence: 84.9] [Reference Citation Analysis (0)] |
| 37. | Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069-1075. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5421] [Cited by in RCA: 5286] [Article Influence: 310.9] [Reference Citation Analysis (0)] |
| 38. | Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest. 2005;115:2679-2688. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1271] [Cited by in RCA: 1326] [Article Influence: 66.3] [Reference Citation Analysis (0)] |
| 39. | Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741-752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2430] [Cited by in RCA: 2817] [Article Influence: 156.5] [Reference Citation Analysis (0)] |
| 40. | Scott RC, Juhász G, Neufeld TP. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol. 2007;17:1-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 468] [Cited by in RCA: 492] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 41. | Botti J, Djavaheri-Mergny M, Pilatte Y, Codogno P. Autophagy signaling and the cogwheels of cancer. Autophagy. 2006;2:67-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 112] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 42. | Gozuacik D, Kimchi A. Autophagy and cell death. Curr Top Dev Biol. 2007;78:217-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 311] [Cited by in RCA: 330] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 43. | Meijer AJ, Codogno P. Signalling and autophagy regulation in health, aging and disease. Mol Aspects Med. 2006;27:411-425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 193] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 44. | Mukhopadhyay S, Panda PK, Sinha N, Das DN, Bhutia SK. Autophagy and apoptosis: where do they meet? Apoptosis. 2014;19:555-566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 378] [Cited by in RCA: 464] [Article Influence: 46.4] [Reference Citation Analysis (0)] |
| 45. | Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425-434. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1755] [Cited by in RCA: 1924] [Article Influence: 96.2] [Reference Citation Analysis (0)] |
| 46. | Jung HS, Chung KW, Won Kim J, Kim J, Komatsu M, Tanaka K, Nguyen YH, Kang TM, Yoon KH, Kim JW, Jeong YT, Han MS, Lee MK, Kim KW, Shin J, Lee MS. Loss of autophagy diminishes pancreatic beta cell mass and function with resultant hyperglycemia. Cell Metab. 2008;8:318-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 503] [Cited by in RCA: 549] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 47. | Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, Stone CD, Brunt EM, Xavier RJ, Sleckman BP, Li E, Mizushima N, Stappenbeck TS, Virgin HW 4th. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259-263. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1256] [Cited by in RCA: 1242] [Article Influence: 73.1] [Reference Citation Analysis (0)] |
| 48. | Cadwell K, Patel KK, Komatsu M, Virgin HW 4th, Stappenbeck TS. A common role for Atg16L1, Atg5 and Atg7 in small intestinal Paneth cells and Crohn disease. Autophagy. 2009;5:250-252. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 192] [Cited by in RCA: 182] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 49. | Lala S, Ogura Y, Osborne C, Hor SY, Bromfield A, Davies S, Ogunbiyi O, Nuñez G, Keshav S. Crohn's disease and the NOD2 gene: a role for paneth cells. Gastroenterology. 2003;125:47-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 388] [Cited by in RCA: 361] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 50. | Ayabe T, Satchell DP, Wilson CL, Parks WC, Selsted ME, Ouellette AJ. Secretion of microbicidal alpha-defensins by intestinal Paneth cells in response to bacteria. Nat Immunol. 2000;1:113-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 776] [Cited by in RCA: 780] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 51. | Wehkamp J, Harder J, Weichenthal M, Schwab M, Schäffeler E, Schlee M, Herrlinger KR, Stallmach A, Noack F, Fritz P, Schröder JM, Bevins CL, Fellermann K, Stange EF. NOD2 (CARD15) mutations in Crohn's disease are associated with diminished mucosal alpha-defensin expression. Gut. 2004;53:1658-1664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 578] [Cited by in RCA: 592] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 52. | Rumio C, Besusso D, Palazzo M, Selleri S, Sfondrini L, Dubini F, Ménard S, Balsari A. Degranulation of paneth cells via toll-like receptor 9. Am J Pathol. 2004;165:373-381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 115] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 53. | Salzman NH, Ghosh D, Huttner KM, Paterson Y, Bevins CL. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature. 2003;422:522-526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 582] [Cited by in RCA: 562] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 54. | Jin M, Zhang Y. Autophagy and Inflammatory Diseases in Autophagy: Biology and Diseases. Le W, Editor. Beijing: Science Press Beijing, 2020: 391-400. |
| 55. | Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;13:722-737. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1278] [Cited by in RCA: 1517] [Article Influence: 126.4] [Reference Citation Analysis (0)] |
| 56. | Hu CA, Hou Y, Yi D, Qiu Y, Wu G, Kong X, Yin Y. Autophagy and tight junction proteins in the intestine and intestinal diseases. Anim Nutr. 2015;1:123-127. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 66] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 57. | Kobayashi T, Siegmund B, Le Berre C, Wei SC, Ferrante M, Shen B, Bernstein CN, Danese S, Peyrin-Biroulet L, Hibi T. Ulcerative colitis. Nat Rev Dis Primers. 2020;6:74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 351] [Cited by in RCA: 978] [Article Influence: 195.6] [Reference Citation Analysis (0)] |
| 58. | Foerster EG, Mukherjee T, Cabral-Fernandes L, Rocha JDB, Girardin SE, Philpott DJ. How autophagy controls the intestinal epithelial barrier. Autophagy. 2022;18:86-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 224] [Article Influence: 74.7] [Reference Citation Analysis (0)] |
| 59. | Nighot PK, Hu CA, Ma TY. Autophagy enhances intestinal epithelial tight junction barrier function by targeting claudin-2 protein degradation. J Biol Chem. 2015;290:7234-7246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 186] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 60. | Wu Y, Tang L, Wang B, Sun Q, Zhao P, Li W. The role of autophagy in maintaining intestinal mucosal barrier. J Cell Physiol. 2019;234:19406-19419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 85] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 61. | Wong M, Ganapathy AS, Suchanec E, Laidler L, Ma T, Nighot P. Intestinal epithelial tight junction barrier regulation by autophagy-related protein ATG6/beclin 1. Am J Physiol Cell Physiol. 2019;316:C753-C765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 62. | Zhang C, Yan J, Xiao Y, Shen Y, Wang J, Ge W, Chen Y. Inhibition of Autophagic Degradation Process Contributes to Claudin-2 Expression Increase and Epithelial Tight Junction Dysfunction in TNF-α Treated Cell Monolayers. Int J Mol Sci. 2017;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 44] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 63. | Tan Y, Guan Y, Sun Y, Zheng C. Correlation of Intestinal Mucosal Healing and Tight Junction Protein Expression in Ulcerative Colitis Patients. Am J Med Sci. 2019;357:195-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 64. | Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R, Anderson CA, Bis JC, Bumpstead S, Ellinghaus D, Festen EM, Georges M, Green T, Haritunians T, Jostins L, Latiano A, Mathew CG, Montgomery GW, Prescott NJ, Raychaudhuri S, Rotter JI, Schumm P, Sharma Y, Simms LA, Taylor KD, Whiteman D, Wijmenga C, Baldassano RN, Barclay M, Bayless TM, Brand S, Büning C, Cohen A, Colombel JF, Cottone M, Stronati L, Denson T, De Vos M, D'Inca R, Dubinsky M, Edwards C, Florin T, Franchimont D, Gearry R, Glas J, Van Gossum A, Guthery SL, Halfvarson J, Verspaget HW, Hugot JP, Karban A, Laukens D, Lawrance I, Lemann M, Levine A, Libioulle C, Louis E, Mowat C, Newman W, Panés J, Phillips A, Proctor DD, Regueiro M, Russell R, Rutgeerts P, Sanderson J, Sans M, Seibold F, Steinhart AH, Stokkers PC, Torkvist L, Kullak-Ublick G, Wilson D, Walters T, Targan SR, Brant SR, Rioux JD, D'Amato M, Weersma RK, Kugathasan S, Griffiths AM, Mansfield JC, Vermeire S, Duerr RH, Silverberg MS, Satsangi J, Schreiber S, Cho JH, Annese V, Hakonarson H, Daly MJ, Parkes M. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42:1118-1125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2110] [Cited by in RCA: 2001] [Article Influence: 133.4] [Reference Citation Analysis (0)] |
| 65. | Wang T, Liu K, Wen L, Yang Y, Yin X, Liu K, Chen Y, He Y, Yang M, Wei Y, Wang B, Chen D. Autophagy and Gastrointestinaş Diseases Autophagy: Biology and Diseases. Le W, Editor. Beijing: Science Press Beijing, 2020: 529-556. |
| 66. | Hugot JP, Chamaillard M, Zouali H, Lesage S, Cézard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M, Binder V, Finkel Y, Cortot A, Modigliani R, Laurent-Puig P, Gower-Rousseau C, Macry J, Colombel JF, Sahbatou M, Thomas G. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599-603. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4223] [Cited by in RCA: 3902] [Article Influence: 162.6] [Reference Citation Analysis (0)] |
| 67. | Sorbara MT, Ellison LK, Ramjeet M, Travassos LH, Jones NL, Girardin SE, Philpott DJ. The protein ATG16L1 suppresses inflammatory cytokines induced by the intracellular sensors Nod1 and Nod2 in an autophagy-independent manner. Immunity. 2013;39:858-873. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 158] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 68. | Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhães JG, Yuan L, Soares F, Chea E, Le Bourhis L, Boneca IG, Allaoui A, Jones NL, Nuñez G, Girardin SE, Philpott DJ. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 916] [Cited by in RCA: 1010] [Article Influence: 63.1] [Reference Citation Analysis (0)] |
| 69. | Patel KK, Stappenbeck TS. Autophagy and intestinal homeostasis. Annu Rev Physiol. 2013;75:241-262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 70] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 70. | Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H, Nieuwenhuis EE, Higgins DE, Schreiber S, Glimcher LH, Blumberg RS. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743-756. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1177] [Cited by in RCA: 1137] [Article Influence: 66.9] [Reference Citation Analysis (0)] |
| 71. | Barmada MM, Brant SR, Nicolae DL, Achkar JP, Panhuysen CI, Bayless TM, Cho JH, Duerr RH. A genome scan in 260 inflammatory bowel disease-affected relative pairs. Inflamm Bowel Dis. 2004;10:513-520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 41] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 72. | Mathiassen SG, De Zio D, Cecconi F. Autophagy and the Cell Cycle: A Complex Landscape. Front Oncol. 2017;7:51. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 148] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 73. | Chen Y, Gibson SB. Three dimensions of autophagy in regulating tumor growth: cell survival/death, cell proliferation, and tumor dormancy. Biochim Biophys Acta Mol Basis Dis. 2021;1867:166265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 74. | Alao JP. The regulation of cyclin D1 degradation: roles in cancer development and the potential for therapeutic invention. Mol Cancer. 2007;6:24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 522] [Cited by in RCA: 653] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 75. | Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer. 2011;11:558-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 962] [Cited by in RCA: 1072] [Article Influence: 76.6] [Reference Citation Analysis (0)] |
| 76. | Wei H, Wei S, Gan B, Peng X, Zou W, Guan JL. Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev. 2011;25:1510-1527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 271] [Cited by in RCA: 324] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 77. | Hamurcu Z, Delibaşı N, Geçene S, Şener EF, Dönmez-Altuntaş H, Özkul Y, Canatan H, Ozpolat B. Targeting LC3 and Beclin-1 autophagy genes suppresses proliferation, survival, migration and invasion by inhibition of Cyclin-D1 and uPAR/Integrin β1/ Src signaling in triple negative breast cancer cells. J Cancer Res Clin Oncol. 2018;144:415-430. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 89] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 78. | Liu PF, Hsu CJ, Tsai WL, Cheng JS, Chen JJ, Huang IF, Tseng HH, Chang HW, Shu CW. Ablation of ATG4B Suppressed Autophagy and Activated AMPK for Cell Cycle Arrest in Cancer Cells. Cell Physiol Biochem. 2017;44:728-740. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 79. | Liu PF, Leung CM, Chang YH, Cheng JS, Chen JJ, Weng CJ, Tsai KW, Hsu CJ, Liu YC, Hsu PC, Pan HW, Shu CW. ATG4B promotes colorectal cancer growth independent of autophagic flux. Autophagy. 2014;10:1454-1465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 68] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 80. | Zheng HY, Zhang XY, Wang XF, Sun BC. Autophagy enhances the aggressiveness of human colorectal cancer cells and their ability to adapt to apoptotic stimulus. Cancer Biol Med. 2012;9:105-110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 33] [Reference Citation Analysis (0)] |
| 81. | Ahn CH, Jeong EG, Lee JW, Kim MS, Kim SH, Kim SS, Yoo NJ, Lee SH. Expression of beclin-1, an autophagy-related protein, in gastric and colorectal cancers. APMIS. 2007;115:1344-1349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 163] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 82. | Mokarram P, Albokashy M, Zarghooni M, Moosavi MA, Sepehri Z, Chen QM, Hudecki A, Sargazi A, Alizadeh J, Moghadam AR, Hashemi M, Movassagh H, Klonisch T, Owji AA, Łos MJ, Ghavami S. New frontiers in the treatment of colorectal cancer: Autophagy and the unfolded protein response as promising targets. Autophagy. 2017;13:781-819. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 120] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 83. | Xiao T, Zhu W, Huang W, Lu SS, Li XH, Xiao ZQ, Yi H. RACK1 promotes tumorigenicity of colon cancer by inducing cell autophagy. Cell Death Dis. 2018;9:1148. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 51] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 84. | Huang W, Zeng C, Hu S, Wang L, Liu J. ATG3, a Target of miR-431-5p, Promotes Proliferation and Invasion of Colon Cancer via Promoting Autophagy. Cancer Manag Res. 2019;11:10275-10285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 85. | Lévy J, Cacheux W, Bara MA, L'Hermitte A, Lepage P, Fraudeau M, Trentesaux C, Lemarchand J, Durand A, Crain AM, Marchiol C, Renault G, Dumont F, Letourneur F, Delacre M, Schmitt A, Terris B, Perret C, Chamaillard M, Couty JP, Romagnolo B. Intestinal inhibition of Atg7 prevents tumour initiation through a microbiome-influenced immune response and suppresses tumour growth. Nat Cell Biol. 2015;17:1062-1073. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 150] [Article Influence: 15.0] [Reference Citation Analysis (0)] |