Published online Jan 14, 2024. doi: 10.3748/wjg.v30.i2.115
Peer-review started: November 20, 2023
First decision: December 11, 2023
Revised: December 21, 2023
Accepted: December 28, 2023
Article in press: December 28, 2023
Published online: January 14, 2024
Processing time: 53 Days and 4.9 Hours
Small nucleolar RNAs (snoRNAs) represent a class of non-coding RNAs that play pivotal roles in post-transcriptional RNA processing and modification, thereby contributing significantly to the maintenance of cellular functions related to protein synthesis. SnoRNAs have been discovered to possess the ability to influence cell fate and alter disease progression, holding immense potential in controlling human diseases. It is suggested that the dysregulation of snoRNAs in cancer exhibits differential expression across various cancer types, stages, metastasis, treatment response and/or prognosis in patients. On the other hand, colorectal cancer (CRC), a prevalent malignancy of the digestive system, is characterized by high incidence and mortality rates, ranking as the third most common cancer type. Recent research indicates that snoRNA dysregulation is associated with CRC, as snoRNA expression significantly differs between normal and cancerous conditions. Consequently, assessing snoRNA expression level and function holds promise for the prognosis and diagnosis of CRC. Nevertheless, current comprehension of the potential roles of snoRNAs in CRC remains limited. This review offers a comprehensive survey of the aberrant regulation of snoRNAs in CRC, providing valuable insights into the discovery of novel biomarkers, therapeutic targets, and potential tools for the diagnosis and treatment of CRC and furnishing critical cues for advancing research into CRC and the judicious selection of therapeutic targets.
Core Tip: Small nucleolar RNAs (snoRNAs) play vital roles in post-transcriptional RNA processing, influencing the cell fate and diverse disease progression. Dysregulated snoRNAs in various cancers, including colorectal cancer (CRC), show promise for improving prognosis and diagnosis. Despite this potential, understanding of snoRNAs’ roles in CRC remains limited, warranting further research into their precise functions.
- Citation: Lan YZ, Wu Z, Chen WJ, Fang ZX, Yu XN, Wu HT, Liu J. Small nucleolar RNA and its potential role in the oncogenesis and development of colorectal cancer. World J Gastroenterol 2024; 30(2): 115-127
- URL: https://www.wjgnet.com/1007-9327/full/v30/i2/115.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i2.115
For many years, small nucleolar RNAs (snoRNAs) have been considered one of the prime examples of non-coding RNAs (ncRNAs). They are small RNA molecules, typically consisting of 60 to 300 nucleotides, primarily located within the nucleolus of the cell[1,2]. The discovery of snoRNAs traces back to the late 1960s when researchers identified a group of distinct low-molecular-weight RNAs in the nucleoplasm and nucleolus of HeLa cells, in length ranging from 100 to 180 base pairs[3]. However, due to technological limitations of the time, the exact nature and precise locations of these small RNAs remained largely unknown. It wasn’t until 1976 that scientists first identified a unique RNA in the cell nucleus, later named snoRNA U3[4], sparking further investigations into snoRNAs.
With the continuous development and application of transcription inhibitors and dynamic labeling techniques, snoRNAs were gradually confirmed to be stable products transcribed by RNA Polymerase II. In the study of mammals, frogs, and other organisms, researchers progressively unveiled the structural features of snoRNA expression, particularly variations like U3, revealing their close association with ribosomal RNA (rRNA)[1,5,6]. Then novel strategies and technologies for analyzing snoRNAs emerged from studies of mRNA splicing[7]. Since then, extensive research has illuminated snoRNA’s primary role in modifying rRNA, which is its pivotal function to guarantee the proper assembly and functioning of ribosomes, the cellular machinery responsible for protein synthesis. Moreover, snoRNAs play a fundamental role in upholding cellular functions related to protein synthesis by actively engaging in the post-transcriptional processing and modification of various RNA molecules[8,9].
However, investigations into the potential roles of snoRNAs in cancer began with a study reporting significant downregulation of snoRNA in meningiomas when compared to normal brain tissue[10]. Recent research has also shown differential expression of snoRNAs in various cancers, such as breast cancer and non-small cell lung cancer (NSCLC), in clinical samples and cell lines, indicating their potential diagnostic and prognostic value[11,12]. These findings underscore the differential expression of snoRNAs in healthy and tumor tissues as well as body fluids, suggesting that snoRNAs might have a broader role as major gene regulatory factors at both the transcriptional and epigenetic levels[13]. Aberrant snoRNA expression has been identified as a hallmark of tumorigenesis, suggesting that these RNAs play a more complex and diverse role in cellular gene regulation[14,15]. Furthermore, changes in snoRNA function and expression levels have been linked to various diseases, particularly cancers[16], highlighting the active and expanding research field exploring the roles of snoRNAs in disease onset and progression.
Colorectal cancer (CRC) is a global health concern, has emerged as the leading cause of digestive system cancer-related deaths, ranking third in new cancer cases and cancer-related deaths, posing a significant threat to human health and quality of life[17,18]. Patients with early-stage CRC often lack noticeable symptoms, with unspecific manifestations such as constipation, diarrhea, rectal bleeding, abdominal pain, fatigue, and weight loss emerging as the disease progresses[19]. Unfortunately, these symptoms typically become apparent only in advanced stages, contributing to a dismal 5-year survival rate of approximately 14% for late-stage CRC patients. Shockingly, around 25% of CRC patients present with metastasis at the time of diagnosis[20]. Furthermore, roughly 50% of treated patients eventually experience metastasis during their lifetime[21,22]. The high mortality rate among CRC patients can be attributed primarily to its significant heterogeneity and proclivity for metastasis. The inherent characteristic poses a significant challenge to early diagnosis and treatment, because patients with CRC can be effectively cured if detected and treated in its early stages. Hence, early detection significantly reduces the mortality associated with CRC[23].
Until now, numerous established diagnostic approaches have contributed to the early management of CRC, thus enhancing patient prognosis. These methodologies encompass screening procedures such as fecal occult blood tests, fecal immunochemical tests, and fecal DNA testing[24]. Additionally, colonoscopies permit the direct sampling of irregular tissues or polyps[25]. Furthermore, blood-based biomarkers, such as carcinoembryonic antigen (CEA) and circulating tumor DNA, have emerged as non-invasive CRC screening techniques, with limitations regarding sensitivity and specificity[26,27]. Additionally, fecal DNA testing lacks comprehensive validation. Furthermore, concerns of a practical nature arise, including the risks associated with invasive colonoscopies, substantial financial outlays, and the need for skilled endoscopists, as well as patient compliance[28]. Hence, the development of innovative and efficient CRC diagnostic biomarkers holds paramount importance in facilitating early detection and mitigating CRC-related mortality. Researchers have identified leukocyte immunoglobulin-like receptor B2 (LILRB2) protein as a distinctive marker of CRC regarding diagnosis and therapeutics, facilitating early screening and precision treatment[29]. It is also shown that the combined assessment of the methylation status of SEPT9, SDC2, and ALX4 in plasma has the potential to enhance the sensitivity of CRC detection[30]. Recent research has shed light on the significant association between snoRNA and CRC, delving into the potential role of snoRNAs in the onset and progression of CRC[31]. Aberrant expression or function of snoRNAs may be associated with the development and progression of CRC, which provides new clues for further research into the pathogenesis and treatment of CRC[2,32]. This article purposed to review the role and mechanisms of snoRNAs in the progression of CRC and explore their clinical applications in the diagnosis and prognosis of CRC.
SnoRNAs are a class of small ncRNAs with lengths ranging from 60 to 300 nucleotides, primarily named due to their localization within the nucleolus and typically playing a crucial role in the modification, maturation, and stability of rRNA by forming complexes with small nucleolar ribonucleoproteins (snoRNPs)[1,7]. Similar to other ncRNAs, snoRNAs lack translation functionality, but what sets them apart is their specific nucleotide sequences and spatial structures[33]. Based on distinct nucleotide motifs and secondary structures, snoRNAs are primarily categorized into C/D box snoRNAs and H/ACA box snoRNAs[34,35]. C/D box snoRNAs guide the 2’-O-methylation of specific nucleotides in the target RNA molecule by binding with four core proteins, namely nucleolar protein 56 (NOP56), NOP58, small nuclear ribonucleoprotein 13 (SNU13), and a methyltransferase fibrillarin (FBL)[36,37], while H/ACA box snoRNAs associated with conserved proteins glycine arginine-rich protein 1 (GAR1), non-histone chromosome protein 2 (NHP2), NOP10, and dyskeratosis congenita 1 (DKC1) guide the conversion of specific uridine nucleotides to pseudouridine[38].
C/D box snoRNAs (SNORDs) are typically 60 nt to 90 nt long and consist of two concise sequence elements, the C box (5’RUGAUGA3’) situated at the 5’ end and the D box (5’CUGA3’) located at the 3’ end. They contain internal structures similar to the C and D boxes, referred to as box C’ and box D’, respectively, with 4-6 nucleotides of reverse complementary sequences at both ends, forming a single hairpin structure[39]. SNORDs act as recognition sites for specific proteins such as NOP56, NOP58, SNU13, and FBL. When these proteins directly bind to SNORDs, they form a scaffold-like structure, enabling interactions with other proteins to execute various biological functions[40]. SNORDs are crucial for maintaining the structural stability of snoRNAs and their localization within the nucleolus, involving in the splicing and processing of rRNA precursors[41]. Most SNORDs contain an upstream segment of more than 21 nucleotides in the C/D box region, referred to as antisense snoRNA[42]. This segment has the ability to bind to target RNA through complementary base pairing. Antisense snoRNAs act as guide RNAs, directing the methylation of specific 2’-O-hydroxyl positions on target molecules, thereby facilitating 2’-O ribose methylation modifications at specific sites on rRNA[43].
H/ACA box snoRNAs (SNORAs) range from 120 nt to 140 nt and exhibit a typical secondary structure known as the “hairpin-hinge-hairpin-tail” structure. The hinge region consists of a single-stranded nucleic acid segment that acts as a bridge connecting two hairpin structures and both the hinge and tail regions contain conserved sequence elements known as the H box (5’ANANNA3’) and ACA box, respectively, with the ACA box usually located at the third nucleotides upstream from the 3’ end[44,45]. Due to the presence of the H box and ACA box, SNORAs can form a double-hairpin structure, and within the pseudouridine pockets of these hairpins, there exists antisense snoRNA that binds to specific sites on target molecules[46]. When antisense snoRNA associates with core proteins, such as GAR1, NHP2, NOP10, and DKC1, it forms a homologous snoRNP complex, guiding the pseudouridylation modification of target RNA[47]. In essence, the majority of SNORAs guide the pseudouridylation modification of specific sites within eukaryotic rRNA and snRNA, converting uridine into pseudouridine[48]. It is revealed that mammalian SNORAs share a similar H/ACA motif with human telomerase RNA, suggesting that H/ACA-like structural domains may be associated with cell proliferation[49,50].
Additionally, small Cajal body-associated RNAs (scaRNAs) are more variable in length, representing a specific subset of snoRNAs[51]. ScaRNAs play critical roles within the Cajal bodies, small membraneless subcompartments in the cell nucleus, and are involved in regulating the modification of snRNAs and post-transcriptional regulation of RNA. Structurally, scaRNAs are similar to snoRNAs, and some scaRNAs contain both C/D box and H/ACA box sequences[52]. Moreover, it has been identified a connection between dysregulated scaRNAs and the progression of certain cancers, including CRC. Notably, SCARNA12[53], SCARNA15[54], and SCARNA22[31] have been observed to have increased expression levels in CRC cell lines and clinical CRC tissue samples. However, there remains a limited understanding of their specific biological functions and the underlying molecular mechanisms within the context of CRC.
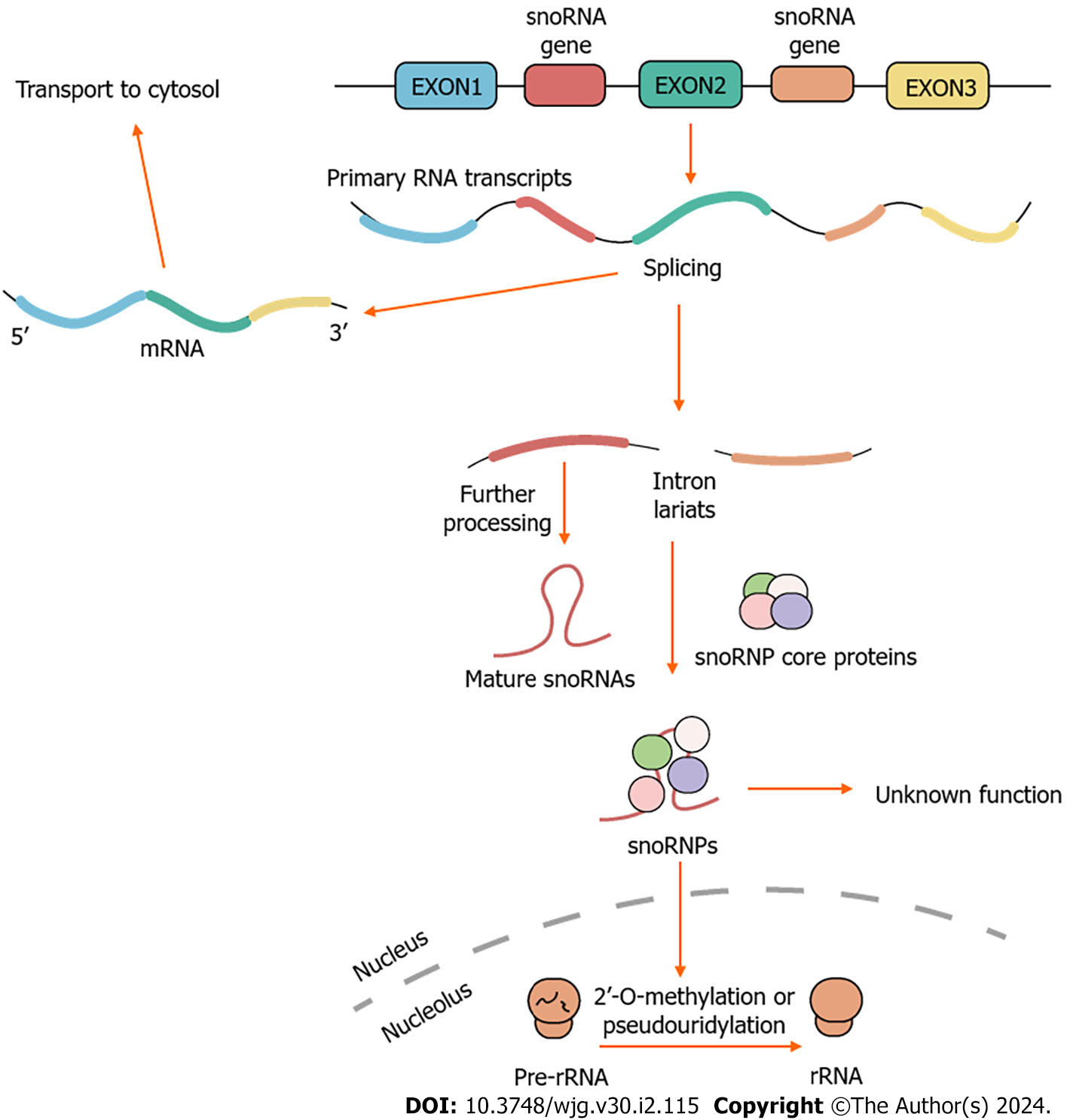
SnoRNAs participate in various biological processes, including the processing of rRNA[55], regulation of RNA splicing and translation[56], as well as responses to oxidative stress[57]. In essence, these post-transcriptional modifications are crucial for the production of efficient and accurate ribosomes, to maintain the intricate cellular functions associated with protein synthesis[58]. Over the past two decades, the understanding of the cellular functions of snoRNAs has significantly expanded, including the production of other regulatory RNAs involved in the regulation of gene expression (e.g., the production of piRNAs[59], the production of miRNAs by shearing of snoRNAs[60,61]), the modulation of mRNA abundance[62], and the regulation of translational efficiency[63]. There have been reports of orphan snoRNAs, but their functions and target actions remain unclear[64]. Therefore, traditional snoRNA classifications may not comprehensively encompass all the functions of snoRNAs (Figure 1).
In normal physiological processes, snoRNAs stably exist by forming snoRNPs through interactions with specific proteins, and they subsequently participate in 2’-O-methylation or pseudouridylation of rRNA[35]. With the completion of the human genome sequencing and the advancement of high-throughput sequencing in the early 21st century, coupled with the deepening research in nucleic acids and proteomics, an increasing number of snoRNAs associated with diseases have been discovered. Currently, the focus of research is to gain a deeper understanding of the functions of snoRNAs under both normal and pathological conditions[65].
The first report of snoRNAs’ role in human diseases came with Prader-Willi syndrome, suggesting that snoRNA may have an important role in human diseases[66,67]. Subsequently, abnormal snoRNA expression has been reported in various human diseases, including congenital heart defects[68], neurodegenerative disorders[69], and cancers[70]. Researchers have observed a significant and pronounced alteration in the expression of snoRNAs in myocardial tissue affected by Tetralogy of Fallot, compared to healthy control myocardium, suggesting that snoRNA dysregulation might have an impact on development[68]. In mammals, especially in humans, approximately 90% of snoRNAs are encoded within introns, often associated with genes related to ribosome biogenesis[71]. This co-localization implies a coordinated expression designed to facilitate their collaborative functions.
Furthermore, it is revealed that SNORD113-1 suppresses the development of hepatocellular carcinoma (HCC). Compared to adjacent non-cancerous tissue, the expression of SNORD113-1 is significantly downregulated in HCC. Moreover, the decreased SNORD113-1 level is notably associated with adverse patient survival outcomes, indicating that SNORD113-1 plays an anti-cancer role in HCC as a potential diagnostic and therapeutic target for this condition[72]. Additionally, functionally aberrant snoRNAs may play a role in the onset and progression of malignant tumors in humans. For instance, SNORA42 has been shown to act as an oncogene in lung tumorigenesis[73]. Furthermore, snoRNAs (SNORD15A, SNORD15B, SNORD22, SNORD17, and SNORD87) and snoRNPs are frequently overexpressed in both mouse and human breast cancer and prostate cancer, demonstrating their association with tumorigenicity and highlighting the significant role of snoRNAs in regulating cancer biology[74].
As snoRNAs can be detected in both plasma and serum reliably, their levels in blood indicate various disease states and correlate with clinical pathological features, highlighting their applicability as potential disease biomarkers[31]. For instance, the expression level of SNORD33 in plasma was found to be correlated with the effectiveness of platinum-based drugs in patients with metastatic triple-negative breast cancer (TNBC), indicating that SNORD33’s expression holds promise as a potential biomarker for predicting the efficacy of platinum drugs in TNBC patients[75]. Furthermore, the elevated expression of SNORD52 in HCC is closely associated with poor clinical prognosis, which promotes HCC development by upregulating CDK1, thus enhancing the stability of the CDK1 protein, indicating that targeting the Upf1/SNORD52/CDK1 pathway has therapeutic potential for HCC treatment[76]. In addition, SNORA38B exhibits high expression in NSCLC tissues and cell lines, correlating with unfavorable prognosis. SNORA38B acts as an oncogene in NSCLC, in part, by directly binding to E2F1 and regulating the GAB2/AKT/mTOR pathway[77].
To sum up, in the process of tumorigenesis, ribosomes, which serve as molecular factories for protein biosynthesis, are typically highly activated to meet the heightened raw material demands of proliferating cancer cells. SnoRNAs play the critical role in the maturation of ribosomes[5,6]. Consequently, alterations in snoRNA expression may have an extensive impact on the mechanisms of cancer development and pathological processes. However, current reports are primarily centered around snoRNA screening and validating their associations with diseases, with an incomplete understanding of their mechanisms, especially their roles in cellular signal transduction pathways.
CRC is one of the most prevalent cancers in humans[19]. Currently, advances in cancer prevention, early detection, and treatment have led to a decrease in mortality rates associated with CRC. However, the 5-year survival rate for this disease still remains relatively low[78]. Despite the utilization of advanced therapeutic approaches including surgical resection, chemotherapy, and radiotherapy, there hasn’t been a notable improvement in the mortality rate of CRC patients, due to the resemblance of early-stage CRC to non-malignant intestinal conditions, a scarcity of distinctive symptoms, and the consequent delayed diagnoses[79]. Consequently, the absence of efficacious molecular biomarkers for early detection remains a substantial challenge in addressing CRC. Increasing evidence suggests that snoRNAs serve as key regulators of gene expression and RNA modification[58]. Defects in the functions of these molecules have become markers of tumorigenesis and cancer development[74]. Differential expression of snoRNAs in CRC may hold diagnostic and prognostic value[80,81]. Therefore, a comprehensive understanding of the mechanisms by which aberrant snoRNAs contribute to the development of CRC is imperative and the involvement of snoRNAs in the progression of CRC is gaining importance as a reliable molecular biomarker (Table 1). It has the potential to greatly enhance early clinical diagnosis and improve prognosis for CRC patients.
| snoRNA | Type | Modification site | Length (nt) | Host gene | Expression | Mechanism | Potential functions | Ref. |
| SNORA5C | H/ACA | 18S:U1238; 18S:U1625 | 137 | TBRG4 | Up-regulation | Unknown | Promote proliferation, and colony formation | [82] |
| SNORA15 | H/ACA | 18S:U1367 | 133 | CCT6A | Up-regulation | Unknown | Exert the oncogenic effect | [83] |
| SNORA21 | H/ACA | 28S:U4401; 28S:U4270 | 132 | RPL23 | Up-regulation | Hippo signaling pathway, and Wnt signaling | Promote cell proliferation, and enhances tumor invasiveness | [84] |
| SNORA23 | H/ACA | 28S:U3737; 28S:U4331 | 189 | IPO7 | Up-regulation | Unknown | Unproved | [54] |
| SNORA24 | H/ACA | 18S:U863; 18S:U609 | 131 | SNHG8 | Up-regulation | P53 | Promote G1/S phase transition, and cell proliferation | [54,85] |
| SNORA41 | H/ACA | 18S:U1643 | 132 | EEF1B2 | Up-regulation | Unknown | Exert the oncogenic effect | [83] |
| SNORA42 | H/ACA | 18S:U572; 18S:U109 | 134 | TTC6 | Up-regulation | Unknown | Enhance cell proliferation, migration, invasion, anoikis resistance, and tumorigenicity | [31] |
| SNORA71A | H/ACA | 18S:U406 | 138 | SNHG17 | Up-regulation | LBP, NF-kappaB, Toll-like receptor | Promote cell proliferation, migration and invasion | [54] |
| SNORD1C | C/D | 28S:G4362 | 78 | SNHG16 | Up-regulation | Wnt/β-catenin pathway | Promote cell proliferation, migration, invasion and enhance cancer cell stemness | [86,87] |
| SNORD12B | C/D | 28S:G3878 | 91 | ZFAS1 | Up-regulation | Unknown | Promote tumorigenesis, proliferation, and metastasis | [88] |
| SNORD12C | C/D | 28S:G3878 | 89 | ZFAS1 | Up-regulation | ZFAS1-NOP58-SNORD12C/78-EIF4A3/LAMC2 | Promote cell proliferation | [13,89] |
| SNORD14E | C/D | Unknown | 85 | HSPA8 | Up-regulation | Unknown | Unproved | [89] |
| SNORD15B | C/D | 28S:A3764 | 146 | RPS3 | Up-regulation | Unknown | Promote proliferation, and colony formation | [82] |
| SNORD17 | C/D | 28S:U3797 | 237 | SNX5 | Up-regulation | Unknown | Unproved | [89] |
| SNORD33 | C/D | 18S:U1326 | 83 | RPL13A | Down-regulation | Unknown | Suppress cell anchorage | [83] |
| SNORD44 | C/D | 18S:A166 | 63 | GAS5 | Down-regulation | Caspase dependent pathway, PI3K/Akt | Inhibit tumor growth, and induce apoptosis | [90] |
| SNORD48 | C/D | 28S:C2279 | 64 | SNHG32 | Up-regulation | Unknown | Unproved | [82] |
| SNORD50A | C/D | 28S:C2848; 28S:G2863 | 75 | SNHG5 | Down-regulation | Methylation of 28S rRNA gene, k-Ras | Promote tumor growth | [91] |
| SNORD50B | C/D | Unknown | 70 | SNHG5 | Down-regulation | Methylation of 28S rRNA gene, k-Ras | Promote tumor growth | [91] |
| SNORD57 | C/D | 18S:A99 | 72 | NOP56 | Up-regulation | Unknown | Unproved | [92] |
| SNORD67 | C/D | U6:C60 | 111 | CKAP5 | Up-regulation | Unknown | Unproved | [89] |
| SNORD76 | C/D | 28S:A2350 | 80 | GAS5 | Up-regulation | Unknown | Unproved | [31] |
| SNORD78 | C/D | 28S:G4593 | 65 | GAS5 | Up-regulation | ZFAS1-NOP58-SNORD12C/78-EIF4A3/LAMC2 | Promote cell proliferation | [13,31] |
| SNORD126 | C/D | Unknown | 77 | CCNB1IP1 | Up-regulation | PI3K/Akt | Promote cell growth | [65] |
| SCARNA12 | scaRNA | U5:U46 | 270 | PHB2 | Up-regulation | PI3K/Akt | Promote proliferation and tumorigenicity | [53] |
| SCARNA15 | scaRNA | U2:U37 | 127 | SNHG21 | Up-regulation | Unknown | Unproved | [54] |
| SCARNA22 | scaRNA | Unknown | 125 | NSD2 | Up-regulation | Unknown | Unproved | [31] |
SNORA21: SNORA21 is a type of H/ACA box snoRNA with a length of 132 nt and its host gene is RPL23. Next-generation sequencing results have suggested that SNORA21 could be a potential early detection and prognostic biomarker for NSCLC[93]. Whereafter, Yoshida et al[84] observed an upregulation in the expression levels of SNORA21 in both colorectal adenomas and CRC through a series of in vitro and in vivo experiments. Using CRISPR/Cas9-mediated manipulation of SNORA21 expression, they demonstrated its regulatory influence on multiple cancer-associated pathways, resulting in reduced cell proliferation and diminished invasion and migration capabilities. In summary, this substantiates the oncogenic function of SNORA21 in CRC, wherein it modulates the cell cycle, consequently inducing alterations in cell proliferation and tumor invasion. Furthermore, SNORA21 has demonstrated the capacity to stimulate the proliferation of CRC cells by influencing pivotal cancer-related pathways, including the Hippo and Wnt signaling pathways, and heightened levels of SNORA21 have exhibited a correlation with distant metastasis in the context of CRC. These findings underscore the pivotal role that snoRNAs may play in cancer biology and suggest SNORA21 as a potential target for CRC therapy[84].
SNORA24: SNORA24, a type of H/ACA box snoRNA, exhibits a length of 131 nt and originates from the host gene SNHG8. It is shown that SNORA24 is downregulated in HCC, and may be a tumor suppressor gene[94]. However, research indicates that SNORA24 is markedly upregulated in various cancers, including CRC. Overexpression of SNORA24 promotes cell proliferation and the growth of xenograft tumors by regulating the cell cycle process, particularly by facilitating the G1/S phase transition. Additionally, silencing SNORA24 induces substantial apoptosis within cells and modulates the stability of p53 protein through the proteasome degradation pathway[95]. These findings elucidate the mechanism by which SNORA24 exerts its oncogenic influence in CRC, in a p53-dependent manner, underscoring its potential utility as a biomarker and therapeutic target for patients with CRC[85].
SNORA42: SNORA42 belongs to the H/ACA box snoRNA, with a length of 134 nt, and its locus is TTC6. Research indicates that SNORA42 is highly expressed in NSCLC tissues and has been experimentally verified for its oncogenic function[73]. Soon, the study from another group suggests that SNORA24 may function as an oncogene in CRC, as its overexpression leads to enhanced cell proliferation, migration, invasion, anoikis resistance, as well as tumorigenicity. Notably, heightened SNORA42 expression levels have been linked to distant metastasis and unfavorable prognosis in individuals afflicted with CRC. Furthermore, available data indicates that the expression of SNORA42 can be used to identify high-risk patients with stage II CRC. Consequently, utilizing SNORA42 expression as a molecular biomarker holds promise in identifying high-risk CRC patients, particularly those with stage II CRC, for postoperative adjuvant therapy to improve prognosis[31].
SNORA71A: SNORA71A is a H/ACA box snoRNA with a total length of 134 nt, located at the genomic locus AL080249.26, and its host gene is SNHG17. The transcription of SNORA71A appears to be synchronized with that of SNHG17. It has been reported that SNHG17 exhibits upregulation in CRC tissues, implying a potential upregulation of SNORA71A in CRC as well[96]. Researchers conducted a comprehensive analysis of snoRNA expression profiles in CRC utilizing small RNA sequencing technology. The investigation revealed a substantial upregulation of SNORA71A in CRC tissues and cells. Furthermore, SNORA71A expression displayed a significant correlation with TNM staging and lymph node metastasis. It demonstrates both high sensitivity and specificity, making it a promising candidate as a biomarker for CRC. SNORA71A seems to play a role within the NF-κB signaling pathway and Toll-like receptor signaling pathway in CRC, suggesting its involvement in the initiation and progression of CRC. And SNORA71A may be necessary for effective proliferation, migration, and invasion of CRC cells, as the results show that cell proliferation, migration, and invasion are significantly inhibited when SNORA71A is knocked down. These findings indicate the potential crucial role of SNORA71A in the malignant transformation, progression, or metastasis of CRC[54].
Other H/ACA box snoRNAs: In addition to the H/ACA box snoRNAs discussed above, there are several other H/ACA box snoRNAs that are closely associated with CRC. For instance, SNORA5C has been linked to promoting the proliferative potential of CRC cells[82], while SNORA15 and SNORA41 have been implicated in the potential oncogenic effects in CRC[83]. Besides SNOR71A, Zhang et al[54] also reported the high abundance of SNORA23 related to CRC, without function evaluation. However, the specific molecular mechanisms and biological functions of these snoRNAs require further investigation.
SNORD1C: SNORD1C is a C/D box snoRNA with a length of 78 nt, and its host gene is SNHG16. It is well-known that ALDH1+ tumor cells are typically characterized by heightened proliferation rates, enhanced self-renewal capabilities, and an increased potential for tumor formation in vivo, factors closely linked to cancer recurrence. Mannoor et al[97] found that SNORD1C exhibits elevated levels in ALDH1+ NSCLC cells in comparison to ALDH1- NSCLC cells, indicating the driving role of SNORD1C in NSCLC[97,98]. However, the precise role of SNORD1C in CRC remains a subject of ongoing investigation. Researchers have observed significant upregulation of SNORD1C in the serum of CRC patients, and this upregulation is associated with low tissue differentiation and high CEA expression. Therefore, the elevated expression of SNORD1C is closely linked to poor outcomes and prognosis in CRC patients[86]. Furthermore, subsequent studies found that knocking down SNORD1C in CRC cell lines led to reduced cell proliferation, colony formation, migration, and invasion, as well as promoting apoptosis. Besides, SNORD1C has been proved to augment cancer cell stemness within the context of CRC, acting through the Wnt/β-catenin pathway. In summary, the suppression of SNORD1C is shown to reduce Wnt/β-catenin pathway expression. These alterations collectively lead to reduced resistance to chemotherapy, ultimately hampering the progression of CRC, which strongly suggests the clinical potential of targeting this specific snoRNA in the treatment of CRC[87].
SNORD12B: SNORD12B is a type of C/D box snoRNA with a length of 91nt and it is hosted by the gene ZFAS1. To explore factors associated with tumor progression and metastasis, Xu et al[88] examined the role of noncoding RNAs in CRC and discovered 32 differentially expressed snoRNAs in metastatic and late-stage CRC tissues, with SNORD12B exhibiting significant upregulation compared to normal samples. Through correlation analysis, they found a significant correlation between SNORD12B and the expression of miRNAs and mRNAs, suggesting its potential role in regulating the expression of other RNAs. Interestingly, the upregulation of SNORD12B was positively associated with hypoxia, tumor metastasis, and upregulated genes in the pentose phosphate pathway, while it was negatively correlated with the expression of tumor suppressor genes. These findings imply that SNORD12B may be involved in diverse aspects related to the tumor occurrence, proliferation, and metastasis[88]. Additionally, its host gene, ZFAS1 is also involved in various human malignancies and associated with many aspects of carcinogenesis, including proliferation, invasion, metastasis, apoptosis, cell cycle regulation, and drug resistance. Besides, SNORD12, and SNORD12C, hosted by ZFAS1, has been also significantly overexpressed in various human malignant tumors, including CRC. This suggests the potential close association between ZFAS1 and its hosted snoRNAs, such as SNORD12B, potentially playing a pivotal role in the malignant evolution, progression, or metastasis of CRC[99].
SNORD44: SNORD44 is a C/D box snoRNA, with a length of 63 nt, located within the exon of the GAS5 gene[100]. Researchers assessed the expression levels of SNORD44 and GAS5 in CRC tissues using qRT-PCR and evaluated their correlation via Pearson correlation analysis. It is observed that both SNORD44 and GAS5 were expressed at low levels in CRC tissues compared to control samples. Additionally, a positive correlation between the expression of SNORD44 and GAS5 in tumor samples was detected. The SNORD44 did not regulate the expression of its host gene, GAS5, and vice versa. The researchers engineered an oncolytic adenovirus (SPDD-UG) to overexpress SNORD44 and GAS5, and demonstrated that SNORD44 and GAS5 exhibited anti-tumor effects in CRC cells, inhibiting cancer growth and inducing apoptosis[90]. It is indicated that SNORD44 exhibited downregulated expression in CRC tissues, and overexpressing SNORD44 inhibited the growth of CRC cells, suggesting that SNORD44 may function as a tumor suppressor in CRC and providing a promising direction for potential CRC therapeutic strategies.
SNORD57: SNORD57 is a C/D box snoRNA with a length of 72 nt, its host gene is NOP56, and its derivative piRNA piR-54265 found in the 5′ end fragment of SNORD57. It is suggested that serum piR-54265 could serve as a biomarker for early detection and clinical monitoring of CRC[101,102]. After detecting SNORD57 and its derivative piRNA piR-54265 in the serum of CRC patients, it is evident that SNORD57 exhibits a higher abundance in serum, thus rendering it a promising non-invasive biomarker for the early detection and clinical monitoring of CRC[92].
SNORD50A/B: SNORD50A, originating from the host gene SNHG5, is a C/D box snoRNA with a length of 75 nt. Similarly, SNORD50B, also derived from the host gene SNHG5, is a C/D box snoRNA with a length of 70 nt. Through a comparative analysis of tumor genomes and normal genomes, researchers have identified that the deletion frequency of SNORD50A and SNORD50B genes in various common cancers, including CRC, ranges from 10% to 40%. This deletion has been correlated with decreased survival rates in patients. It has been observed that SNORD50A and SNORD50B binds to K-Ras and inhibits K-Ras oncogenic function in various tumor types. Subsequent experiments have revealed that the absence of SNORD50A and SNORD50B in tumors augments the activities of K-Ras and its associated signaling pathway and an increased risk of tumorigenesis. These research findings suggest a potential synergistic interaction between mutations in the KRAS gene and the deletion of SNORD50A and SNORD50B in cancer[103]. Notably, KRAS plays a crucial role in the development, signaling, and proliferation of CRC. Mutations in the KRAS gene result in the permanent activation of the K-Ras protein, which is pivotal for tumor growth and heterogeneity[104]. Therefore, the loss of SNORD50A and SNORD50B may play a significant role in the pathogenesis of CRC, at least partially through interacting with KRAS gene mutations to promote tumorigenesis and progression, providing valuable insights into the mechanisms of CRC and offer new targets for potential therapeutic strategies.
SNORD12C/78: SNORD12C, arising from the host gene ZFAS1, is a C/D box snoRNA with a length of 89 nt. And SNORD78, originating from the host gene GAS5, is a C/D box snoRNA with a length of 65 nt. It is reported that SNORD12C/78 participate in a novel signaling pathway, ZFAS1-NOP58-SNORD12C/78-EIF4A3/LAMC2 axis, influencing the development and progression of CRC. This sheds light on new molecular mechanisms that underlie the regulation of CRC initiation and pathogenesis by ZFAS1. ZFAS1 assumes a critical oncogenic role in CRC, with its expression and function are indispensable for the development and maintenance of CRC cells and tissues. Mechanically, ZFAS1 recruits NOP58 as a scaffold protein to maintain the methylation function of SNORD12C and SNORD78, thereby facilitating the activities of SNORD12C and SNORD78 mediated 28S rRNA 2’-O-methylation at specific sites. This leads substantially heightened stability and translational activity of downstream target genes, such as EIF4A3, and LAMC2, ultimately fostering the proliferation of colon cancer cells. Inhibition of SNORD12C or SNORD78 results in diminished 2’-O-methylation modifications under the regulation of ZFAS1, thereby suppressing the stability and translational activity of downstream target genes. Thus, this unveils fresh possibilities for the potential application of snoRNA and cellular 2’-O-methylation-dependent translation networks in the prevention and treatment of CRC[13].
Other C/D box snoRNAs: In addition to the C/D box snoRNAs discussed above, several other C/D box snoRNAs have been show a close correlation with diverse characteristics of CRC. The elevated expression of SNORD15B[82] and SNORD126[65] has found to be associated with the proliferative potential of CRC cells. Conversely, reduced expression of SNORD33 can inhibit the anchoring function of CRC cells[83]. However, the potential roles of SNORD14E[89], SNORD17[89], SNORD48[82], SNORD67[89], SNORD76[31], and others in CRC remain unclear, requiring further investigation to uncover their precise molecular mechanisms and biological functions.
CRC is a disease characterized by a multitude of genetic and epigenetic features[105,106], with its onset attributable to various intrinsic and extrinsic factors, including the accumulation of genetic mutations, susceptibility alleles associated with family history, and chronic or sustained inflammation[107]. Based on the source of mutations, CRC can be categorized into three types, sporadic (70%), hereditary (5%), and familial (25%). The pathogenic mechanisms leading to this condition may involve three types, namely chromosomal instability, microsatellite instability, and CpG island methylator phenotype[108,109]. While the molecular mechanisms of CRC have been extensively studied, there are still many details that remain unclear.
One of the primary reasons for the poor 5-year overall survival rate in CRC is late detection, missing the window for effective treatment[17]. Early diagnosis is the critical means of reducing mortality in CRC patients, and biomarker detection plays a pivotal role in early diagnosis[78]. Recently, the emergence of liquid biopsy techniques has brought new hope for early disease diagnosis. This method allows the detection of nucleic acids, peptides, proteins, and intact tumor cells in bodily fluids. Minimally invasive techniques like liquid biopsy are preferred for diagnosing and assessing patient prognosis, significantly improving clinical management in cancer patients[110,111]. SnoRNAs, due to their location within the nucleolus, are typically unaffected by hemolysis and remain relatively stable in the bloodstream as they form complexes with specific proteins. The application of high-quality microarrays and high-throughput sequencing has unequivocally demonstrated differential snoRNA expression in clinical samples and cell lines, implying diagnostic and prognostic potential in CRC[54,92]. While mounting evidence supports the functional role of snoRNAs in CRC development and their potential as biomarkers, there remains a paucity of research on their clinical significance and functional roles in CRC progression. Overall, snoRNAs possess advantages such as non-invasiveness, high sensitivity, and specificity, rendering them ideal biomarkers for early cancer screening and diagnosis in the current context. Moreover, they may exhibit functions akin to non-invasive biomarkers, resembling miRNAs[112,113].
Currently, the treatment for CRC primarily involves surgery, adjuvant chemotherapy, radiation therapy, and immunotherapy[21]. Given their abundance and detectability in both solid tumors and blood, snoRNAs may emerge as central targets in cancer therapy[16]. Although current understanding of the molecular mechanisms of snoRNAs in tumorigenesis remains limited, some of their characteristics make them ideal candidates for therapeutic interventions[114]. For instance, studies indicate that manipulating snoRNA expression, both in vitro and in vivo, can enhance the tumorigenic and invasive properties of CRC, suggesting that targeting snoRNA expression represents a novel approach for CRC prevention and treatment[31,84]. Furthermore, snoRNAs involved in transcriptional gene silencing pathways may offer high therapeutic benefits[115]. Moreover, certain snoRNAs seem to have the potential for protein binding or dependence on secondary structures, providing a novel means of therapeutic intervention[116], while oncolytic adenovirus vectors have gained extensive use in cancer therapy[117]. The vectors exhibit selective infectivity towards tumor cells, leading to oncolysis while causing minimal toxicity to normal tissues[118,119]. Some investigations have explored the deployment of oncolytic adenovirus vectors for delivering snoRNAs and their host genes for cancer treatment, demonstrating selective replicability and potential cytotoxicity in cancer cells[90].
Nonetheless, the primary obstacle to implementing snoRNA-based therapeutic strategies in cancer patients primarily arises from a dearth of understanding regarding their functional roles in tumorigenesis and their specific downstream gene targets or pathways. As technological limitations are surmounted, snoRNAs may emerge as potential targets for treatment. While this field appears to be in its nascent stages, it should be currently witnessing the potential of snoRNAs in cancer therapy. In the near future, it is useful to harness their potential successfully.
An increasing body of research on snoRNAs in CRC has expanded the understanding of their molecular mechanisms and biological processes[2]. Specifically, the dysregulation of snoRNA function appears to be closely associated with the onset and development of CRC[31]. However, the functional roles of the majority of snoRNAs in CRC remain uncertain. Their roles in cell signaling pathways, molecular mechanisms, and regulation, require further clarification, and more research is needed to delve into the snoRNAs present in CRC and their respective targets. As the knowledge of novel snoRNA functions deepens, research based on snoRNAs has the potential to reshape the landscape of tumor biology and genetics, possibly uncovering new pathways that drive CRC.
In summary, the study of snoRNAs in the context of CRC is a continually expanding field, and future discoveries may provide deeper insights into the disease, utilizing snoRNAs as therapeutic targets, as well as diagnostic and prognostic markers. Current literature underscores the role of snoRNA dysregulation in promoting CRC, emphasizing the significance of snoRNA regulation in CRC biology and clinical outcomes. Therefore, understanding the roles of snoRNAs may enhance our comprehension of CRC and potentially harness the robust potential of snoRNAs in the clinical diagnosis and treatment of CRC in the future.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Teramoto-Matsubara OT, Mexico S-Editor: Chen YL L-Editor: A P-Editor: Yu HG
| 1. | Maxwell ES, Fournier MJ. The small nucleolar RNAs. Annu Rev Biochem. 1995;64:897-934. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 413] [Cited by in RCA: 454] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 2. | Zhang X, Wang C, Xia S, Xiao F, Peng J, Gao Y, Yu F, Chen X. The emerging role of snoRNAs in human disease. Genes Dis. 2023;10:2064-2081. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 28] [Reference Citation Analysis (0)] |
| 3. | Weinberg RA, Penman S. Small molecular weight monodisperse nuclear RNA. J Mol Biol. 1968;38:289-304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 394] [Cited by in RCA: 400] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 4. | Zieve G, Penman S. Small RNA species of the HeLa cell: metabolism and subcellular localization. Cell. 1976;8:19-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 301] [Cited by in RCA: 326] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 5. | Kedersha NL, Rome LH. Isolation and characterization of a novel ribonucleoprotein particle: large structures contain a single species of small RNA. J Cell Biol. 1986;103:699-709. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 227] [Cited by in RCA: 258] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 6. | Lübben B, Marshallsay C, Rottmann N, Lührmann R. Isolation of U3 snoRNP from CHO cells: a novel 55 kDa protein binds to the central part of U3 snoRNA. Nucleic Acids Res. 1993;21:5377-5385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 51] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 7. | Tollervey D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 1987;6:4169-4175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 184] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 8. | Ni J, Tien AL, Fournier MJ. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565-573. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 366] [Cited by in RCA: 394] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 9. | Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol. 2007;8:209-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 653] [Cited by in RCA: 590] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 10. | Chang LS, Lin SY, Lieu AS, Wu TL. Differential expression of human 5S snoRNA genes. Biochem Biophys Res Commun. 2002;299:196-200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 43] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 11. | Schulten HJ, Bangash M, Karim S, Dallol A, Hussein D, Merdad A, Al-Thoubaity FK, Al-Maghrabi J, Jamal A, Al-Ghamdi F, Choudhry H, Baeesa SS, Chaudhary AG, Al-Qahtani MH. Comprehensive molecular biomarker identification in breast cancer brain metastases. J Transl Med. 2017;15:269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 69] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 12. | Liao J, Yu L, Mei Y, Guarnera M, Shen J, Li R, Liu Z, Jiang F. Small nucleolar RNA signatures as biomarkers for non-small-cell lung cancer. Mol Cancer. 2010;9:198. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 190] [Cited by in RCA: 216] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 13. | Wu H, Qin W, Lu S, Wang X, Zhang J, Sun T, Hu X, Li Y, Chen Q, Wang Y, Zhao H, Piao H, Zhang R, Wei M. Long noncoding RNA ZFAS1 promoting small nucleolar RNA-mediated 2'-O-methylation via NOP58 recruitment in colorectal cancer. Mol Cancer. 2020;19:95. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 96] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 14. | Deogharia M, Majumder M. Guide snoRNAs: Drivers or Passengers in Human Disease? Biology (Basel). 2018;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 15. | Dsouza VL, Adiga D, Sriharikrishnaa S, Suresh PS, Chatterjee A, Kabekkodu SP. Small nucleolar RNA and its potential role in breast cancer - A comprehensive review. Biochim Biophys Acta Rev Cancer. 2021;1875:188501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 16. | Toden S, Zumwalt TJ, Goel A. Non-coding RNAs and potential therapeutic targeting in cancer. Biochim Biophys Acta Rev Cancer. 2021;1875:188491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 199] [Article Influence: 49.8] [Reference Citation Analysis (0)] |
| 17. | Morgan E, Arnold M, Gini A, Lorenzoni V, Cabasag CJ, Laversanne M, Vignat J, Ferlay J, Murphy N, Bray F. Global burden of colorectal cancer in 2020 and 2040: incidence and mortality estimates from GLOBOCAN. Gut. 2023;72:338-344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 511] [Cited by in RCA: 923] [Article Influence: 461.5] [Reference Citation Analysis (1)] |
| 18. | Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 9950] [Article Influence: 4975.0] [Reference Citation Analysis (2)] |
| 19. | Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394:1467-1480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1570] [Cited by in RCA: 3025] [Article Influence: 504.2] [Reference Citation Analysis (3)] |
| 20. | Miller KD, Nogueira L, Devasia T, Mariotto AB, Yabroff KR, Jemal A, Kramer J, Siegel RL. Cancer treatment and survivorship statistics, 2022. CA Cancer J Clin. 2022;72:409-436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 621] [Cited by in RCA: 1556] [Article Influence: 518.7] [Reference Citation Analysis (0)] |
| 21. | Biller LH, Schrag D. Diagnosis and Treatment of Metastatic Colorectal Cancer: A Review. JAMA. 2021;325:669-685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 1440] [Article Influence: 360.0] [Reference Citation Analysis (0)] |
| 22. | Vatandoust S, Price TJ, Karapetis CS. Colorectal cancer: Metastases to a single organ. World J Gastroenterol. 2015;21:11767-11776. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 232] [Cited by in RCA: 232] [Article Influence: 23.2] [Reference Citation Analysis (2)] |
| 23. | Beniwal SS, Lamo P, Kaushik A, Lorenzo-Villegas DL, Liu Y, MohanaSundaram A. Current Status and Emerging Trends in Colorectal Cancer Screening and Diagnostics. Biosensors (Basel). 2023;13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 22] [Reference Citation Analysis (0)] |
| 24. | Yao T, Sun Q, Xiong K, Su Y, Zhao Q, Zhang C, Zhang L, Li X, Fang H. Optimization of screening strategies for colorectal cancer based on fecal DNA and occult blood testing. Eur J Public Health. 2023;33:336-341. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Reference Citation Analysis (0)] |
| 25. | Chan SCH, Liang JQ. Advances in tests for colorectal cancer screening and diagnosis. Expert Rev Mol Diagn. 2022;22:449-460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 45] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 26. | Ferrari A, Neefs I, Hoeck S, Peeters M, Van Hal G. Towards Novel Non-Invasive Colorectal Cancer Screening Methods: A Comprehensive Review. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 27. | Issa IA, Noureddine M. Colorectal cancer screening: An updated review of the available options. World J Gastroenterol. 2017;23:5086-5096. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 434] [Cited by in RCA: 392] [Article Influence: 49.0] [Reference Citation Analysis (11)] |
| 28. | Binefa G, Rodríguez-Moranta F, Teule A, Medina-Hayas M. Colorectal cancer: from prevention to personalized medicine. World J Gastroenterol. 2014;20:6786-6808. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 245] [Cited by in RCA: 256] [Article Influence: 23.3] [Reference Citation Analysis (3)] |
| 29. | Wang QQ, Zhou L, Qin G, Tan C, Zhou YC, Yao SK. Leukocyte immunoglobulin-like receptor B2 overexpression as a promising therapeutic target and noninvasive screening biomarker for colorectal cancer. World J Gastroenterol. 2023;29:5313-5326. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 3] [Cited by in RCA: 4] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 30. | Li Y, Li B, Jiang R, Liao L, Zheng C, Yuan J, Zeng L, Hu K, Zhang Y, Mei W, Hong Z, Xiao B, Kong L, Han K, Tang J, Jiang W, Pan Z, Zhang S, Ding P. A novel screening method of DNA methylation biomarkers helps to improve the detection of colorectal cancer and precancerous lesions. Cancer Med. 2023;12:20626-20638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 31. | Okugawa Y, Toiyama Y, Toden S, Mitoma H, Nagasaka T, Tanaka K, Inoue Y, Kusunoki M, Boland CR, Goel A. Clinical significance of SNORA42 as an oncogene and a prognostic biomarker in colorectal cancer. Gut. 2017;66:107-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 108] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 32. | Weng M, Wu D, Yang C, Peng H, Wang G, Wang T, Li X. Noncoding RNAs in the development, diagnosis, and prognosis of colorectal cancer. Transl Res. 2017;181:108-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 73] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 33. | Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861-874. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3151] [Cited by in RCA: 3684] [Article Influence: 263.1] [Reference Citation Analysis (0)] |
| 34. | Balakin AG, Smith L, Fournier MJ. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823-834. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 307] [Cited by in RCA: 352] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 35. | Henras AK, Dez C, Henry Y. RNA structure and function in C/D and H/ACA s(no)RNPs. Curr Opin Struct Biol. 2004;14:335-343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 101] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 36. | Khoshnevis S, Dreggors RE, Hoffmann TFR, Ghalei H. A conserved Bcd1 interaction essential for box C/D snoRNP biogenesis. J Biol Chem. 2019;294:18360-18371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 37. | Paul A, Tiotiu D, Bragantini B, Marty H, Charpentier B, Massenet S, Labialle S. Bcd1p controls RNA loading of the core protein Nop58 during C/D box snoRNP biogenesis. RNA. 2019;25:496-506. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 38. | Filipowicz W, Pogacić V. Biogenesis of small nucleolar ribonucleoproteins. Curr Opin Cell Biol. 2002;14:319-327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 306] [Cited by in RCA: 299] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 39. | Watkins NJ, Bohnsack MT. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA. 2012;3:397-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 331] [Cited by in RCA: 358] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 40. | Samarsky DA, Fournier MJ, Singer RH, Bertrand E. The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J. 1998;17:3747-3757. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 160] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 41. | Kiss-László Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077-1088. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 577] [Cited by in RCA: 641] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 42. | Bachellerie JP, Michot B, Nicoloso M, Balakin A, Ni J, Fournier MJ. Antisense snoRNAs: a family of nucleolar RNAs with long complementarities to rRNA. Trends Biochem Sci. 1995;20:261-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 125] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 43. | Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 575] [Cited by in RCA: 628] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 44. | Watkins NJ, Gottschalk A, Neubauer G, Kastner B, Fabrizio P, Mann M, Lührmann R. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA. 1998;4:1549-1568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 173] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 45. | Hamma T, Reichow SL, Varani G, Ferré-D'Amaré AR. The Cbf5-Nop10 complex is a molecular bracket that organizes box H/ACA RNPs. Nat Struct Mol Biol. 2005;12:1101-1107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 89] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 46. | Yu YT, Meier UT. RNA-guided isomerization of uridine to pseudouridine--pseudouridylation. RNA Biol. 2014;11:1483-1494. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 103] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 47. | Ganot P, Bortolin ML, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799-809. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 438] [Cited by in RCA: 477] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 48. | Kiss T, Fayet-Lebaron E, Jády BE. Box H/ACA small ribonucleoproteins. Mol Cell. 2010;37:597-606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 191] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 49. | Pogacić V, Dragon F, Filipowicz W. Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol Cell Biol. 2000;20:9028-9040. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 174] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 50. | MacNeil DE, Bensoussan HJ, Autexier C. Telomerase Regulation from Beginning to the End. Genes (Basel). 2016;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 62] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 51. | Deryusheva S, Gall JG. scaRNAs and snoRNAs: Are they limited to specific classes of substrate RNAs? RNA. 2019;25:17-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 52. | Marnef A, Richard P, Pinzón N, Kiss T. Targeting vertebrate intron-encoded box C/D 2'-O-methylation guide RNAs into the Cajal body. Nucleic Acids Res. 2014;42:6616-6629. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 44] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 53. | Zhang H, Liu X, Zhang W, Deng J, Lin C, Qi Z, Li Y, Gu Y, Wang Q, Shen L, Wang Z. Oncogene SCARNA12 as a potential diagnostic biomarker for colorectal cancer. Mol Biomed. 2023;4:37. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 54. | Zhang Z, Tao Y, Hua Q, Cai J, Ye X, Li H. SNORA71A Promotes Colorectal Cancer Cell Proliferation, Migration, and Invasion. Biomed Res Int. 2020;2020:8284576. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 55. | Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999;18:5399-5410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 447] [Cited by in RCA: 494] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 56. | Huang C, Shi J, Guo Y, Huang W, Huang S, Ming S, Wu X, Zhang R, Ding J, Zhao W, Jia J, Huang X, Xiang AP, Shi Y, Yao C. A snoRNA modulates mRNA 3' end processing and regulates the expression of a subset of mRNAs. Nucleic Acids Res. 2017;45:8647-8660. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 79] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 57. | Li S, Yang Q, Zhou Z, Fu M, Yang X, Hao K, Liu Y. SNHG3 cooperates with ELAVL2 to modulate cell apoptosis and extracellular matrix accumulation by stabilizing SNAI2 in human trabecular meshwork cells under oxidative stress. Environ Toxicol. 2021;36:1070-1079. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 58. | Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem Sci. 2002;27:344-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 471] [Cited by in RCA: 495] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 59. | Zhong F, Zhou N, Wu K, Guo Y, Tan W, Zhang H, Zhang X, Geng G, Pan T, Luo H, Zhang Y, Xu Z, Liu J, Liu B, Gao W, Liu C, Ren L, Li J, Zhou J. A SnoRNA-derived piRNA interacts with human interleukin-4 pre-mRNA and induces its decay in nuclear exosomes. Nucleic Acids Res. 2015;43:10474-10491. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 78] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 60. | Scott MS, Ono M. From snoRNA to miRNA: Dual function regulatory non-coding RNAs. Biochimie. 2011;93:1987-1992. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 178] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 61. | Brameier M, Herwig A, Reinhardt R, Walter L, Gruber J. Human box C/D snoRNAs with miRNA like functions: expanding the range of regulatory RNAs. Nucleic Acids Res. 2011;39:675-686. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 218] [Cited by in RCA: 243] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 62. | Bergeron D, Laforest C, Carpentier S, Calvé A, Fafard-Couture É, Deschamps-Francoeur G, Scott MS. SnoRNA copy regulation affects family size, genomic location and family abundance levels. BMC Genomics. 2021;22:414. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 63. | Bratkovič T, Božič J, Rogelj B. Functional diversity of small nucleolar RNAs. Nucleic Acids Res. 2020;48:1627-1651. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 160] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 64. | Bratkovič T, Rogelj B. The many faces of small nucleolar RNAs. Biochim Biophys Acta. 2014;1839:438-443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 68] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 65. | Fang X, Yang D, Luo H, Wu S, Dong W, Xiao J, Yuan S, Ni A, Zhang KJ, Liu XY, Chu L. SNORD126 promotes HCC and CRC cell growth by activating the PI3K-AKT pathway through FGFR2. J Mol Cell Biol. 2017;9:243-255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 66. | Sahoo T, del Gaudio D, German JR, Shinawi M, Peters SU, Person RE, Garnica A, Cheung SW, Beaudet AL. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat Genet. 2008;40:719-721. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 495] [Cited by in RCA: 431] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 67. | Gallagher RC, Pils B, Albalwi M, Francke U. Evidence for the role of PWCR1/HBII-85 C/D box small nucleolar RNAs in Prader-Willi syndrome. Am J Hum Genet. 2002;71:669-678. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 88] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 68. | O'Brien JE Jr, Kibiryeva N, Zhou XG, Marshall JA, Lofland GK, Artman M, Chen J, Bittel DC. Noncoding RNA expression in myocardium from infants with tetralogy of Fallot. Circ Cardiovasc Genet. 2012;5:279-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 94] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 69. | Lester E, Ooi FK, Bakkar N, Ayers J, Woerman AL, Wheeler J, Bowser R, Carlson GA, Prusiner SB, Parker R. Tau aggregates are RNA-protein assemblies that mislocalize multiple nuclear speckle components. Neuron. 2021;109:1675-1691.e9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 132] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 70. | Romano G, Veneziano D, Acunzo M, Croce CM. Small non-coding RNA and cancer. Carcinogenesis. 2017;38:485-491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 346] [Cited by in RCA: 324] [Article Influence: 40.5] [Reference Citation Analysis (0)] |
| 71. | Tycowski KT, Shu MD, Steitz JA. A mammalian gene with introns instead of exons generating stable RNA products. Nature. 1996;379:464-466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 230] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 72. | Xu G, Yang F, Ding CL, Zhao LJ, Ren H, Zhao P, Wang W, Qi ZT. Small nucleolar RNA 113-1 suppresses tumorigenesis in hepatocellular carcinoma. Mol Cancer. 2014;13:216. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 91] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 73. | Mei YP, Liao JP, Shen J, Yu L, Liu BL, Liu L, Li RY, Ji L, Dorsey SG, Jiang ZR, Katz RL, Wang JY, Jiang F. Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene. 2012;31:2794-2804. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 186] [Cited by in RCA: 214] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 74. | Su H, Xu T, Ganapathy S, Shadfan M, Long M, Huang TH, Thompson I, Yuan ZM. Elevated snoRNA biogenesis is essential in breast cancer. Oncogene. 2014;33:1348-1358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 153] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 75. | Wang B, Zhao Y, Li Y, Xu Y, Chen Y, Jiang Q, Yao D, Zhang L, Hu X, Fu C, Zhang S, Chen S. A plasma SNORD33 signature predicts platinum benefit in metastatic triple-negative breast cancer patients. Mol Cancer. 2022;21:22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 76. | Li C, Wu L, Liu P, Li K, Zhang Z, He Y, Liu Q, Jiang P, Yang Z, Liu Z, Yuan Y, Chang L. The C/D box small nucleolar RNA SNORD52 regulated by Upf1 facilitates Hepatocarcinogenesis by stabilizing CDK1. Theranostics. 2020;10:9348-9363. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 77. | Zhuo Y, Li S, Hu W, Zhang Y, Shi Y, Zhang F, Zhang J, Wang J, Liao M, Chen J, Qian H, Li D, Sun C. Targeting SNORA38B attenuates tumorigenesis and sensitizes immune checkpoint blockade in non-small cell lung cancer by remodeling the tumor microenvironment via regulation of GAB2/AKT/mTOR signaling pathway. J Immunother Cancer. 2022;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 33] [Article Influence: 11.0] [Reference Citation Analysis (1)] |
| 78. | Welch HG, Robertson DJ. Colorectal Cancer on the Decline--Why Screening Can't Explain It All. N Engl J Med. 2016;374:1605-1607. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 151] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 79. | Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011;6:479-507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1163] [Cited by in RCA: 1271] [Article Influence: 90.8] [Reference Citation Analysis (1)] |
| 80. | Jiang MC, Ni JJ, Cui WY, Wang BY, Zhuo W. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am J Cancer Res. 2019;9:1354-1366. [PubMed] |
| 81. | Liang J, Wen J, Huang Z, Chen XP, Zhang BX, Chu L. Small Nucleolar RNAs: Insight Into Their Function in Cancer. Front Oncol. 2019;9:587. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 187] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 82. | Shen L, Lu W, Huang Y, He J, Wang Q, Zheng X, Wang Z. SNORD15B and SNORA5C: Novel Diagnostic and Prognostic Biomarkers for Colorectal Cancer. Biomed Res Int. 2022;2022:8260800. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 83. | Yang X, Li Y, Li L, Liu J, Wu M, Ye M. SnoRNAs are involved in the progression of ulcerative colitis and colorectal cancer. Dig Liver Dis. 2017;49:545-551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 84. | Yoshida K, Toden S, Weng W, Shigeyasu K, Miyoshi J, Turner J, Nagasaka T, Ma Y, Takayama T, Fujiwara T, Goel A. SNORA21 - An Oncogenic Small Nucleolar RNA, with a Prognostic Biomarker Potential in Human Colorectal Cancer. EBioMedicine. 2017;22:68-77. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 64] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 85. | Shen L, Lin C, Lu W, He J, Wang Q, Huang Y, Zheng X, Wang Z. Involvement of the oncogenic small nucleolar RNA SNORA24 in regulation of p53 stability in colorectal cancer. Cell Biol Toxicol. 2023;39:1377-1394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 86. | Liu Y, Zhao C, Sun J, Wang G, Ju S, Qian C, Wang X. Overexpression of small nucleolar RNA SNORD1C is associated with unfavorable outcome in colorectal cancer. Bioengineered. 2021;12:8943-8952. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 87. | Liu Y, Zhao C, Wang G, Chen J, Ju S, Huang J, Wang X. SNORD1C maintains stemness and 5-FU resistance by activation of Wnt signaling pathway in colorectal cancer. Cell Death Discov. 2022;8:200. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 23] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 88. | Xu L, Ziegelbauer J, Wang R, Wu WW, Shen RF, Juhl H, Zhang Y, Rosenberg A. Distinct Profiles for Mitochondrial t-RNAs and Small Nucleolar RNAs in Locally Invasive and Metastatic Colorectal Cancer. Clin Cancer Res. 2016;22:773-784. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 89. | Huang L, Liang XZ, Deng Y, Liang YB, Zhu X, Liang XY, Luo DZ, Chen G, Fang YY, Lan HH, Zeng JH. Prognostic value of small nucleolar RNAs (snoRNAs) for colon adenocarcinoma based on RNA sequencing data. Pathol Res Pract. 2020;216:152937. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 90. | Yuan S, Wu Y, Wang Y, Chen J, Chu L. An Oncolytic Adenovirus Expressing SNORD44 and GAS5 Exhibits Antitumor Effect in Colorectal Cancer Cells. Hum Gene Ther. 2017;28:690-700. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 91. | Siprashvili Z, Webster DE, Johnston D, Shenoy RM, Ungewickell AJ, Bhaduri A, Flockhart R, Zarnegar BJ, Che Y, Meschi F, Puglisi JD, Khavari PA. The noncoding RNAs SNORD50A and SNORD50B bind K-Ras and are recurrently deleted in human cancer. Nat Genet. 2016;48:53-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 141] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 92. | Tosar JP, García-Silva MR, Cayota A. Circulating SNORD57 rather than piR-54265 is a promising biomarker for colorectal cancer: common pitfalls in the study of somatic piRNAs in cancer. RNA. 2021;27:403-410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 93. | Gao L, Ma J, Mannoor K, Guarnera MA, Shetty A, Zhan M, Xing L, Stass SA, Jiang F. Genome-wide small nucleolar RNA expression analysis of lung cancer by next-generation deep sequencing. Int J Cancer. 2015;136:E623-E629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 72] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 94. | McMahon M, Contreras A, Holm M, Uechi T, Forester CM, Pang X, Jackson C, Calvert ME, Chen B, Quigley DA, Luk JM, Kelley RK, Gordan JD, Gill RM, Blanchard SC, Ruggero D. A single H/ACA small nucleolar RNA mediates tumor suppression downstream of oncogenic RAS. Elife. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 109] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 95. | Zhang D, Zhou J, Gao J, Wu RY, Huang YL, Jin QW, Chen JS, Tang WZ, Yan LH. Targeting snoRNAs as an emerging method of therapeutic development for cancer. Am J Cancer Res. 2019;9:1504-1516. [PubMed] |
| 96. | Bian Z, Zhou M, Cui K, Yang F, Cao Y, Sun S, Liu B, Gong L, Li J, Wang X, Li C, Yao S, Yin Y, Huang S, Fei B, Huang Z. SNHG17 promotes colorectal tumorigenesis and metastasis via regulating Trim23-PES1 axis and miR-339-5p-FOSL2-SNHG17 positive feedback loop. J Exp Clin Cancer Res. 2021;40:360. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 38] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 97. | Mannoor K, Shen J, Liao J, Liu Z, Jiang F. Small nucleolar RNA signatures of lung tumor-initiating cells. Mol Cancer. 2014;13:104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 76] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 98. | Jiang F, Qiu Q, Khanna A, Todd NW, Deepak J, Xing L, Wang H, Liu Z, Su Y, Stass SA, Katz RL. Aldehyde dehydrogenase 1 is a tumor stem cell-associated marker in lung cancer. Mol Cancer Res. 2009;7:330-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 598] [Cited by in RCA: 622] [Article Influence: 38.9] [Reference Citation Analysis (0)] |
| 99. | Jiang X, Yang Z, Li Z. Zinc finger antisense 1: A long noncoding RNA with complex roles in human cancers. Gene. 2019;688:26-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 100. | Bai B, Yegnasubramanian S, Wheelan SJ, Laiho M. RNA-Seq of the nucleolus reveals abundant SNORD44-derived small RNAs. PLoS One. 2014;9:e107519. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 101. | Mai D, Ding P, Tan L, Zhang J, Pan Z, Bai R, Li C, Li M, Zhou Y, Tan W, Zhou Z, Li Y, Zhou A, Ye Y, Pan L, Zheng Y, Su J, Zuo Z, Liu Z, Zhao Q, Li X, Huang X, Li W, Wu S, Jia W, Zou S, Wu C, Xu RH, Zheng J, Lin D. PIWI-interacting RNA-54265 is oncogenic and a potential therapeutic target in colorectal adenocarcinoma. Theranostics. 2018;8:5213-5230. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 126] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 102. | Mai D, Zheng Y, Guo H, Ding P, Bai R, Li M, Ye Y, Zhang J, Huang X, Liu D, Sui Q, Pan L, Su J, Deng J, Wu G, Li R, Deng S, Bai Y, Ligu Y, Tan W, Wu C, Wu T, Zheng J, Lin D. Serum piRNA-54265 is a New Biomarker for early detection and clinical surveillance of Human Colorectal Cancer. Theranostics. 2020;10:8468-8478. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 64] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 103. | Su X, Feng C, Wang S, Shi L, Gu Q, Zhang H, Lan X, Zhao Y, Qiang W, Ji M, Hou P. The noncoding RNAs SNORD50A and SNORD50B-mediated TRIM21-GMPS interaction promotes the growth of p53 wild-type breast cancers by degrading p53. Cell Death Differ. 2021;28:2450-2464. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 35] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 104. | Krens LL, Baas JM, Gelderblom H, Guchelaar HJ. Therapeutic modulation of k-ras signaling in colorectal cancer. Drug Discov Today. 2010;15:502-516. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 30] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 105. | Jung G, Hernández-Illán E, Moreira L, Balaguer F, Goel A. Epigenetics of colorectal cancer: biomarker and therapeutic potential. Nat Rev Gastroenterol Hepatol. 2020;17:111-130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 542] [Cited by in RCA: 522] [Article Influence: 104.4] [Reference Citation Analysis (0)] |
| 106. | Puccini A, Berger MD, Naseem M, Tokunaga R, Battaglin F, Cao S, Hanna DL, McSkane M, Soni S, Zhang W, Lenz HJ. Colorectal cancer: epigenetic alterations and their clinical implications. Biochim Biophys Acta Rev Cancer. 2017;1868:439-448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 45] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 107. | Lichtenstern CR, Ngu RK, Shalapour S, Karin M. Immunotherapy, Inflammation and Colorectal Cancer. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 202] [Article Influence: 40.4] [Reference Citation Analysis (0)] |
| 108. | Dienstmann R, Vermeulen L, Guinney J, Kopetz S, Tejpar S, Tabernero J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat Rev Cancer. 2017;17:79-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 491] [Cited by in RCA: 595] [Article Influence: 74.4] [Reference Citation Analysis (0)] |
| 109. | Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW, Kane MF, Kolodner RD, Vogelstein B, Kunkel TA, Baylin SB. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A. 1998;95:6870-6875. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1343] [Cited by in RCA: 1373] [Article Influence: 50.9] [Reference Citation Analysis (0)] |
| 110. | Vacante M, Ciuni R, Basile F, Biondi A. The Liquid Biopsy in the Management of Colorectal Cancer: An Overview. Biomedicines. 2020;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 111. | Marcuello M, Vymetalkova V, Neves RPL, Duran-Sanchon S, Vedeld HM, Tham E, van Dalum G, Flügen G, Garcia-Barberan V, Fijneman RJ, Castells A, Vodicka P, Lind GE, Stoecklein NH, Heitzer E, Gironella M. Circulating biomarkers for early detection and clinical management of colorectal cancer. Mol Aspects Med. 2019;69:107-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 194] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 112. | Zhang Z, Zhang J, Diao L, Han L. Small non-coding RNAs in human cancer: function, clinical utility, and characterization. Oncogene. 2021;40:1570-1577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 45] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 113. | Zygulska AL, Pierzchalski P. Novel Diagnostic Biomarkers in Colorectal Cancer. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 135] [Article Influence: 45.0] [Reference Citation Analysis (0)] |
| 114. | Abel Y, Rederstorff M. SnoRNAs and the emerging class of sdRNAs: Multifaceted players in oncogenesis. Biochimie. 2019;164:17-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 115. | Mannoor K, Liao J, Jiang F. Small nucleolar RNAs in cancer. Biochim Biophys Acta. 2012;1826:121-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 98] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 116. | Li Q, Xie B, Chen X, Lu B, Chen S, Sheng X, Zhao Y. SNORD6 promotes cervical cancer progression by accelerating E6-mediated p53 degradation. Cell Death Discov. 2023;9:192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 117. | Toth K, Doronin K, Tollefson AE, Wold WS. A multitasking oncolytic adenovirus vector. Mol Ther. 2003;7:435-437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 118. | Xu HN, Huang WD, Cai Y, Ding M, Gu JF, Wei N, Sun LY, Cao X, Li HG, Zhang KJ, Liu XR, Liu XY. HCCS1-armed, quadruple-regulated oncolytic adenovirus specific for liver cancer as a cancer targeting gene-viro-therapy strategy. Mol Cancer. 2011;10:133. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 20] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 119. | Zhao L, Gu J, Dong A, Zhang Y, Zhong L, He L, Wang Y, Zhang J, Zhang Z, Huiwang J, Qian Q, Qian C, Liu X. Potent antitumor activity of oncolytic adenovirus expressing mda-7/IL-24 for colorectal cancer. Hum Gene Ther. 2005;16:845-858. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 101] [Article Influence: 5.1] [Reference Citation Analysis (0)] |