INTRODUCTION
Hepatocellular carcinoma (HCC) accounts for 80%-90% of all liver tumors and its incidence has increased in recent decades[1,2]. The risk factors are numerous, including hepatitis B or C infection, alcoholic and non-alcoholic fatty liver disease, and diabetes. In general, these pathological conditions lead to liver fibrosis, which develops into cirrhosis and eventually HCC. This tumor is extremely heterogeneous and aggressive, has a high recurrence rate and is often resistant to chemotherapy. For this reason, survival rates for HCC are poor despite improvements in diagnostic and therapeutic approaches[3]. Understanding the molecular networks underlying the development and progression of HCC is important for new biomarkers and therapeutic goals.
Non-coding RNAs (ncRNAs) account for 97% of total transcriptional output. Advancing technologies in the enrichment and sequencing of ncRNAs shed light on their active role in the regulation of biological processes and made them increasingly attractive. The ncRNAs can be classified according to their function, length and shape[4]. The most important classes are: (1) MicroRNAs (miRNAs): Small ncRNAs with a length of about 20 nucleotides (nt). Their primary function is the inhibition of mRNA translation by binding sequences with incomplete complementarity in the 3’-untranslated regions of the target mRNAs[5]; (2) PIWI-interacting RNAs (piRNAs): Small ncRNAs (mainly 24-35 nt long)[6,7], mainly found in germline cells. As the name suggests, they interact with proteins belonging to the PIWI family and regulate gene expression at transcriptional (through epigenetic chromatin modification) and post-transcriptional level (through RNA and protein interactions)[8]; (3) Long ncRNAs (lncRNAs): NcRNAs generated from pre-mRNAs. They are more than 200 nt long and are characterized by complex secondary/tertiary structures designated for interactions with their molecular targets (proteins or DNA/RNA). LncRNAs regulate many biological processes by acting as miRNA sponges, as recruiters of transcription factors to target promoters and as scaffolding meant to facilitate protein interactions[9]; and (4) Circular RNAs (circRNAs): NcRNAs which are similar in length and function to lncRNAs, except for their distinct ring-like shape. CircRNAs are formed by the circularization of exons, introns, or other ncRNAs by a non-canonical splicing event, so-called “back-splicing”[10].
Since ncRNAs play a crucial role in various biological processes, it is not surprising that evidence of their massive involvement in cancer is accumulating[11,12]. Accordingly, studying the biogenesis and functions of ncRNAs could be a valuable tool in deepening our knowledge of cancer. The editorial by Papadopoulos and Trifylli[13] in the World Journal of Gastroenterology discussed the role of exosomal circRNAs in HCC and proposed them as new therapeutic targets. In this editorial, we will extend the discussion to piRNAs, another class of ncRNAs, and their potential utility for the clinical treatment of HCC.
PIRNA
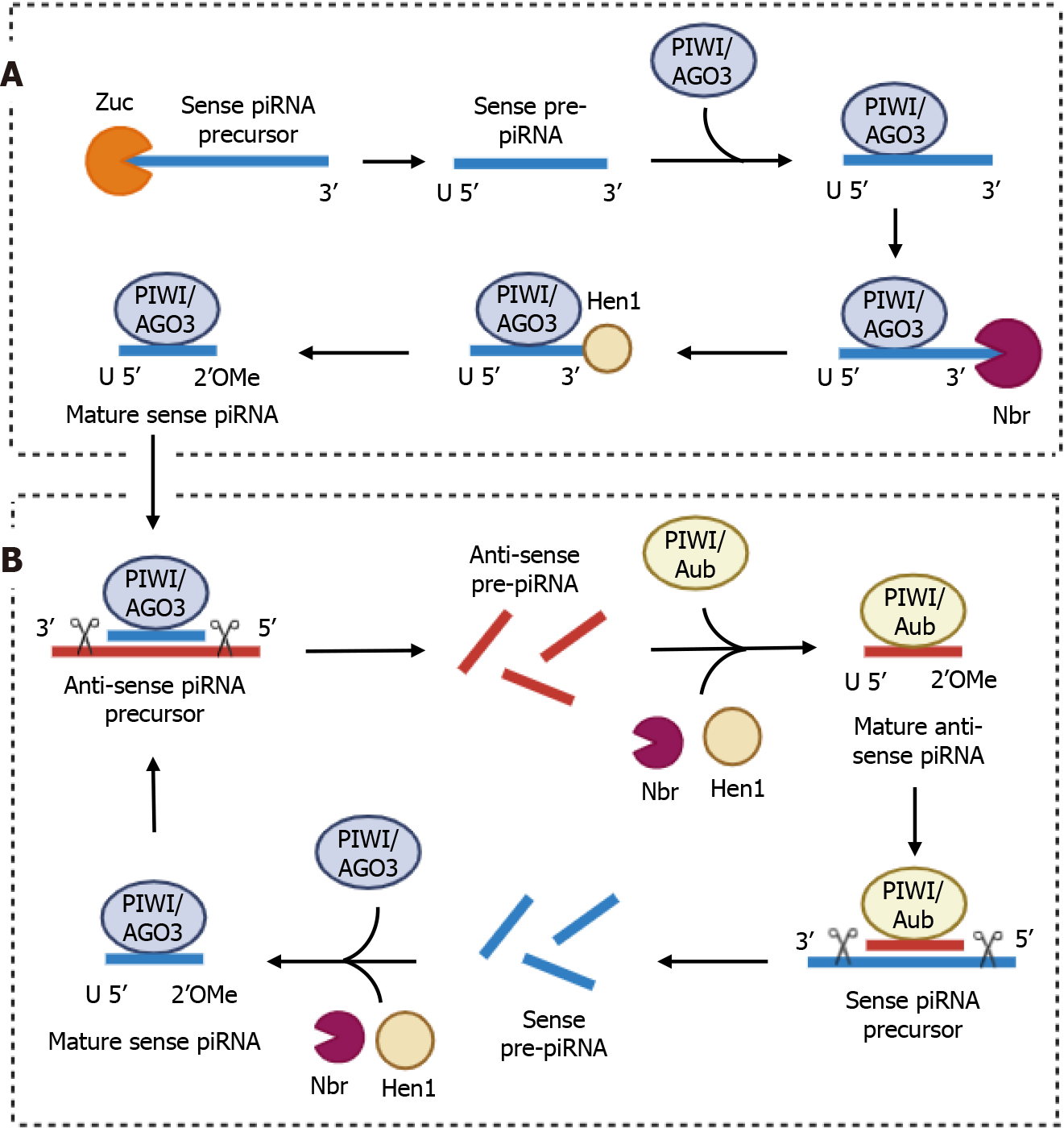
A brief note on the biology of piRNA: These ncRNAs are transcribed from specific loci, enriched with transposons, the so-called “piRNA clusters”, which can be divided into single-stranded (ss) and double-stranded (ds) clusters. The ss-cluster generates a piRNA precursor with a 3’-polyadenylated tail and a 5’-methylguanosine-cap, similar to the canonical transcription of mRNAs. The ds-cluster can instead generate two non-polyadenylated piRNA precursors (sense and anti-sense) by means of a protein complex, including RNA polymerase II. Regardless of their origin, the piRNA precursors are exported to the cytoplasm, in particular to the outer membrane of the mitochondria, where the proteins involved in their maturation are located. The piRNA precursor is first cleaved at the 5’-end by an endonuclease called Zucchini to form a 5’-monophosphorylated pre-piRNA, which, in turn, is loaded into a PIWI protein (in a 1:1 ratio) and further cleaved at the 3’-end by the 3’-to-5’ exonuclease Nibbler. The new, truncated 3’-terminal end is finally methylated at the 2’-oxygen by Hen1, a 2’-O-methyltransferase, presumably to increase the stability of the piRNA. These primary mature piRNAs can trigger a secondary piRNA maturation cycle, called “ping-pong cycle”, in order to exponentially increase the mature cellular piRNA pool. Basically, a mature anti-sense piRNA is loaded into the Aub protein to cleave the sense piRNA precursor into multiple sense piRNA precursors, which are fully processed and then finally loaded into the Ago3 protein. The mature sense piRNA-Ago3 complex cleaves the anti-sense piRNA precursor into multiple anti-sense piRNA precursors, which are loaded into Aub proteins to restart the cycle[14] (Figure 1). It is important to emphasise that our knowledge of the biogenesis of piRNA is still incomplete, and comes mainly from Drosophila melanogaster and mice-related studies. Despite the high degree of evolutionary conservation of this pathway, a deeper understanding of piRNA biogenesis, especially in humans, is crucial to assessing its importance for basic and translational research.
Figure 1 Schematic diagrams of the biosynthesis of PIWI-interacting RNAs.
A and B: The primary (A) and secondary (B) cycles (also known as the “ping-pong cycle”) are shown. Note that the PIWI-interacting RNA (piRNA) precursor that triggers primary biosynthesis can be either a sense or an anti-sense piRNA precursor. Image created by Biorender. Zuc: Zucchini; piRNA: PIWI-interacting RNA; Nbr: Nibbler.
Functionally, the main role of piRNAs is to silence gene expression at the transcriptional level by recruiting DNA methyltransferase and histone methyltransferase to the target promoter and target histone, respectively. In addition, piRNAs protect the cells from transposons and prevent their amplification and mobilisation. The piRNA-mediated regulation of gene expression can also be observed at a post-transcriptional level: piRNA can inhibit mRNA translation and trigger the degradation of pseudogenic transcripts and lncRNAs by interacting with complementary sequences at the 5’-end. Finally, it has been reported that many piRNAs can act as scaffolding to facilitate interaction with multiple proteins, similar to circRNAs and lncRNAs[15]. In recent decades, increasing number of studies have reported the possible involvement of piRNA dysregulation in the development and progression of cancer. We provide a brief chronological overview of articles dealing with HCC-associated piRNAs.
PIRNA IN HCC
According to PubMed research, the paper by Law et al[16] is the first article to evaluate the clinical benefit of piRNAs in HCC. Using Illumina sequencing, they identified a novel piRNA, called piR-Hep1, which was significantly overexpressed in the tumour tissue of 73 HCC patients compared to adjacent normal tissue. Furthermore, inhibition of piR-Hep1 in HCC cell lines (HKCI-4 and HKCI-8) led to a reduction in cell viability, invasiveness, and phosphorylation levels of AKT, indicating a possible involvement of piR-Hep1 in the regulation of the AKT signalling pathway[16].
In 2016, Rizzo et al[17] used next-generation sequencing techniques to characterise the piRNA expression profile of the different pathological stages underlying the diagnosis and progression of HCC: (1) Cirrhotic nodules (CN); (2) Low-grade dysplastic nodules (LGDN); (3) High-grade dysplastic nodules (HGDN); (4) Early HCC (eHCC); and (5) Progressed HCC (pHCC). For this purpose, they obtained resection samples from 17 HCC patients with multiple nodules: (1) 17 CN; (2) 9 LGDN; (3) 6 HGDN; (4) 6 eHCC; and (5) 23 pHCC. Sequencing revealed a 125 piRNA expression signature specific for eHCC and pHCC, and a 24 piRNA expression signature specific for LDGN and HDGN[17]. Similar results were also obtained in 2018 by Koduru et al[18] through database screening and informatic analysis. The RNA-seq datasets analysed were from the NIH Short Read Archive and were from 14 CN, 9 LDGN, 6 HDGN, 6 eHCC, and 20 pHCC samples. They found a specific piRNA expression profile for each HCC-associated pathological stage (number of specific dysregulated piRNAs in round brackets): (1) CN (75); (2) LDGN (60); (3) HGDGN (49); (4) eHCC (56); and (5) pHCC (128). These results indicate dynamic changes in piRNoma during disease progression, which complicates their use as clinical tools.
In 2018, Tang et al[19] also investigated the role of piRNAs in those liver pathologies which will lead to the development of HCC in mice. They found that piR-823 was highly expressed in primary hepatic stellate cells (HSCs) and that its inhibition suppressed the activation of HSCs. In contrast, overexpression of piR-823 increased the proliferation of HSCs and the production of alfa-smooth muscle actin, collagen type I alpha 1, and transforming growth factor beta 1 (crucial genes for liver fibrosis progression). Accordingly, piR-823 could be an early therapeutic target to prevent liver fibrosis and, subsequently, the progression of HCC.
In 2023, Wu et al[20] found that piR-017724 was significantly downregulated in 45 HCC tissues compared to adjacent normal tissues and that its downregulation was correlated with poor prognosis and advanced tumour stage. The oncosuppressive role of this piRNA was confirmed in HCC cell lines (SMMC-7721 and PLC/PRF/5), where silencing of piR-017724 resulted in the inhibition of cell proliferation and invasiveness but not apoptosis. Functional analyses suggested that piR-017724 could inhibit the expression of PLIN3, a member of the abdominal lipoprotein family involved in lipid droplet homeostasis, and plays a crucial role in the regulation of gene expression, protein degradation, signalling and membrane trafficking, particularly in the liver[20].
Rui et al[21] conducted a comprehensive characterisation of serum exosome piRNA levels in 125 HCC patients and 44 healthy controls in 2023, to assess their suitability as diagnostic biomarkers. They found 253 dysregulated piRNA in exosomes of tumour patients, compared to healthy controls. Then, Rui et al[21] selected the five most upregulated piRNA in HCC exosomes (piR-1029, piR-15254, novel-piR-35395, novel-piR-32132, and novel-piR-43597) and validated them in two different cohorts, confirming their overexpression in the tumour and, thus, their diagnostic potential.
CONCLUSION
As with the other classes of ncRNAs, the discovery of disease-associated piRNAs is highly dependent on building the pipeline of identification methods. Nowadays, piRNA identification methods include two approaches: Experimental and computational. The first method is based on common techniques for the isolation of protein-associated RNAs, such as RNA immunoprecipitation sequencing or cross-linking immunoprecipitation and high-throughput sequencing. However, these techniques are expensive, time-consuming, and not very sensitive for low expressed piRNAs. The computational approach to piRNA identification has attracted much attention in recent years as piRNA data has increased significantly. Essentially, it involves the application of various bioinformatics tools to specific piRNA databases [such as: (1) piRBase; (2) piRNAclusterDB; or (3) piRNAtarget] to identify novel piRNAs (e.g., 2L-piRNA), piRNA clusters (e.g. protract), piRNA targets (e.g. miRanda), and disease-associated piRNAs (e.g., WGCNA)[22]. Despite the promising performance, these computational methods have several issues that need to be addressed, such as standardising piRNA nomenclature, developing a gold standard for bioinformatics analysis pipeline and improving the quality and quantity of data in the current piRNA databases (especially regarding the data on healthy controls)[23]. Nevertheless, more and more studies show the involvement of piRNAs in the development and progression of pathological diseases in humans. It is expected that there will be a corresponding increase in high-throughput technologies for piRNA studies in the coming years.
In HCC, research into piRNAs is still in its infancy, and much still needs to be clarified about their involvement in tumour progression. The importance of piRNA mechanisms in HCC is also emphasised by the involvement of PIWI proteins. In 2015, Xie et al[24] demonstrated that Hiwi (or PIWIL1), a member of the PIWI family, is overexpressed at both mRNA and protein levels in HCC cell lines (MHCC97L and MHCC97H) compared to normal liver cell lines (L02), and in tumour tissues from 60 HCC patients compared to normal controls from 48 peritumour samples. Functional analysis showed that the silencing of Hiwi in HCC cell lines led to a reduction in cell proliferation and invasiveness. These results suggest that Hiwi is a potentially useful HCC biomarker and a valuable new therapeutic target[24]. Recently, Hammad et al[25] investigated the mRNA expression levels of PIWI family members [(1) PIWIL1; (2) PIWIL2; (3) PIWIL3; and (4) PIWIL4] in both tissues and serum of HCC patients. Their results showed significant overexpression of these mRNAs in 50 tumour tissues compared to adjacent normal tissues and in 50 HCC sera compared to those of 25 healthy controls. Furthermore, a significant correlation was found between PIWIL1-4 overexpression and advanced tumour stage, at least as far as the tissue samples are concerned[25]. With high-throughput technologies, it is now possible to extend the investigations to a large number of patients, as well as to deepen the molecular mechanisms in cellular models. All in all, these results suggest that piRNAs are attractive candidates in HCC research with the aim of expanding our knowledge of HCC development and progression, as well as opening up new potential applications in the screening and treatment of patients.
ACKNOWLEDGEMENTS
We would like to thank Mrs Helen Ager and Mr Robin G Ager for reviewing our manuscript and for their useful suggestions.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country of origin: Italy
Peer-review report’s classification
Scientific Quality: Grade B
Novelty: Grade B
Creativity or Innovation: Grade B
Scientific Significance: Grade B
P-Reviewer: Liu S, China S-Editor: Wang JJ L-Editor: A P-Editor: Yuan YY