Published online Jan 14, 2024. doi: 10.3748/wjg.v30.i2.170
Peer-review started: November 8, 2023
First decision: December 7, 2023
Revised: December 15, 2023
Accepted: December 26, 2023
Article in press: December 26, 2023
Published online: January 14, 2024
Processing time: 65 Days and 3.7 Hours
Deep learning provides an efficient automatic image recognition method for small bowel (SB) capsule endoscopy (CE) that can assist physicians in diagnosis. How
To propose a novel and effective classification and detection model to automatically identify various SB lesions and their bleeding risks, and label the lesions accurately so as to enhance the diagnostic efficiency of physicians and the ability to identify high-risk bleeding groups.
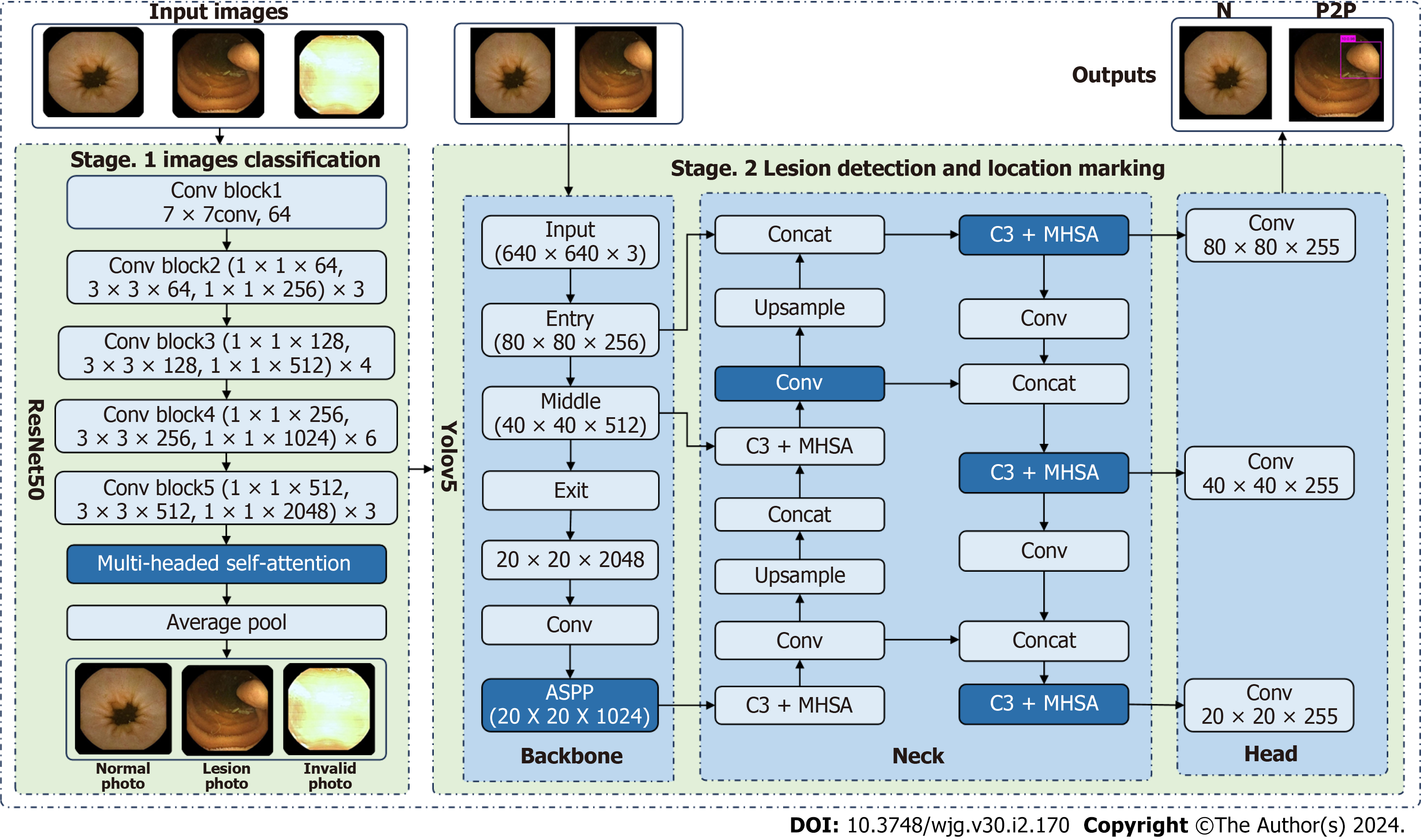
The proposed model represents a two-stage method that combined image classification with object detection. First, we utilized the improved ResNet-50 classification model to classify endoscopic images into SB lesion images, normal SB mucosa images, and invalid images. Then, the improved YOLO-V5 detection model was utilized to detect the type of lesion and its risk of bleeding, and the location of the lesion was marked. We constructed training and testing sets and compared model-assisted reading with physician reading.
The accuracy of the model constructed in this study reached 98.96%, which was higher than the accuracy of other systems using only a single module. The sen
The deep learning model of image classification combined with object detection exhibits a satisfactory diagnostic effect on a variety of SB lesions and their bleeding risks in CE images, which enhances the diagnostic efficiency of physicians and improves the ability of physicians to identify high-risk bleeding groups.
Core Tip: In clinical practice, capsule endoscopy is often used to detect small bowel (SB) lesions and find the cause of bleeding. Here, we have proposed a classification and detection model to automatically identify various SB lesions and their bleeding risks, and label the lesions accurately. This model can enhance the diagnostic efficiency of physicians and improve the ability of physicians to identify high-risk bleeding groups.
- Citation: Zhang RY, Qiang PP, Cai LJ, Li T, Qin Y, Zhang Y, Zhao YQ, Wang JP. Automatic detection of small bowel lesions with different bleeding risks based on deep learning models. World J Gastroenterol 2024; 30(2): 170-183
- URL: https://www.wjgnet.com/1007-9327/full/v30/i2/170.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i2.170
Capsule endoscopy (CE), introduced in 2000, has successfully solved the problem pertaining to visualizing the small intestine and revolutionized the medical field. CE can be utilized as the preferred method for the diagnosis of small bowel (SB) diseases[1-3]. At present, obscure gastrointestinal bleeding is the most common indication for CE[4,5]. Therefore, physicians dedicate more attention to the bleeding risks of SB lesions in CE examination. However, CE reading is time-consuming and complicated[6-9], and abnormal parts account for only a small proportion. Thus, it is easy to miss the diagnosis, which affects the detection of lesions and assessment of bleeding risks. In addition, when there is a large amount of bile, food debris or air bubbles in the gastrointestinal tract, numerous invalid pictures will appear which will seriously affect the diagnostic efficiency[10].
In recent years, deep learning models have been widely utilized in automatic recognition of digestive endoscopic images[11,12]. Deep learning is characterized by processing large amounts of data with better experience and high per
Although CNNs achieve excellent performance, they still have some limitations. First, a CNN cannot effectively focus on the important part of the image, which is easily affected by the organs and tissues around the area to be detected, resulting in limited accuracy of the model. Second, most of the existing research is related to the development of the classification model, while the image classification diagnosis system only adopts the binary classification method, which cannot distinguish two or more types of lesions in the image. Moreover, image classification cannot determine the specific location of the lesion. Therefore, its practicability still needs to be further improved. Third, most of the existing methods utilize spatial pyramid pooling (SPP), which has a lightweight characteristic of the backbone network and reduces the parameters. Although SPP improves the detection speed, it consequently suffers from a reduction in detection accuracy. It is worth emphasizing that existing studies have not evaluated the bleeding risks of SB lesions, but in clinical work, we urgently need to pay attention to the bleeding risks of lesions and try to find the cause of bleeding.
To solve the aforementioned problems, we have made the following efforts. We first added a multi-head self-attention (MHSA) mechanism to increase efficacy of the model’s focus on important regions, improving the overall performance of the model. We next utilized a two-stage method to classify and detect various SB lesions. This two-stage method com
Specifically, ResNet-50 utilizes a deep residual network to solve the gradient disappearance problem, and the increased number of network layers further enhances the image representation ability. Therefore, ResNet-50 was selected as the backbone of the classification model. On this basis, we added an MHSA mechanism, allowing the model to effectively focus on lesions and facilitating image classification. Meanwhile, we also combined YOLO-V5 as the detection model backbone to allow for identification of multiple lesions in the image simultaneously. The accuracy of YOLO exceeds that of general object detection algorithms while maintaining a fast speed, and it is currently one of the most popular algo
In summary, the contributions from our work are as follows: (1) To the best of our knowledge, this is the first time that image classification combined with an object detection model is used to automatically identify a variety of SB lesions and evaluate their bleeding risks; and (2) The model based on deep learning has high accuracy, high sensitivity and high specificity, which improves the diagnostic efficiency of doctors and the ability to identify high-risk bleeding populations. Its diagnostic performance has good potential for clinical application.
A total of 701 patients who underwent SB CE in Shanxi Provincial People's Hospital and Shanxi Provincial Hospital of Traditional Chinese Medicine from 2013 to 2023 were included in this study. Two different capsule types were used at our two centers: PillCam SB2 and SB3 systems (Medtronic, Minneapolis, MN, United States) and MiroCam system (Intromedic, Seoul, South Korea). All patient-generated videos were reviewed, collected, screened, and labeled by three expert gastroenterologists (who had read more than 200 CE studies). The inclusion and final labeling of images were contingent on the agreement of at least two of the three experts.
The lesions included in the pictures were divided into three bleeding risk levels according to Saurin classification[21]: No bleeding risk (P0); Uncertain bleeding risk (P1); And high bleeding risk (P2). We finally divided the included images into the following 12 types: N (normal); P0Lk (lymphangiectasia); P0Lz (lymphoid follicular hyperplasia); Xanthomatosis (P0X); Erosion (P1E); Ulcer smaller than 2 cm (P1U); Protruding lesion smaller than 1 cm (P1P); Ulcer larger than 2 cm (P2U); Protruding lesion larger than 1 cm (P2P); Vascular lesion (P2V); Blood (B); And invalid picture (I).
We selected a total of 111861 images, and randomly divided the training set and test set images into 74574 and 37287 images, respectively, according to the 2:1 ratio. Written informed consent was obtained from all patients. Patient data were anonymized, and any personal identifying information was omitted. This study was approved by the Ethics Com
All image preprocessing algorithms were run on a standard computer using a 64-bit Windows 10 operating system and a Python laboratory environment provided by Anaconda 2.5.0. All experiments using deep learning for model training were conducted on an RTX 3060(GPU) and i7 processor, and the computational resources and computer-aided tools met the experimental requirements. The deep learning models applied in the experiments were provided by the Pytorch framework, which offers a variety of deep learning algorithms and pretrained models.
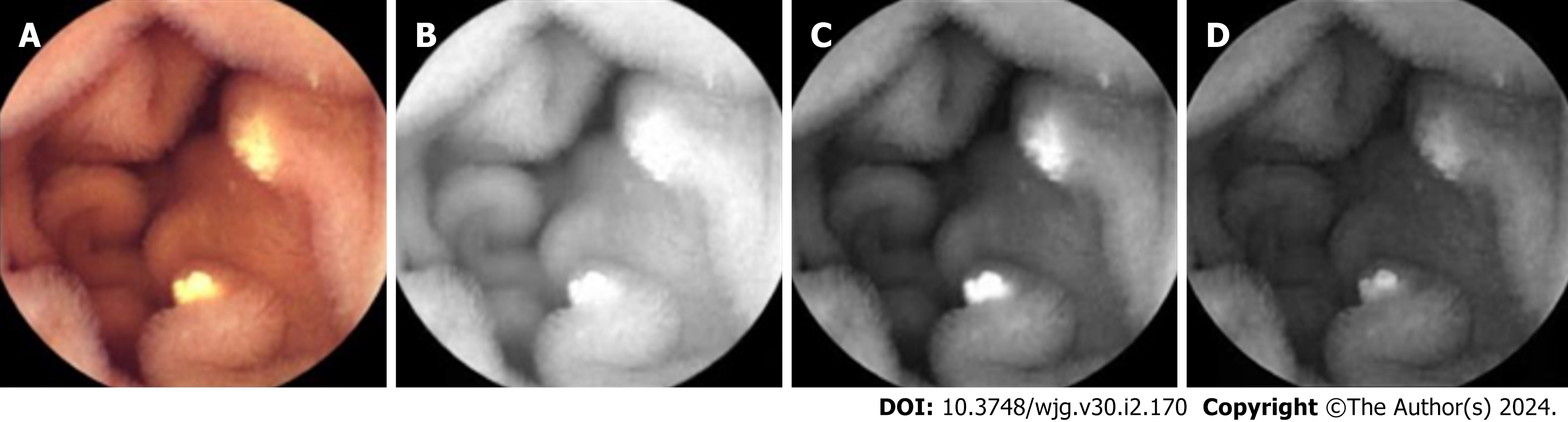
The collected visible light images were decomposed into R, G, and B channels as the network’s input (representative example is depicted in Figure 1).
Herein, two stages were utilized to identify and label SB lesions and their bleeding risks. In the first stage (Stage 1), all input images were entered into the improved ResNet-50 classification model, and the images were divided into small intestinal lesion images, normal small intestinal mucosa images, and invalid images according to whether lesions existed. The main purpose of this stage was to filter invalid images. In the second stage (Stage 2), the images of normal small intestinal mucosa and lesions classified in Stage 1 were entered into the improved YOLO-V5 model, and the lesions were detected, assessed for bleeding risk, and labeled for location. This task can be formally defined as follows: For a given data set 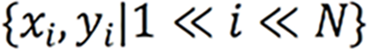 , the research goal was to create a mapping function
, the research goal was to create a mapping function 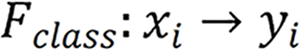 , where
, where 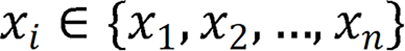 denotes the endoscopic image, and output image
denotes the endoscopic image, and output image 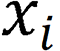 corresponds to the disease category
corresponds to the disease category 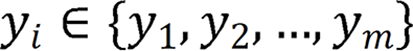 . The model diagram is de
. The model diagram is de
Building the Stage 1 classification model: Stage 1 utilized ResNet-50 as the network backbone and incorporated three branches to learn the features of the three RGB channels. The global average pooling layer was connected into the network, which can reduce the number and complexity of the neural network and simultaneously extract the global information of image features. Thus, the classification task was performed optimally. In addition, after the network’s fifth convolutional block, we introduced a MHSA mechanism to fuse features at different levels, which can automatically capture the relationship between different locations or features. Thus, we captured the context information and crucial features in the image in an optimal manner. Finally, the fused features were fed into a softmax layer, which received a vector of scores from each category of the model and transformed these scores into a probability distribution representing the probabilities of each category. Specifically, the softmax function normalized the raw scores to a value between 0 and 1 and ensured that the sum was 1.
The ResNet-50 network model included one convolutional block, four residual blocks, and one output layer. 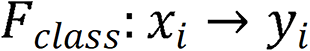 comprised two components: A nonlinear feature mapping structure and a classifier. During training, a two-dimensional image was mapped into a one-dimensional vector, which was then entered into the classifier for judgment:
comprised two components: A nonlinear feature mapping structure and a classifier. During training, a two-dimensional image was mapped into a one-dimensional vector, which was then entered into the classifier for judgment: 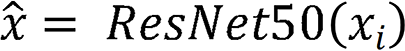 . Here,
. Here, 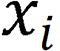 represents the input image, and
represents the input image, and 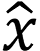 represents the image vector after ResNet-50 feature extraction.
represents the image vector after ResNet-50 feature extraction.
In addition, the attention module was a simulation of the attention module associated with the human brain. Since individuals’ eyes move to the place of interest and subsequently focus on a certain place, when the attention module was introduced, the proposed model focused on the place of feature focus distribution during training, as depicted: 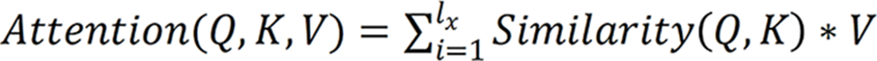 , where Q, K and V are the feature vectors
, where Q, K and V are the feature vectors 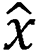 of the input
of the input 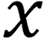 , respectively. Given Q, the correlation between Q and its different K values can be calculated, i.e. the weight coefficients of different V values in K. The weighted average result of V can be used as the attention value. The specific calculation process is shown.
, respectively. Given Q, the correlation between Q and its different K values can be calculated, i.e. the weight coefficients of different V values in K. The weighted average result of V can be used as the attention value. The specific calculation process is shown.
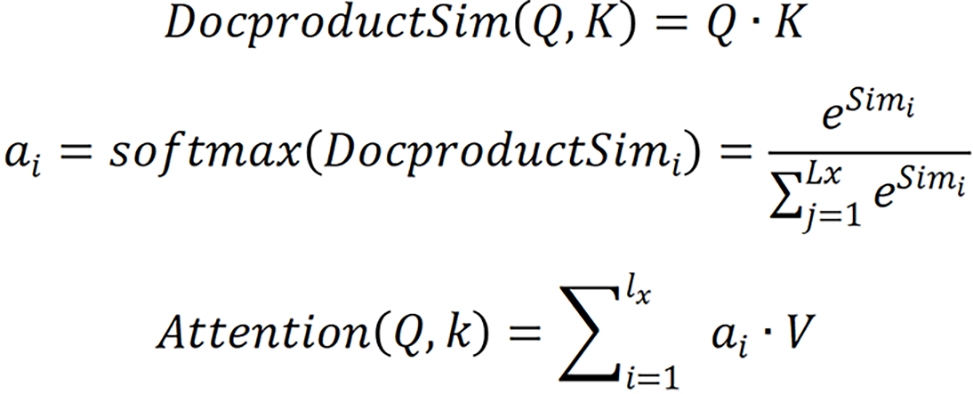
Specifically, the correlation between Q and different K was calculated by dot product, i.e. the weight system of each part in the image was calculated. The output of the previous stage was then normalized to map the range of values between 0 and 1. Finally, the results of multiplication of the value and the corresponding weight of each value were accumulated to obtain the attention value.
The loss function of the model was the cross-entropy loss function, and the calculation process is shown.
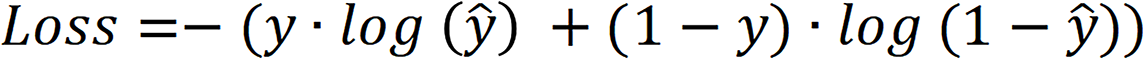
Here, 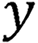 is the actual category, and
is the actual category, and 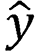 is the predicted category.
is the predicted category.
Building the stage 2 lesion detection model: Stage 2 adopted YOLO-V5 as the network backbone, adopted three branches to learn the features of three RGB channels respectively, and introduced a parallel network, the feature erase module, ASPP, and MHSA mechanism for detection. YOLO-V5 is composed of the following four components: Input layer; backbone network; middle layer; and prediction layer. In the input layer, the input image was scaled, the data were enhanced, and the optimal anchor value was calculated. The backbone network was composed of a convolutional network to extract the main features. In the middle layer, the feature pyramid network and path aggregation network were utilized to extract more complex features. The prediction layer was utilized to predict the location and category of the target.
YOLO-V5 adopted the SPP module. To enhance the feature extraction capability of the backbone network, we replaced the SPP module with the ASPP module. The dilated convolution adopted by ASPP differs from the ordinary convolution in that it introduces the "rate" parameter, which represents the number of intervals between points in the convolution kernel. By adjusting the expansion rate, the receptive field size of the convolution operation can be controlled without having to reduce the resolution of the feature map. This enabled the ASPP module to effectively capture information in a wider range while maintaining a high resolution, thereby enhancing the feature extraction performance of the backbone network.
Considering the correlation between the three channels, features were extracted from three channels of the same image, followed by the fusion of these extracted features that was achieved through a parallel network to capture both inde
Meanwhile, to enable the model to focus on the crucial image components and enhance the ability of the model to extract features, we fused the MHSA mechanism in the C3 module of Neck to enhance the detection accuracy, and finally performed the detection in the Head layer.
The model’s loss function was complete intersection over union loss, and the calculation process is shown:
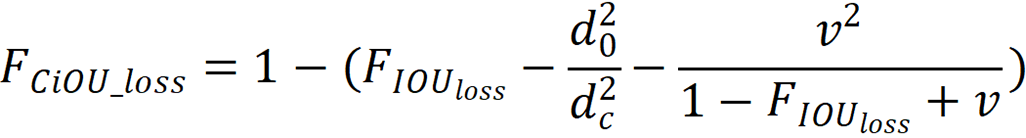
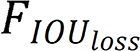 is the intersection over union loss function,
is the intersection over union loss function, 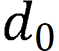 is the Euclidean distance between the target box and the center point of the prediction box,
is the Euclidean distance between the target box and the center point of the prediction box, 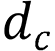 is the diagonal distance of the target box, and
is the diagonal distance of the target box, and 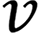 is the parameter measuring the aspect ratio.
is the parameter measuring the aspect ratio.
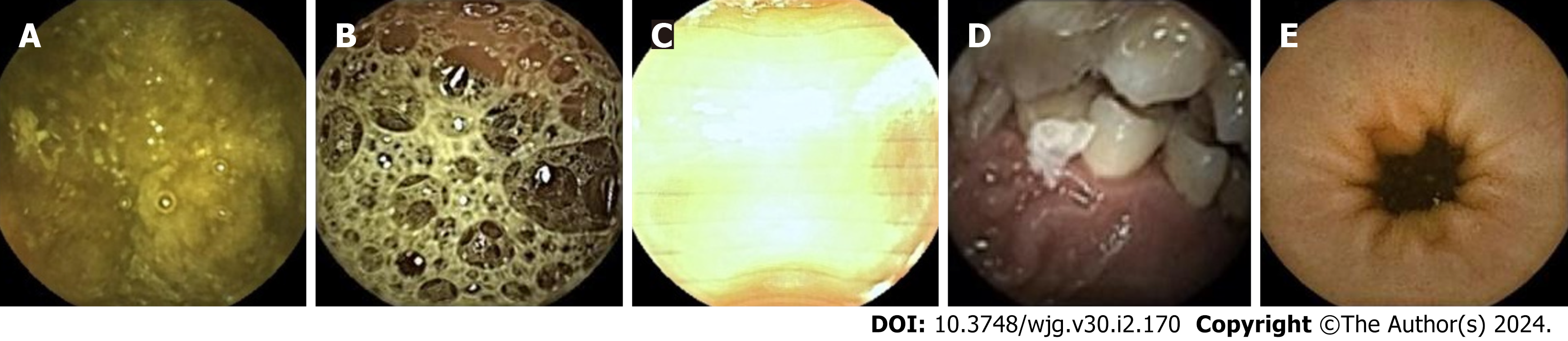
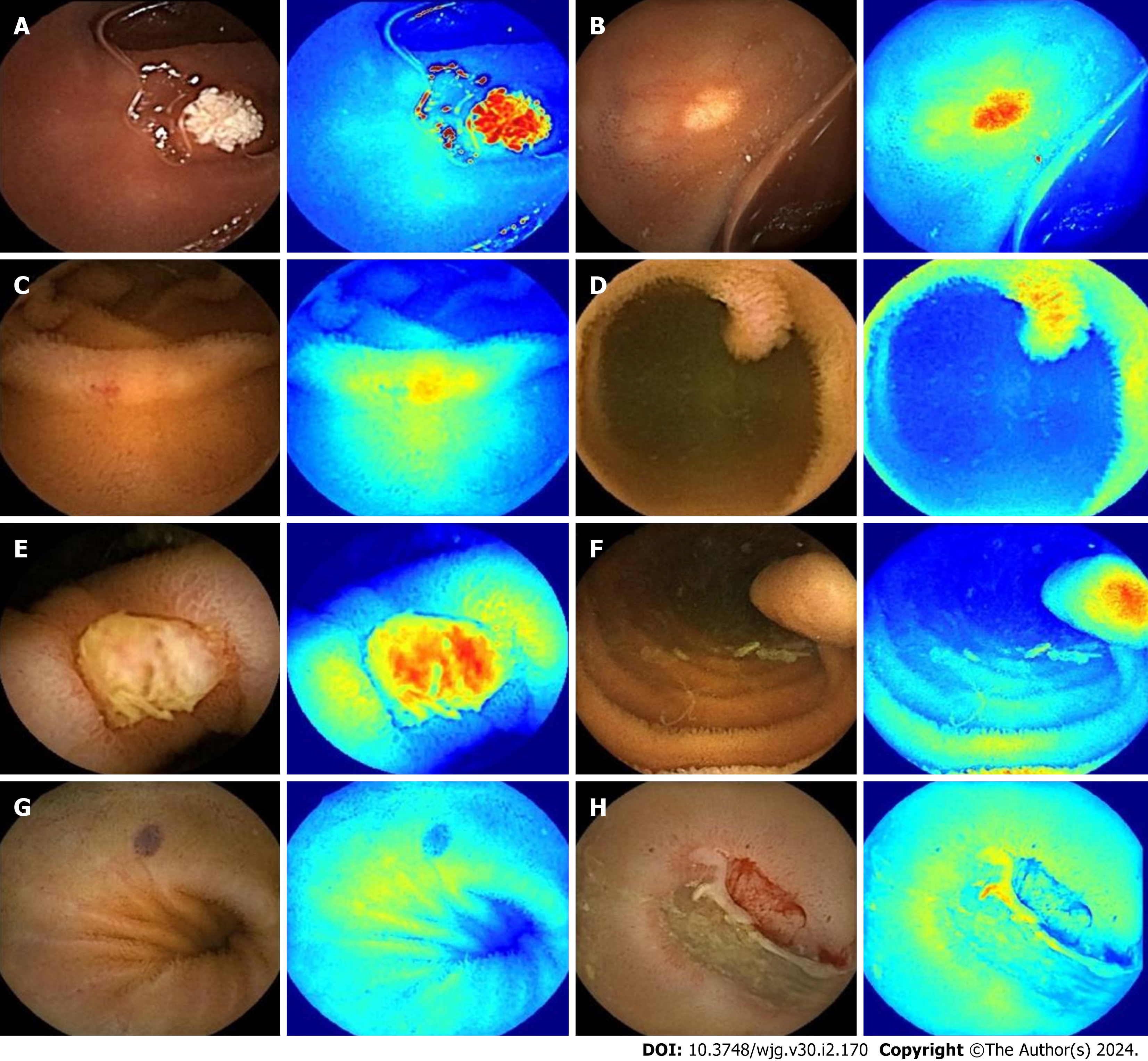
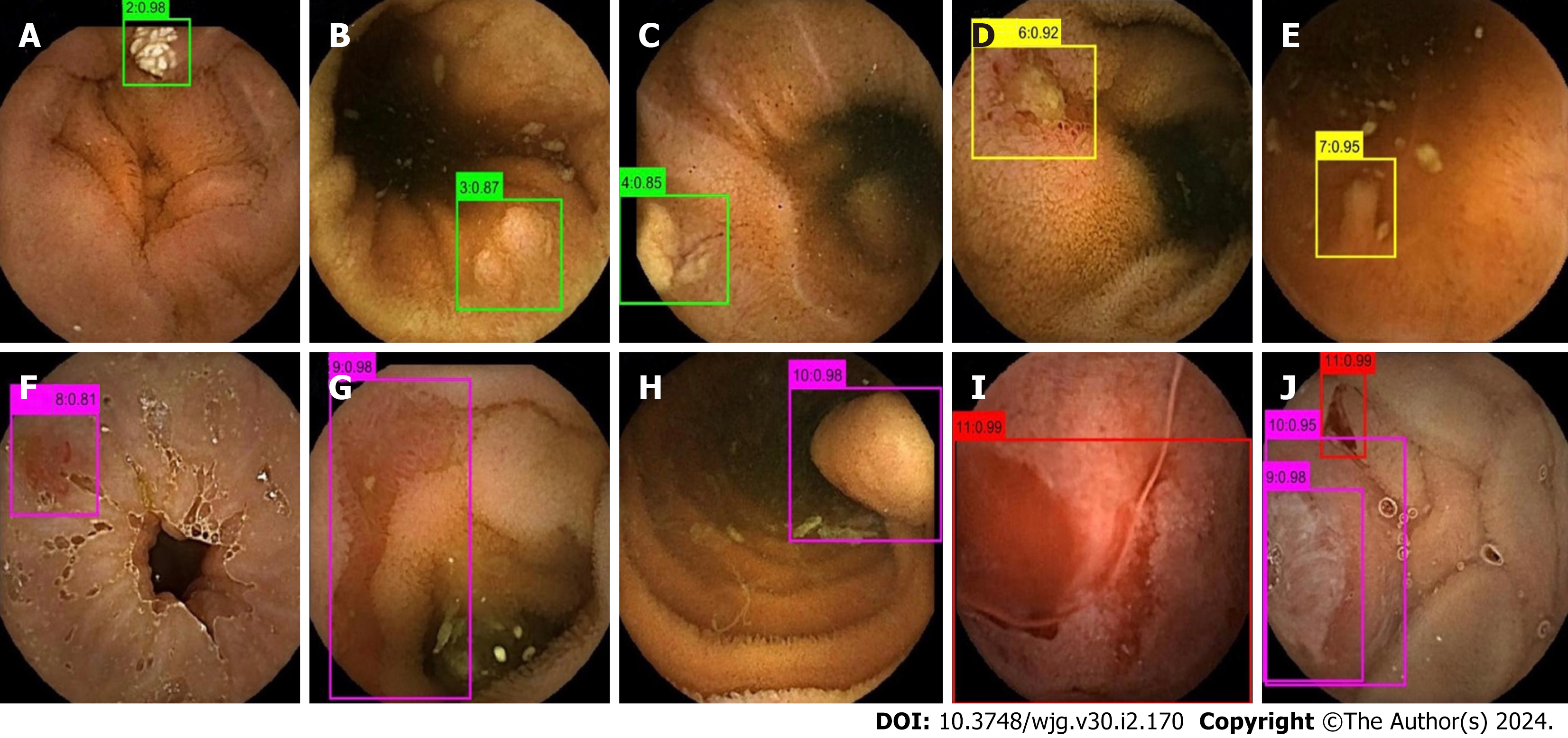
The images in the test set were read by three physicians (gastroenterologists who read less than 10 CE examinations) through physician reading (A) and model-assisted reading (B). Process A randomly assigned 37287 images to the three physicians for reading. In process B, 37287 CE images were first entered into the model. After model classification and detection, the new image package was randomly assigned to the three physicians for a secondary review. Stage 1 of the model divided images into small intestinal lesion images, normal small intestinal mucosa images, and invalid images, and filtered out many invalid images. Figure 3 depicts a representative example of invalid images and normal small intestinal mucosa images. Then, Stage 2 detected the images of 11 types (normal SB mucosa, lymphangiectasia, lymphoid follicular hyperplasia, xanthoma, erosion, ulcer smaller than 2 cm, protruding lesion smaller than 1 cm, ulcer larger than 2 cm, protruding lesion larger than 1 cm, vascular lesions, and blood), assessed their risk of bleeding, and labeled the lesions. The effect of the proposed model structure on CE recognition is depicted in Figure 4. All the physicians determined a diagnosis of each frame of the picture through independent reading and model-assisted reading. If the diagnosis of physician reading was consistent with model-assisted reading, no further evaluation was conducted. If the final diagnosis was inconsistent and/or different lesions were observed, the diagnosis of three experts was assumed to be the gold standard.
In the image pool (n = 74574) of the training dataset, there were 52310 negative pictures (normal small intestinal mucosa pictures and invalid pictures) and 22264 positive pictures. In the image pool of the test dataset (n = 37287), there were 26155 negative pictures and 11132 positive pictures. The distribution of specific types of images is depicted in Table 1.
| Set | Type of CE | Type of pictures | ||||||||||||
| N | P0Lk | P0Lz | P0X | P1E | P1U | P1P | P2U | P2P | P2V | B | I | Total | ||
| Training set | PillCam | 14026 | 2086 | 1505 | 551 | 1851 | 3867 | 687 | 1564 | 689 | 930 | 1529 | 17935 | 47220 |
| MiroCam | 13452 | 612 | 817 | 494 | 1091 | 2062 | 180 | 341 | 205 | 421 | 782 | 6897 | 27354 | |
| Test set | PillCam | 9221 | 245 | 435 | 122 | 235 | 1261 | 119 | 2608 | 109 | 139 | 1703 | 8446 | 24643 |
| MiroCam | 4518 | 112 | 418 | 153 | 68 | 415 | 68 | 1581 | 68 | 92 | 1181 | 3970 | 12644 | |
The test set of pictures were respectively passed through the two processes of physician reading and model-assisted reading, and the final diagnosis was compared with the diagnosis provided by the expert analysis, which was the gold standard. The primary outcome measures included sensitivity, specificity, and accuracy.
Qualitative analysis: Representative examples of the heat maps generated by the model for 10 lesion types and of the results of the model system are depicted in Figures 5 and 6.
Quantitative analysis: (1) Physician reading. The sensitivity, specificity, and accuracy of physician reading for all pictures were 93.80%, 99.38%, and 98.92%, respectively. The sensitivity, specificity, and accuracy of physician reading for positive pictures were 89.95%, 99.80%, and 99.51%, respectively. The sensitivity, specificity, and accuracy for different types of image recognition are depicted in Table 2.
| Type of CE | Mode of reading | Sensitivity, % | Specificity, % | Accuracy, % |
| I | P | 95.96 | 98.10 | 97.39 |
| M | 99.85 | 99.57 | 99.66 | |
| N | P | 94.96 | 94.25 | 94.52 |
| M | 98.84 | 99.77 | 99.43 | |
| P0Lk | P | 84.31 | 99.89 | 99.75 |
| M | 96.92 | 99.97 | 99.94 | |
| P0Lz | P | 78.66 | 99.71 | 99.23 |
| M | 97.30 | 99.90 | 99.83 | |
| P0X | P | 65.45 | 99.87 | 99.62 |
| M | 93.82 | 99.98 | 99.94 | |
| P1E | P | 67.33 | 99.55 | 99.28 |
| M | 91.75 | 99.96 | 99.90 | |
| P1U | P | 74.88 | 99.68 | 98.57 |
| M | 98.39 | 99.94 | 99.87 | |
| P1P | P | 64.71 | 99.78 | 99.61 |
| M | 94.12 | 99.94 | 99.91 | |
| P2U | P | 98.35 | 99.94 | 99.76 |
| M | 99.69 | 99.99 | 99.95 | |
| P2P | P | 88.14 | 99.71 | 99.65 |
| M | 100 | 99.96 | 99.96 | |
| P2V | P | 52.38 | 99.92 | 99.62 |
| M | 97.40 | 99.97 | 99.96 | |
| B | P | 100 | 100 | 100 |
| M | 100 | 100 | 100 |
(2) Performance of the model. In Stage 1 we utilized ablation experiments, and the results indicated (Tables 3 and 4) that the accuracy, sensitivity, and specificity of the multimodal system model with three-channel RGB were more optimal than those of the R channel, G channel, and B channel. The accuracy, sensitivity, and specificity of the MHSA mechanism were more optimal than those of the spatial attention mechanism and the channel attention mechanism. The RGB mul
| Method | Color channel module | Accuracy, % | Sensitivity, % | Specificity, % | |||
| R | G | B | RGB | ||||
| Method 1 | √ | × | × | × | 98.32 | 98.29 | 98.36 |
| Method 2 | × | √ | × | × | 96.97 | 96.99 | 96.93 |
| Method 3 | × | × | √ | × | 99.04 | 99.02 | 99.08 |
| Method 4 | × | × | × | √ | 99.08 | 99.05 | 99.12 |
| Method | Attention module | Accuracy, % | Sensitivity, % | Specificity, % | ||
| SA | CA | MHSA | ||||
| Method 1 | √ | × | × | 98.79 | 98.75 | 98.86 |
| Method 2 | × | √ | × | 98.82 | 98.66 | 99.06 |
| Method 3 | × | × | √ | 99.08 | 99.05 | 99.12 |
In Stage 2, we also utilized ablation experiments, and the results indicated (Tables 5 and 6) that the accuracy and AUC of the model with a parallel network were further enhanced and that the equal error rate was reduced. After the parallel network, the feature erase module was used to find that the RGB multimodal model was more conducive to enhancing the performance of the overall diagnostic model. The addition of ASPP and MHSA mechanism was more conducive to improving the performance of the overall diagnostic model. Therefore, RGB multi-channel, parallel network, feature erase module, ASPP and the MHSA mechanism were used to access the detection model backbone.
| Module | Accuracy, % | EER, % | AUC, % |
| PN, √ | 98.96 | 0.24 | 98.86 |
| PN, × | 96.38 | 0.29 | 95.02 |
| ASPP, √ | 98.96 | 0.24 | 98.86 |
| ASPP, × | 96.01 | 0.28 | 96.47 |
| MHSA, √ | 98.96 | 0.24 | 98.86 |
| MHSA, × | 96.22 | 0.29 | 95.68 |
| Random number | Accuracy, % | EER, % | AUC, % |
| 001 | 97.91 | 0.29 | 98.49 |
| 010 | 97.92 | 0.28 | 98.47 |
| 100 | 97.91 | 0.29 | 98.50 |
| 011 | 98.58 | 0.24 | 98.63 |
| 101 | 98.37 | 0.25 | 98.66 |
| 110 | 98.27 | 0.25 | 98.67 |
| 111 | 98.96 | 0.24 | 98.86 |
(3) Model auxiliary reading. We utilized the optimal model to assist physician reading. The study indicated that the sensitivity, specificity, and accuracy of model-assisted reading for all pictures were 99.17%, 99.92%, and 99.86%, res
(4) Comparison with existing models. The proposed model was compared with the existing research models, and the experimental results are depicted in Table 7. Generally, the specificity and accuracy of the proposed model for the recognition of ulcers, protruding lesions, vascular lesions, and bleeding pictures and the sensitivity for the recognition of ulcers and bleeding pictures were higher than those of the other three methods. The sensitivity of the proposed algorithm in identifying protruding lesions and vascular lesions is slightly lower than that of Ding et al[22].
| Ref. | Year of publication | Application | Algorithm | Sensitivity, % | Specificity, % | Accuracy, % |
| Aoki et al[29] | 2019 | Erosion/ulcer | CNN system based on SSD | 88.2 | 90.9 | 90.8 |
| Ding et al[22] | 2019 | Ulcer | ResNet-152 | 99.7 | 99.9 | 99.8 |
| Bleeding | 99.5 | 99.9 | 99.9 | |||
| Vascular lesion | 98.9 | 99.9 | 99.2 | |||
Aoki et al[30] | 2020 | Protruding lesion | ResNet-50 | 100 | 99.9 | 99.9 |
| Bleeding | 96.6 | 99.9 | 99.9 | |||
| Current study | 2023 | Ulcer( P1U + P2U ) | Improved ResNet-50 + YOLO-V5 | 99.7 | 99.9 | 99.9 |
| Vascular lesion | 97.4 | 99.9 | 99.9 | |||
| Protruding lesion (P1P + P2P) | 98.1 | 99.9 | 99.9 | |||
| Bleeding | 100 | 100 | 100 |
(5) Time calculation. The average processing time of physicians was 0.40 ± 0.24 s/image, and the image processing time of the improved model system was 48.00 ± 7.00 ms/image. The processing time of the system was significantly different from that of the clinicians (P < 0.001).
After 20 years of development, CE has continuously expanded its application depth and breadth and has become a crucial examination method for gastrointestinal diseases[23-25]. However, CE examination is a tedious task in clinical work due to its long reading time. With the wide application of artificial intelligence (AI)[26], the reading time of CE has been immensely shortened. While reducing the reading time, it is more crucial to enhance the performance of the system. Initial research was limited to the identification of one lesion. For example, Tsuboi et al[27] developed a CNN model for the automatic identification of small intestinal vascular lesions and Ribeiro et al[28] developed a CNN model to automatically identify protruding lesions in the small intestine while Ferreira et al[16] developed a CNN system that can automatically identify ulcers and mucosal erosions. With continued research, CNNs have been developed to identify a variety of lesions. For example, Ding et al[22] conducted a multicenter retrospective study that included 77 medical centers and collected 6970 cases undergoing SB CE. A CNN model based on ResidualNet 152 that can automatically detect 10 small intestinal lesions (inflammation, ulcer, polyp, lymphangiectasia, hemorrhage, vascular disease, protrusion lesions, lymphoid follicular hyperplasia, diverticulum, and parasites) was developed[22]. Their system exhibited high-level performance, and this result indicates the potential of AI models for multi-lesion detection. However, CNN cannot effectively focus on the important part of the image, limiting the ability of the model to identify lesions. Moreover, the current research is based on the network model of image classification, which cannot distinguish two or more types of lesions in the image, let alone determine the specific location of the lesions, and its practicability is poor. More importantly, existing models do not identify the bleeding risks of SB lesions.
In this study, we explored the image classification and object detection model to facilitate the evaluation of CE images. Ablation experiments were also conducted on multiple modules to improve ResNet-50 and YOLO-V5, ultimately ob
This study exhibited considerable novelty. First, this was based upon the pioneering AI diagnostic system for clinical CE to automatically detect SB lesions and their bleeding risks. Second, our model had high accuracy (98.96%) and high sensitivity (99.17%) when assisting physician reading. Especially for SB vascular lesions, the sensitivity of physician reading was only 52.38%, which indicated that nearly half of the lesions will be missed, and SB vascular lesions exhibited a high risk of bleeding, which is a common causative factor for SB bleeding. The network model immensely enhanced the sensitivity for such lesions (97.40%), which bears immense significance for physicians tasked with enhancing the diagnosis of bleeding and identifying high-risk bleeding populations. Meanwhile, the model was time-efficient (48.00 ± 7.00 ms for each image compared with 0.40 ± 0.24 s for clinicians). Its diagnostic performance exhibited potential for clinical application.
In general, the proposed model outperforms the existing models in the identification of a variety of lesions (ulcers, luminal protrusion lesions, vascular lesions and bleeding), which can effectively improve the ability of physicians to identify lesions and evaluate bleeding. However, the sensitivity of the proposed algorithm for the recognition of intraluminal protruding lesions and vascular lesions was lower than that of Ding et al[22] (100/98.1, 98.9/97.4), which may be related to the following factors. First, the sample size of the dataset was small, and the model did not fully learn the relevant discriminative features. Second, other existing models only perform the task of picture classification, while the proposed model not only classifies and detects lesions, providing an the accurate location of the lesions, it also evaluates the bleeding risk of lesions. The effective completion of these tasks may affect the sensitivity of the model, however.
Several limitations of this study should be considered when interpreting the results. First, diverticulum and parasite images were not included in the study due to the limited number of images available for training. Future studies should be directed toward enrollment and multicenter collaboration so that the aforementioned issues can be effectively addressed. Second, as an experimental evaluation and first-step investigation, the system was developed and tested on still images; it failed to perform real-time detection and result interpretation on videos. Thus, future studies evaluating the real-time utilization of AI in CE are warranted.
The trained deep learning model based on image classification combined with object detection exhibited satisfactory performance in identifying SB lesions and their bleeding risk, which enhanced the diagnostic efficiency of physicians and improved the ability of physicians to identify high-risk bleeding groups. This system highlighted its future application potential as an AI diagnostic system.
Deep learning provides an efficient automatic image recognition method for small bowel (SB) capsule endoscopy (CE) that can assist physicians in diagnosis. However, the existing deep learning models present some unresolved challenges.
CE reading is time-consuming and complicated. Abnormal parts account for only a small proportion of CE images. Therefore, it is easy to miss the diagnosis, which affects the detection of lesions and assessment of their bleeding risk. Also, both image classification and object detection have made significant progress in the field of deep learning.
To propose a novel and effective classification and detection model to automatically identify various SB lesions and their bleeding risks, and label the lesions accurately, so as to enhance the diagnostic efficiency of physicians and their ability to identify high-risk bleeding groups.
The proposed model was a two-stage method that combined image classification with object detection. First, we utilized the improved ResNet-50 classification model to classify endoscopic images into SB lesion images, normal SB mucosa images, and invalid images. Then, the improved YOLO-V5 detection model was utilized to detect the type of lesion and the risk of bleeding, and the location of the lesion was marked. We constructed training and testing sets and compared model-assisted readings with physician readings.
The accuracy of the model constructed in this study reached 98.96%, which was higher than the accuracy of other systems using only a single module. The sensitivity, specificity, and accuracy of the model-assisted reading detection of all images were 99.17%, 99.92%, and 99.86%, which were significantly higher than those of the endoscopists’ diagnoses. The image processing time of the model was 48 ms/image, and the image processing time of the physicians was 0.40 ± 0.24 s/image (P < 0.001).
The deep learning model of image classification combined with object detection exhibits a satisfactory diagnostic effect on a variety of SB lesions and their bleeding risks in CE images, which enhances the diagnostic efficiency of physicians and improves their ability to identify high-risk bleeding groups.
We utilized a two-stage combination method and added multiple modules to identify normal SB mucosa images, invalid images, and various SB lesions (lymphangiectasia, lymphoid follicular hyperplasia, xanthoma, erosion, ulcer smaller than 2 cm, protruding lesion smaller than 1 cm, ulcer larger than 2 cm, protruding lesion larger than 1 cm, vascular lesions, and blood). The bleeding risk was evaluated and classified.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Mijwil MM, Iraq S-Editor: Fan JR L-Editor: A P-Editor: Xu ZH
| 1. | Sami SS, Iyer PG, Pophali P, Halland M, di Pietro M, Ortiz-Fernandez-Sordo J, White JR, Johnson M, Guha IN, Fitzgerald RC, Ragunath K. Acceptability, Accuracy, and Safety of Disposable Transnasal Capsule Endoscopy for Barrett's Esophagus Screening. Clin Gastroenterol Hepatol. 2019;17:638-646.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 2. | Kim SH, Lim YJ. Artificial Intelligence in Capsule Endoscopy: A Practical Guide to Its Past and Future Challenges. Diagnostics (Basel). 2021;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 3. | Jiang B, Pan J, Qian YY, He C, Xia J, He SX, Sha WH, Feng ZJ, Wan J, Wang SS, Zhong L, Xu SC, Li XL, Huang XJ, Zou DW, Song DD, Zhang J, Ding WQ, Chen JY, Chu Y, Zhang HJ, Yu WF, Xu Y, He XQ, Tang JH, He L, Fan YH, Chen FL, Zhou YB, Zhang YY, Yu Y, Wang HH, Ge KK, Jin GH, Xiao YL, Fang J, Yan XM, Ye J, Yang CM, Li Z, Song Y, Wen MY, Zong Y, Han X, Wu LL, Ma JJ, Xie XP, Yu WH, You Y, Lu XH, Song YL, Ma XQ, Li SD, Zeng B, Gao YJ, Ma RJ, Ni XG, He CH, Liu YP, Wu JS, Liu J, Li AM, Chen BL, Cheng CS, Sun XM, Ge ZZ, Feng Y, Tang YJ, Li ZS, Linghu EQ, Liao Z; Capsule Endoscopy Group of the Chinese Society of Digestive Endoscopy. Clinical guideline on magnetically controlled capsule gastroscopy (2021 edition). J Dig Dis. 2023;24:70-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 15] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 4. | Nennstiel S, Machanek A, von Delius S, Neu B, Haller B, Abdelhafez M, Schmid RM, Schlag C. Predictors and characteristics of angioectasias in patients with obscure gastrointestinal bleeding identified by video capsule endoscopy. United European Gastroenterol J. 2017;5:1129-1135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 5. | Liao Z, Gao R, Xu C, Li ZS. Indications and detection, completion, and retention rates of small-bowel capsule endoscopy: a systematic review. Gastrointest Endosc. 2010;71:280-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 561] [Cited by in RCA: 476] [Article Influence: 31.7] [Reference Citation Analysis (0)] |
| 6. | Rondonotti E, Pennazio M, Toth E, Koulaouzidis A. How to read small bowel capsule endoscopy: a practical guide for everyday use. Endosc Int Open. 2020;8:E1220-E1224. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 7. | Beg S, Card T, Sidhu R, Wronska E, Ragunath K; UK capsule endoscopy users’ group. The impact of reader fatigue on the accuracy of capsule endoscopy interpretation. Dig Liver Dis. 2021;53:1028-1033. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 8. | Leenhardt R, Koulaouzidis A, McNamara D, Keuchel M, Sidhu R, McAlindon ME, Saurin JC, Eliakim R, Fernandez-Urien Sainz I, Plevris JN, Rahmi G, Rondonotti E, Rosa B, Spada C, Toth E, Houdeville C, Li C, Robaszkiewicz M, Marteau P, Dray X. A guide for assessing the clinical relevance of findings in small bowel capsule endoscopy: analysis of 8064 answers of international experts to an illustrated script questionnaire. Clin Res Hepatol Gastroenterol. 2021;45:101637. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 9. | ASGE Technology Committee; Wang A, Banerjee S, Barth BA, Bhat YM, Chauhan S, Gottlieb KT, Konda V, Maple JT, Murad F, Pfau PR, Pleskow DK, Siddiqui UD, Tokar JL, Rodriguez SA. Wireless capsule endoscopy. Gastrointest Endosc. 2013;78:805-815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 194] [Article Influence: 16.2] [Reference Citation Analysis (2)] |
| 10. | Chu Y, Huang F, Gao M, Zou DW, Zhong J, Wu W, Wang Q, Shen XN, Gong TT, Li YY, Wang LF. Convolutional neural network-based segmentation network applied to image recognition of angiodysplasias lesion under capsule endoscopy. World J Gastroenterol. 2023;29:879-889. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 1] [Cited by in RCA: 10] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 11. | Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5683] [Cited by in RCA: 5360] [Article Influence: 670.0] [Reference Citation Analysis (0)] |
| 12. | Hassan C, Spadaccini M, Iannone A, Maselli R, Jovani M, Chandrasekar VT, Antonelli G, Yu H, Areia M, Dinis-Ribeiro M, Bhandari P, Sharma P, Rex DK, Rösch T, Wallace M, Repici A. Performance of artificial intelligence in colonoscopy for adenoma and polyp detection: a systematic review and meta-analysis. Gastrointest Endosc. 2021;93:77-85.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 361] [Cited by in RCA: 308] [Article Influence: 77.0] [Reference Citation Analysis (1)] |
| 13. | Mijwil MM, Al-Mistarehi AH, Abotaleb M, El-kenawy EM, Ibrahim A, Abdelhamid6 AA, Eid MM. From Pixels to Diagnoses: Deep Learning's Impact on Medical Image Processing-A Survey. Wasit J Comput Mathematics Sci. 2023;2:9-15. [DOI] [Full Text] |
| 14. | Mijwil MM. Deep Convolutional Neural Network Architecture to Detect COVID-19 from Chest X-Ray Images. Iraqi J Sci. 2023;64:2561-2574. [DOI] [Full Text] |
| 15. | Mascarenhas Saraiva MJ, Afonso J, Ribeiro T, Ferreira J, Cardoso H, Andrade AP, Parente M, Natal R, Mascarenhas Saraiva M, Macedo G. Deep learning and capsule endoscopy: automatic identification and differentiation of small bowel lesions with distinct haemorrhagic potential using a convolutional neural network. BMJ Open Gastroenterol. 2021;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 16. | Ferreira JPS, de Mascarenhas Saraiva MJDQEC, Afonso JPL, Ribeiro TFC, Cardoso HMC, Ribeiro Andrade AP, de Mascarenhas Saraiva MNG, Parente MPL, Natal Jorge R, Lopes SIO, de Macedo GMG. Identification of Ulcers and Erosions by the Novel Pillcam™ Crohn's Capsule Using a Convolutional Neural Network: A Multicentre Pilot Study. J Crohns Colitis. 2022;16:169-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 40] [Article Influence: 13.3] [Reference Citation Analysis (1)] |
| 17. | Mascarenhas Saraiva M, Ribeiro T, Afonso J, Andrade P, Cardoso P, Ferreira J, Cardoso H, Macedo G. Deep Learning and Device-Assisted Enteroscopy: Automatic Detection of Gastrointestinal Angioectasia. Medicina (Kaunas). 2021;57. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 18. | Zhou JX, Yang Z, Xi DH, Dai SJ, Feng ZQ, Li JY, Xu W, Wang H. Enhanced segmentation of gastrointestinal polyps from capsule endoscopy images with artifacts using ensemble learning. World J Gastroenterol. 2022;28:5931-5943. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 19. | Li P, Li Z, Gao F, Wan L, Yu J. Convolutional neural networks for intestinal hemorrhage detection in wireless capsule endoscopy images. Proceedings of the 2017 IEEE International Conference on Multimedia and Expo (ICME); 2017 Jul 10–14; Hong Kong, China. ICME, 2017: 1518–1523. |
| 20. | Jiang PY, Ergu DJ, Liu FY, Cai Y, Ma B. A Review of Yolo Algorithm Developments. Procedia Comput Sci. 2022;199:1066-1073. |
| 21. | Saurin JC, Delvaux M, Gaudin JL, Fassler I, Villarejo J, Vahedi K, Bitoun A, Canard JM, Souquet JC, Ponchon T, Florent C, Gay G. Diagnostic value of endoscopic capsule in patients with obscure digestive bleeding: blinded comparison with video push-enteroscopy. Endoscopy. 2003;35:576-584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 253] [Cited by in RCA: 315] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 22. | Ding Z, Shi H, Zhang H, Meng L, Fan M, Han C, Zhang K, Ming F, Xie X, Liu H, Liu J, Lin R, Hou X. Gastroenterologist-Level Identification of Small-Bowel Diseases and Normal Variants by Capsule Endoscopy Using a Deep-Learning Model. Gastroenterology. 2019;157:1044-1054.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 208] [Article Influence: 34.7] [Reference Citation Analysis (0)] |
| 23. | Ahmed I, Ahmad A, Jeon G. An IoT-Based Deep Learning Framework for Early Assessment of Covid-19. IEEE Internet Things J. 2021;8:15855-15862. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 24. | Xia J, Xia T, Pan J, Gao F, Wang S, Qian YY, Wang H, Zhao J, Jiang X, Zou WB, Wang YC, Zhou W, Li ZS, Liao Z. Use of artificial intelligence for detection of gastric lesions by magnetically controlled capsule endoscopy. Gastrointest Endosc. 2021;93:133-139.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 41] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 25. | Konishi M, Shibuya T, Mori H, Kurashita E, Takeda T, Nomura O, Fukuo Y, Matsumoto K, Sakamoto N, Osada T, Nagahara A, Ogihara T, Watanabe S. Usefulness of flexible spectral imaging color enhancement for the detection and diagnosis of small intestinal lesions found by capsule endoscopy. Scand J Gastroenterol. 2014;49:501-505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 26. | Maeda M, Hiraishi H. Efficacy of video capsule endoscopy with flexible spectral imaging color enhancement at setting 3 for differential diagnosis of red spots in the small bowel. Dig Endosc. 2014;26:228-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 27. | Tsuboi A, Oka S, Aoyama K, Saito H, Aoki T, Yamada A, Matsuda T, Fujishiro M, Ishihara S, Nakahori M, Koike K, Tanaka S, Tada T. Artificial intelligence using a convolutional neural network for automatic detection of small-bowel angioectasia in capsule endoscopy images. Dig Endosc. 2020;32:382-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 103] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 28. | Ribeiro T, Saraiva MM, Ferreira JPS, Cardoso H, Afonso J, Andrade P, Parente M, Jorge RN, Macedo G. Artificial intelligence and capsule endoscopy: automatic detection of vascular lesions using a convolutional neural network. Ann Gastroenterol. 2021;34:820-828. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 29. | Aoki T, Yamada A, Aoyama K, Saito H, Tsuboi A, Nakada A, Niikura R, Fujishiro M, Oka S, Ishihara S, Matsuda T, Tanaka S, Koike K, Tada T. Automatic detection of erosions and ulcerations in wireless capsule endoscopy images based on a deep convolutional neural network. Gastrointest Endosc. 2019;89:357-363.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 175] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 30. | Aoki T, Yamada A, Kato Y, Saito H, Tsuboi A, Nakada A, Niikura R, Fujishiro M, Oka S, Ishihara S, Matsuda T, Nakahori M, Tanaka S, Koike K, Tada T. Automatic detection of blood content in capsule endoscopy images based on a deep convolutional neural network. J Gastroenterol Hepatol. 2020;35:1196-1200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 72] [Article Influence: 14.4] [Reference Citation Analysis (0)] |