Published online Apr 28, 2024. doi: 10.3748/wjg.v30.i16.2195
Revised: March 5, 2024
Accepted: April 10, 2024
Published online: April 28, 2024
Processing time: 91 Days and 7.1 Hours
As a highly invasive malignancy, esophageal cancer (EC) is a global health issue, and was the eighth most prevalent cancer and the sixth leading cause of cancer-related death worldwide in 2020. Due to its highly immunogenic nature, emer-ging immunotherapy approaches, such as immune checkpoint blockade, have demonstrated promising efficacy in treating EC; however, certain limitations and challenges still exist. In addition, tumors may exhibit primary or acquired resistance to immunotherapy in the tumor immune microenvironment (TIME); thus, understanding the TIME is urgent and crucial, especially given the im-portance of an immunosuppressive microenvironment in tumor progression. The aim of this review was to better elucidate the mechanisms of the suppressive TIME, including cell infiltration, immune cell subsets, cytokines and signaling pathways in the tumor microenvironment of EC patients, as well as the downregulated expression of major histocompatibility complex molecules in tumor cells, to obtain a better understanding of the differences in EC patient responses to immunotherapeutic strategies and accurately predict the efficacy of immunotherapies. Therefore, personalized treatments could be developed to maximize the advantages of immunotherapy.
Core Tip: Esophageal cancer (EC) is a significant global health issue, and immunotherapy holds promise for treating this disease. However, resistance to immunotherapy may occur, and is usually associated with the tumor immune microenvironment (TIME). Understanding the TIME, especially the suppressive TIME, is crucial. The aim of this review is to elucidate the underlying mechanisms of the suppressive TIME in EC, including cell infiltration, immune cell subsets, cytokines and signaling pathways, as well as the downregulated expression of major histocompatibility complex molecules in tumor cells. This summary may help predict EC patient responses to immunotherapies and facilitate personalized treatments to optimize immunotherapeutic outcomes.
- Citation: Zhang XJ, Yu Y, Zhao HP, Guo L, Dai K, Lv J. Mechanisms of tumor immunosuppressive microenvironment formation in esophageal cancer. World J Gastroenterol 2024; 30(16): 2195-2208
- URL: https://www.wjgnet.com/1007-9327/full/v30/i16/2195.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i16.2195
According to the Global Cancer Statistics 2020 database (https://gco.iarc.fr/), approximately 20000000 people are diagnosed with cancer each year, and approximately 10000000 people die from cancer worldwide[1]. Esophageal cancer (EC) accounts for 3.1% of all new cancer cases and ranks eighth in incidence among all cancer types; however, EC accounts for 5.5% of all cancer-related deaths and ranks sixth in mortality[1]. There are two main histological types of EC: Esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC)[2]. Although nearly 90% of EC cases are ESCC, the incidence and mortality rates of EAC are gradually increasing and even surpass those of ESCC in some regions of North America and Europe[3,4]. The main risk factors for EAC include gastroesophageal reflux disease and obesity, and ESCC is associated with chemical carcinogen exposure, cigarette smoking and alcohol consumption, a diet with low amounts of fruits or vegetables, high consumption of pickled vegetables or processed meat, hot drinks, etc[3,5]. The five-year survival rate of patients with EC is usually between 20% and 30%, and mainly depends on the tumor stage at initial diagnosis and the therapeutic strategy, such as surgery combined with neoadjuvant therapy (radiotherapy and chemotherapy)[6-8]. Since conventional treatments have limited efficacy and potential adverse effects, more effective therapeutic strategies are urgently needed to improve the prognosis of patients with EC[9].
Cancer development is closely related to the accumulation of gene mutations, and researchers have focused on changes in cancer cells for quite a long time[10]. Recently, the tumor microenvironment (TME) has become a hot topic, and the regulation of immune cells in the TME has drawn much attention[11]. The immune system can recognize and eliminate tumor cells expressing specific antigens, a process known as cancer immunosurveillance[12]; while, cancer cells can escape or suppress attacks from the immune system by various mechanisms, including decreasing antigen presentation, upregulating the expression of apoptotic inhibitors, increasing the expression of inhibitory molecules on the cell surface, and enhancing the secretion of certain cytokines or recruitment of regulatory cells to create an immunosuppressive microenvironment[12]. As an important component of the TME, the tumor immune microenvironment (TIME) refers to the microenvironment involving interactions between host immune agents and tumor cells[13,14]; tumors may confront host immune systems by gradually forming immunosuppressive conditions, and the presence of protumor and antitumor factors in the TIME may determine cancer progression and response to treatments[14-16]. Therefore, a comprehensive understanding of the interactions between tumor cells and various immune cells or other immune components in the TIME is vital for further elucidating the mechanisms of EC immunotherapy[17-20].
In this review, we mainly summarize the mechanisms of immunosuppression in the TIME of EC, including immune cells, immune checkpoints, immunosuppressive cells and tumor cell-related immunosuppressive factors, to provide evidence for the maintenance of an immune-activated state in the TIME of EC, with the goal of improving immunotherapeutic efficacy.
Esophageal epithelial tumor cells are the main constituents of EC and express tumor-associated antigens (TAAs)[19]. TAAs are a class of overexpressed molecules that are present mainly on the membrane of tumor cells, and are usually expressed at lower levels or undetected in normal cells[21]. T lymphocytes may recognize and bind the TAA peptides presented by major histocompatibility complex (MHC) molecules on tumor cells through the T-cell receptor, thereby initiating an immune response and triggering an attack on tumor cells[21]. In addition, natural killer (NK) lymphocytes and B lymphocytes play important roles in the regulation of immunoreactivity in EC[22,23]. For example, as a class of TAAs associated with 276 genes in more than 70 gene families, the antigen families formed by cancer-testis antigens (CTAs) are expressed mainly in ovarian granulosa cells and testicular germ cells, and are barely expressed in normal tissues[24-26]. Certain CTAs, such as New York ESCC 1 (NY-ESO-1) and melanoma-associated antigen-A (MAGE-A), have been reported to be highly expressed in EC, and specific immune responses targeting MAGE-A and NY-ESO-1 have been observed in EC patients[27-30]. MAGE-A3-specific CD8+ T cells may kill HLA-A2+/MAGE-A3+ tumor cells in ESCC patients, and functional MAGE-C2-specific CD8+ T cells may independently affect the prognosis of EC patients[27,31].
Since EC cells possess high immunogenicity, partially because of the presence of numerous antigens, these molecules could be potential targets for immunotherapy, and immunotherapy has been shown to be more effective in EC patients with an immuno-activated TME, leading to an improved prognosis[32]. However, current immunotherapeutic strategies have several limitations, e.g., accompanying adverse effects and drug resistance cannot be avoided[33]. Therefore, a comprehensive understanding of the underlying mechanisms of the TIME in EC, especially the suppressive TIME, is pivotal and urgent for the management of EC patients.
A suppressive TIME is usually accompanied by the reduced infiltration or exhaustion of immune cells, and is correlated mainly with the presence of immunosuppressive cells and coinhibitory signals[34]. Herein, we focused on the reduced infiltration and exhaustion of T cells and NK cells, which play important roles in the TIME. In addition, immunosuppressive cells, such as suppressive macrophages (M2 macrophages) and myeloid-derived suppressor cells (MDSCs), can inhibit the activities of immune cells through various mechanisms to participate in balancing immune reactions in the TIME[35], and their presence may influence immunotherapeutic efficacy in cancers. Thus, elucidating the underlying molecular mechanisms is highly important for improving the therapeutic efficacy of agents for cancer treatment.
T cells are the major component of infiltrated immune cells in most solid tumors, and CD8+ cytotoxic T cells (CTLs) and CD4+ T helper cells (Ths) play crucial roles in eliminating tumor cells[36,37]. Specifically, activated CTLs may exert a cytotoxic effect on tumor cells by releasing cytotoxic substances, and Ths can promote or suppress host immune activities targeting tumor cells[36-38].
According to the single-cell sequencing results, the percentage of exhausted CD8+ T cells positive for C-X-C motif chemokine ligand 13 (CXCL13) increased, as these cells are the main T-cell type in the TME of EAC patients[39]. In ESCC, the infiltration and proliferation of T-cell clones have also been observed, and an exhausted CD8+ T-cell cluster (CD8-C7-TIGIT) and pre-exhausted CD8-C5-CCL5 and CD8-C6-STMN1 clusters accounted for high proportions of CD8+ T-cell clusters[22]. The expression level of the E3 ubiquitin ligase MARCH7 in ESCC tissues has been shown to be significantly greater than that in nontumor tissues, and was negatively correlated with tumor-infiltrating immune cells, such as CD8+ T cells[40]. Moreover, a subpopulation of CD8+ T cells expressing SPRY1 has been found in ESCC tissues after neoadjuvant immune checkpoint blockade, and these cells may possess certain progenitor cell characteristics and exhibit an exhausted phenotype[41]. Additionally, fibroblast growth factor 2 derived from tumor fibroblasts can induce the expression of SPRY1 in infiltrating T cells and participate in T-cell exhaustion in EC[42].
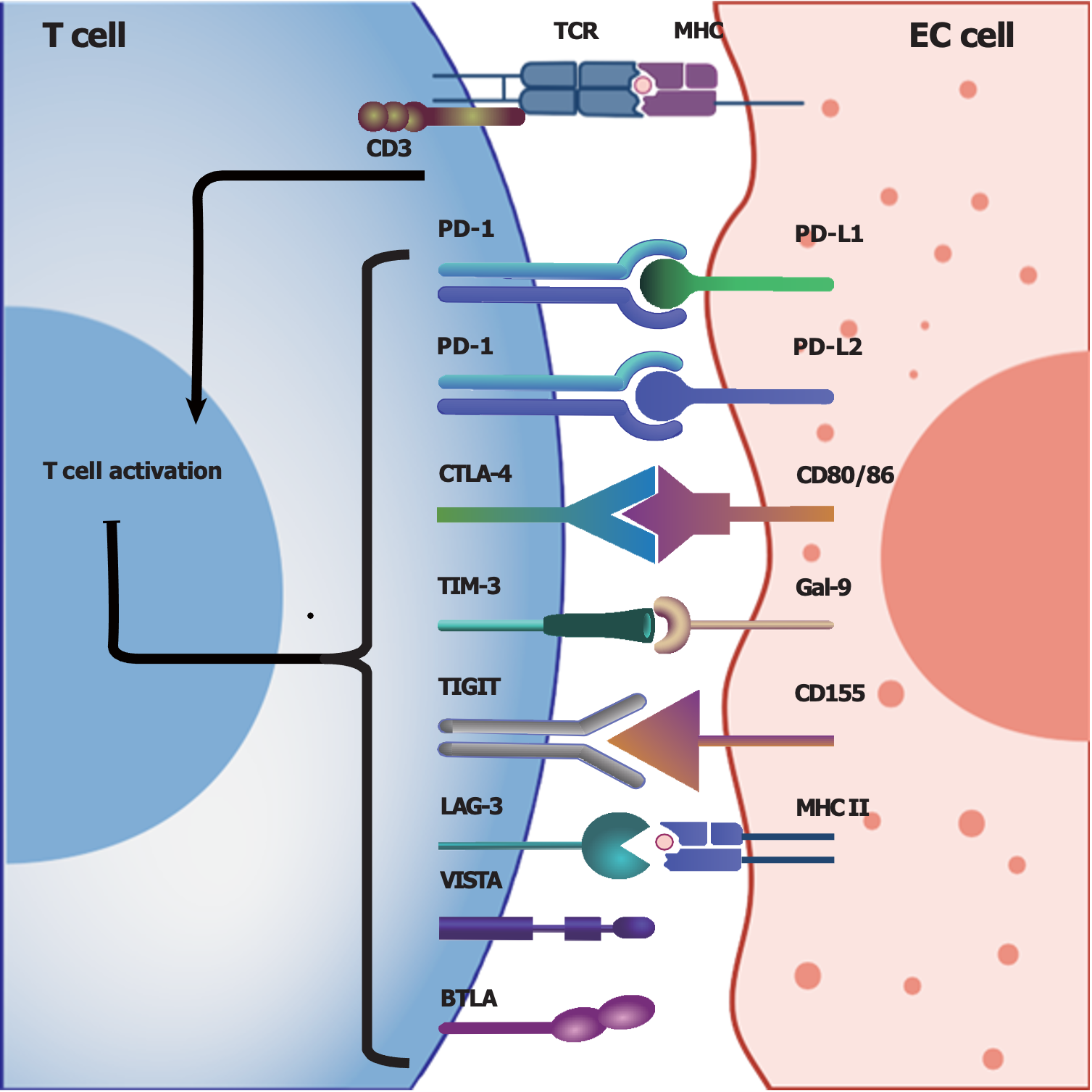
Immune checkpoints: Activated T cells may express various inhibitory receptors, known as immune checkpoints, to prevent excessive immune responses, aiming to maintain an immunologic balance; however, tumor cells may exploit these checkpoints to induce coinhibitory signals in the TME and create an immunosuppressive TME, which plays a pivotal role in tumor immune escape[43,44]. Thus, medications such as immune checkpoint inhibitors have been investigated for their ability to block these checkpoints, subsequently enhancing the ability of the immune system to attack tumor cells[34,45].
Programmed cell death protein 1 (PD-1), which is expressed on the T-cell membrane, is a classic immune checkpoint that can transmit immune inhibitory signals when it interacts with its corresponding ligand programmed cell death ligand 1 (PD-L1), which is expressed on tumor cells[46,47]. These interactions can inhibit the cytotoxic activities of T cells and allow tumor cells to escape immune surveillance and attack, accounting for one of the mechanisms of tumor immune escape[46,47]. For instance, EC patients with high PD-L1/PD-L2 expression, particularly patients in advanced stages, may have a poor prognosis[20]. Therefore, inhibiting PD-1/PD-L1 by blocking their interaction may restore the vigor and cytotoxicity of T cells in the TIME[18]. In recent years, immunotherapy involving checkpoint blockade targeting PD-1/PD-L1 has developed rapidly, becoming a first-line treatment for many cancers[17,48], but the efficacy of PD-1/PD-L1 blockade largely depends on the expression levels of PD-1/PD-L1 in the TME[19,49,50].
The interaction between CD28 on T cells and B7-1 (CD80)/B7-2 (CD86) on antigen-presenting cells or target cells can provide costimulatory activating signals to T cells, and subsequently boost T-cell activation[51]. Cytolytic T lymphocyte-associated antigen-4 (CTLA-4), another important regulatory molecule primarily expressed on regulatory T cells (Tregs) and activated T cells, can competitively bind B7 and inhibit cellular signal transduction for T-cell activation, subsequently suppressing immune responses[52,53]. Therefore, CTLA-4 is also considered an immune checkpoint molecule, and CTLA-4 blockade could effectively enhance immune responses against tumor cells[52]. However, the efficacy and safety of CTLA-4 blockade in EC patients require further investigation due to the limited number of related clinical trials.
In addition to the coinhibitory molecules mentioned above[18,52,54,55], researchers have identified various other immune checkpoints, such as T-cell immunoglobulin (Ig) and mucin domain-containing protein-3 (TIM-3)[56,57], lymphocyte activation gene-3[57-59] and T-cell Ig and ITIM domain[60-63]; detailed information about the potential immune checkpoints involved in EC in Figure 1[63-66].
Regulatory T lymphocytes: Tregs are CD4+CD25+Foxp3+ T cells that play an important role in suppressing the host immune response in the TME[67-70]. The infiltration of Tregs may be correlated with tumor invasion, progression, metastasis and poor survival after chemotherapy[68-71], and the infiltration of Tregs has also been shown to be negatively correlated with antitumor effector cells such as CTLs and NK cells in ESCC[72]. In addition, the hypomethylation-induced chemokine CCL20 in the TIME could affect the immune balance and promote the progression of EC, possibly contributing to the infiltration of Tregs in ESCC[73]. Moreover, Han et al[74] showed that Tregs may have the highest interleukin (IL)-32 expression in the TME of ESCC patients, and this expression is positively correlated with that of Foxp3, potentially promoting tumor progression; in addition, IL-32 may also induce interferon (IFN)-γ secretion by CD8+ T cells and facilitate antitumor immunity. Additionally, an imbalance in Th17/Treg cells has also been reported to occur during the development of Barrett’s esophagus, the precursor of EAC, through the regulation of the release of certain inflammatory cytokines[75].
Generally, T cells may experience functional loss or exhaustion in the TIME through interactions with various coinhibitory factors, and Tregs could play a crucial role in immunosuppression in the TME. Therefore, elucidating the functions and interactions of T cells with other cells in the TIME and understanding the mechanism of Treg-mediated tumor immune escape could provide valuable insights into the mechanisms of tumor immune escape, and thus further provide important evidence for novel immunotherapeutic strategies aimed at overcoming tumor immune escape.
NK cells are another type of tumor cell-killing lymphocyte that has garnered significant attention in cancer immunotherapy[76]. Previous preclinical and clinical studies have shown promising results for NK cell-related immunotherapy, and provided a novel perspective on immunotherapeutic strategies for NK cell-related treatments[77]. However, NK cells often experience a reduction or exhaustion in the immunosuppressive TME similar to that of T cells, which may also limit their antitumor effects[76,78].
The number of NK cells has been shown to be significantly lower in ESCC tissues than in adjacent nontumor tissues; in addition, a specific subset of cells, NK-C3-KLRC1 has been shown to differentiate from NK-C1-NCR3, and the number of NK-C2-STMN1 cells was significantly increased in ESCC[22]. The NK-C1-NCR3 subset has been shown to express relatively high levels of NCR3, CD266, NKG7 and LAMP1, and the NK-C3-KLRC1 and NK-C2-STMN1 subsets have been shown to express relatively high levels of KLRC1 and ITGA1[22]. As a cell surface receptor primarily expressed on NK cells and some types of T cells, NK group 2 member D (NKG2D) can interact with its ligands (NKG2DLs) to activate NK cells and T cells, and subsequently enhance immune surveillance and the clearance of tumor cells or infected cells[79,80]. Researchers have shown that the expression of NKG2DLs is significantly higher in ESCC tissues than in control tissues, and ESCC cells exhibit increased NKG2DL expression, thus providing a potential therapeutic target for ESCC via the use of NK cells[78]. Moreover, the inhibitory receptor NKG2A has been shown to be upregulated in NK cells in ESCC tissues compared to adjacent nontumor tissues[22], and a higher level of TIM-3 in tumor-infiltrating NK cells has been shown to be correlated with functional impairment and related to tumor invasion, lymph node metastasis and advanced stages in EC patients[56]. Notably, the expression of CD16brightCD56dim may significantly decrease in NK cells in ESCC, leading to a weakened antibody-dependent cell-mediated cytotoxicity response mediated by cetuximab, which binds to the CD16 receptor on NK cells and targets the epidermal growth factor receptor (EGFR)[81,82].
Furthermore, numerous cytokines may also participate in regulating the immuno-activation of NK cells. For instance, transforming growth factor (TGF)-β partially contributes to the downregulation of CD16 expression on NK cells, resulting in impaired NK cell function[81]. A lack of IL-18 in ESCC tissues may induce the production of IFN-γ in NK cells and CD8+ T cells, and potentially promote the clearance of tumor cells and improve the TME in patients with EC[83]. The expression level of IL-6, an important cytokine secreted by ESCC cells in the TME, has been shown to be higher in tumor tissues and blood circulation in ESCC patients, and may significantly upregulate the expression of CD39 on NK cells and impair the functions of NK cells, as well as be related to the poor prognosis of ESCC patients[84]. Another clinical study reported that IL-6 and IL-8 secreted by ESCC cells may downregulate the expression of certain activating receptors on NK cells and impair the function of NK cells by activating the signaling transducers and activators of transcription 3 (STAT3) signaling pathway[85]. Taken together, the above results demonstrate the decreased number and dysfunction of NK cells, effects that may disrupt immune surveillance in cancer patients, and pose a challenge for the investigation and clinical application of NK cell-related immunotherapy in ESCC patients.
Macrophages: Macrophages are important components of the innate immune system, and play pivotal roles in recognizing and removing damaged cells, pathogens and other foreign matter, as well as regulating adaptive immune responses by secreting various cytokines and chemokines[86]. Based on their functions and phenotypes, tumor-associated macrophages (TAMs) can be classified into two types: M1 and M2 macrophages[86,87]. M1 macrophages have proinflammatory properties and primarily participate in clearing pathogens, whereas M2 macrophages promote cell proliferation and tissue repair[86,87]. M1 macrophages in the tumor stroma are involved mainly in inhibiting the migration and invasion of ESCC cells, and serve as good prognostic factors for ESCC patients[88,89].
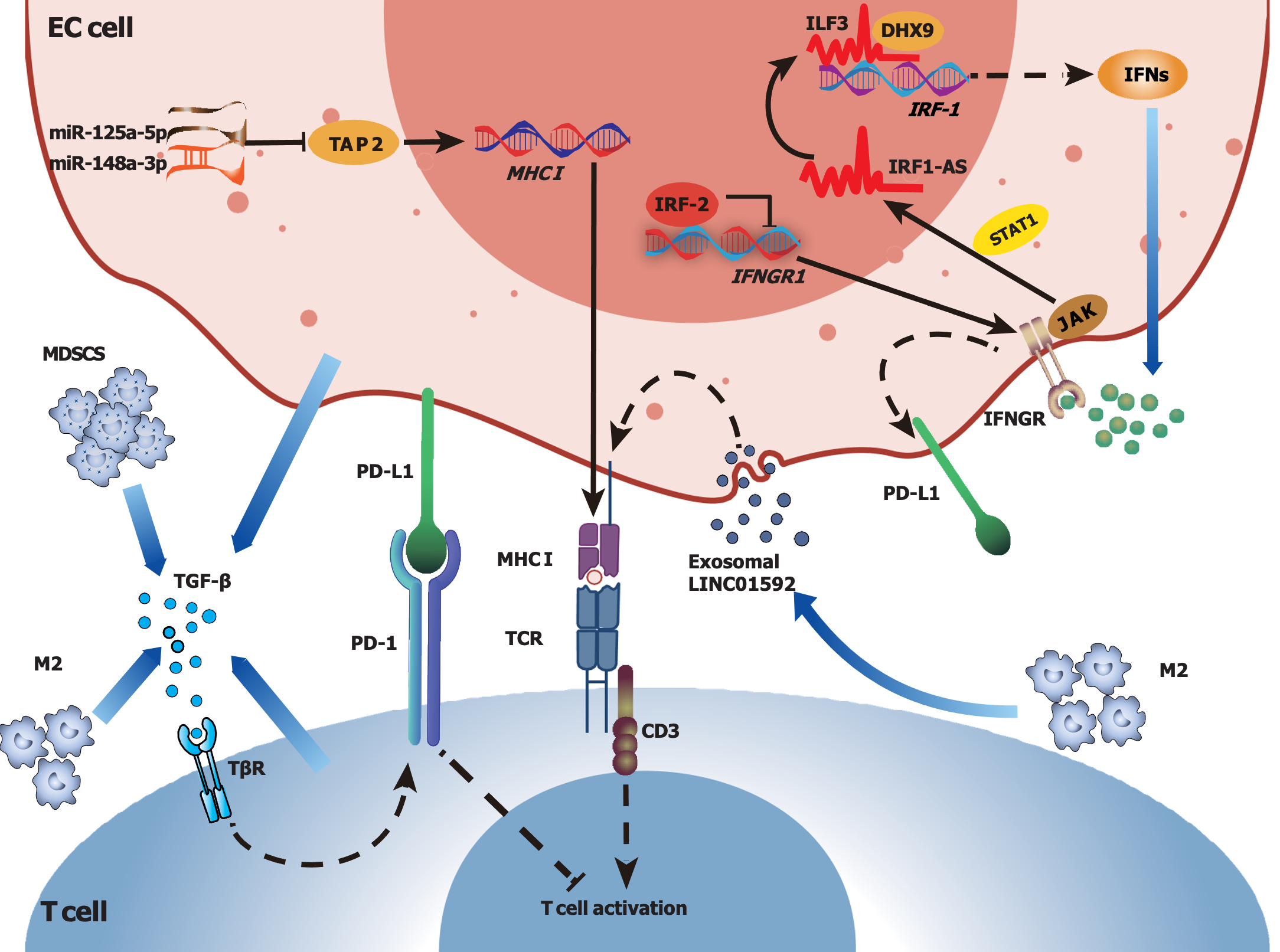
As an element of immunosuppression, M2 macrophages enriched in the TME of ESCC may suppress cell-mediated immune responses, secrete immunosuppressive factors and promote tumor angiogenesis[22]. M2 polarization may increase the expression of PD-L2 in ESCC cells, and lead to tumor immune escape and progression via PD-1-related signaling pathways[89]. In addition, Lu et al[90] reported that the upregulation of S100A7, a member of the S100 superfamily, could promote macrophage infiltration and M2 polarization, facilitating tumor angiogenesis by enhancing the activation of the p-ErK and p-FAK signaling pathways in the TME of ESCC. IL-32, which is highly secreted by Tregs, may promote the formation of an immunosuppressive TME; in addition, researchers have shown that IL-32, which is secreted from ESCC cells via extracellular vesicles, may shuttle into macrophages to promote M2 polarization via the FAK-STAT3 signaling pathway, further contributing to ESCC metastasis[91]. Moreover, Wang et al[92] reported that ESCC FOXO1+ cells may promote M2 polarization and recruitment to the TME in ESCC through the transcriptional regulation of CCL20 and CSF-1, and FOXO1+ tumor-induced M2 macrophages could promote tumor proliferation through FAK-PI3K-AKT signaling, which could be blocked by the blockade of PI3K[92]. In a rodent ESCC model, researchers found that CCL18, a chemokine secreted by TAMs, may promote tumor cell proliferation through the Janus-activated kinase 2 (JAK2)/STAT3 signaling pathway, and higher CCL18 levels are correlated with poor prognosis in ESCC patients[93]. To investigate the potential therapeutic efficacy of CCL18, researchers synthesized a CCL18-blocking peptide (Pep3) and found that it could inhibit the proliferation of EC-109 cells, suggesting potential targets through which CCL18 represses the progression of ESCC[93]. CCL22, another chemokine produced by TAMs in ESCC, may activate the FAK/AKT pathway and facilitate the malignant progression of ESCC cells[94]. Moreover, M2 macrophages may transmit the long noncoding RNA (lncRNA) AFAP1-AS1 to ESCC cells via secreted exosomes, downregulating miR-26a expression and upregulating ATF2 expression, thereby promoting tumor cell invasion and metastasis in EC[95]. Furthermore, a recent study showed that exosomes secreted by M2 macrophages carrying LINC01592 could be transferred to EC cells, resulting in a decrease in MHC-I expression, thereby allowing tumor cells to escape from attacks by CD8+ CTLs[96]. When the E2F6/NBR1/MHC-I signaling pathway was disrupted by small interfering RNAs or corresponding blocking antibodies, the tumor-promoting effects induced by LINC01592, as well as M2-driven tumor growth, were significantly inhibited[96]. In summary, M2 macrophages play an inhibitory role in the TIME of EC and can be recognized as key regulators of cancer occurrence, progression and metastasis. Therefore, targeting M2 macrophages and related signaling pathways may provide a promising perspective on therapeutic strategies for EC management.
MDSCs: MDSCs are widely accepted as a population of immature bone marrow cells, that can be classified into granulocyte-like MDSCs (G-MDSCs) and monocyte-like MDSCs (M-MDSCs)[97]. Both G-MDSCs and M-MDSCs play important roles in inhibiting immune cell activities in the TME, thus promoting tumor growth and metastasis[97]. It has been reported that the proportions of MDSCs and Tregs are significantly greater in EC patients than in controls, further suggesting an immunosuppressive role of MDSCs in EC[98]. Therefore, inhibiting the recruitment of MDSCs to the TME might be a promising approach for treating EC via immunotherapy. For example, TGF-β secreted by MDSCs in the TME may induce the phosphorylation of Smad2/Smad3, and contribute to the increased expression of the cancer/testis-associated gene Maelstrom (MAEL) in EC cells[99]. MAEL may be correlated with increased IL-8 expression by regulating the Akt1/RelA signaling pathway, and IL-8, in turn, may guide the recruitment of MDSCs into the TME of ESSCs[99]. In addition, the expression of MAEL in ESCC cells has been shown to be associated with recurrence and poor prognosis[99]. Moreover, it has been shown that the gene developmentally downregulated 9, which is critical for maintaining the stemness phenotype of ESCC cells, can regulate the expression of CXCL8 through the ERK signaling pathway, thereby contributing to the recruitment of MDSCs to the TME[100].
In addition to focusing on the recruitment of MDSCs, inhibiting MDSC function in the TME might be another important strategy. MDSCs with higher CD38 expression have been shown to be better able to inhibit activated T cells and promote tumor growth than MDSCs with lower CD38 expression[101]. This enhanced immunosuppressive capacity of CD38high MDSCs may be attributed to their increased production of inducible nitric oxide synthase (iNOS), since the upregulated iNOS may act as an immunosuppressive molecule to suppress the immune responses of T cells and contribute to tumor immune escape[101]. Moreover, EC patients exhibit increased numbers of MDSCs and Th17 cells in the peripheral circulation, as well as increased levels of plasma Arg1 and iNOS mRNA in peripheral blood mononuclear cells[102]. Additionally, the expression of myeloid cell markers in ESCC may be positively correlated with the increased expression of certain immune checkpoints, such as PD1, TIM3 and V-domain Ig suppressor of T-cell activation, as well as the development of ESCC[103]. However, the depletion of Gr1+ MDSCs may reduce the number of MDSCs, decrease the expression levels of immune checkpoint molecules, and inhibit tumor growth, suggesting the potential roles of MDSCs in the immunosuppression and progression of ESCC[103]. Furthermore, another fundamental study reported higher levels of lnc-17Rik in MDSCs derived from the peripheral blood of EC patients, and indicated that lnc-17Rik may enhance tumor immunosuppression by increasing the expression and enhancing the activation of certain key genes involved in MDSC differentiation, such as arginase 1, cyclooxygenase 2, NOS2, and NADPH oxidase 2[104]. These findings highlight the significance of elucidating the functions of MDSCs in the TIME, and suggest potential targets for therapeutic interventions aimed at overcoming immunosuppression and improving therapeutic efficacy in patients with EC.
Although EC exhibits strong immune responsiveness, as previously mentioned, it may still achieve immune escape in the immunosuppressive TME through various mechanisms (Figure 2), including the downregulation of MHC expression, the secretion of immunosuppressive factors and alterations in tumor metabolism.
Tumor immune escape is often accompanied by a decrease in or loss of MHC molecules, which play crucial roles in the recognition and killing of tumor cells by immune cells[105]. Notably, the expression of HLA-ABC molecules is usually decreased or even absent in ESSC tissues[106]. Specifically, a previous study reported that approximately 41% of EC patients had no HLA-ABC expression, more than half of the EC patients had weak expression, and only approximately 3% of the EC patients had strong HLA-ABC expression[106]. In addition, the reduced or absent expression of HLA-ABC in ESCC may be strongly correlated with the expression of certain molecules that participate in antigen processing, such as b2m, ATP binding cassette subfamily B member 1 (TAP1), TAP2, LMP2 and LMP7[107]. Moreover, allelic loss in the 6p21.3 region, observed in approximately 46.9% of ESCC patients in a Chinese study, has been shown to be associated with the downregulation of HLA class I antigens[108,109], and DNA hypermethylation may result in deficient expression of HLA class I genes in ESCC[110]. Numerous ncRNAs, such as miR-125a-5p and miR-148a-3p, may downregulate the expression of TAP2 and HLA-I to affect the antigen presentation process[111], and exosomal LINC01592 released from TAMs may also downregulate the expression of MHC-I in EC cells and promote malignant EC progression[96]. Downregulation of the expression of MHC molecules in the TME hampers antigen processing and presentation processes, thereby enabling tumor immune evasion in patients with EC. Investigating these underlying mechanisms is crucial for advancing innovative cancer immunotherapy focused on these molecules.
An immunosuppressive TME is partially generated by immunosuppressive factors secreted by tumor cells, immune cells and stromal cells[112], and these factors play crucial roles in tumor proliferation, angiogenesis and invasion, as well as in EC progression[113]. Some classic immunosuppressive cytokines, such as TGF-β and IFN-γ, may inhibit the functions of immune cells, thereby weakening the ability of the immune system to attack tumor cells[113,114].
The TGF-β signaling pathway could play a dual role in cancer development depending on the stage of disease[114]. Under pathological conditions, the overexpression of TGF-β may lead to epithelial mesenchymal transition, extracellular matrix deposition and the formation of cancer-associated fibroblasts, resulting in fibrotic diseases and cancers[115]. In addition, TGF-β can restrict the infiltration of T cells to the TME and decrease antitumor immunoactivity[116]. Moreover, TGF-β derived from MDSCs in the TME of ESCC may increase PD-1 expression in CD8+ T cells, leading to resistance to immunotherapy via PD-1/PD-L1[27]. Furthermore, the combination of TGF-β and PD-L1 blockade has been shown to significantly increase the number of tumor-infiltrating T cells and reduce the tumor burden in EAC patients[116].
The IFN signaling pathway also plays a dual role in the TME. On the one hand, IFN-γ acts as a cytotoxic cytokine and induces tumor cell apoptosis, thus exerting antitumor effects[117]. On the other hand, IFN-γ may contribute to immunosuppression in the TIME by promoting the synthesis of immune checkpoint-related factors, such as PD-L1, thus allowing tumors to escape immune surveillance[117,118]. Notably, interferon regulatory factors (IRFs) play important roles in regulating the effects of IFN-γ: IRF-1 is generally considered a tumor suppressor, whereas IRF-2 is regarded as an oncogenic factor[119,120]. In addition, IRF-1 expression has been shown to be decreased, and IRF-2 expression has been shown to be increased in EC, contributing to the suppression of immune responses[121]. Most importantly, IFN-γ can interact with various factors. For instance, an IFN-induced lncRNA, IRF1-AS, has been shown to activate IRF-1 transcription by interacting with IL enhancer binding factor 3 and DExH-box helicase 9, thereby activating the IFN response[119]. However, IRF-2 may inhibit the transcription of IFN-γ receptor 1 (IFNGR1) by binding to specific motifs in the IFNGR1 promoter, thereby reducing the sensitivity of EC cells to IFN-γ and enhancing the resistance of EC cells to IFN-γ[120]. IFNs can regulate the JAK-STAT signaling pathway, and the activation of STATs often facilitates tumor progression[122,123]. MAGE-C3 may enhance the interaction between IFNGR1 and STAT1 by binding to IFNGR1, which can activate IFN-γ signaling and upregulate PD-L1 expression, thus contributing to immunosuppression[118]. Moreover, the overexpression of MAGE-C3 may be associated with lymph node metastasis and poor survival in ESCC patients[118]. Therefore, various factors have been suggested to participate in the immunosuppression mediated by IFN-γ in EC, but the underlying mechanisms urgently need to be elucidated.
The interplay of cytokines and signaling pathways in the TIME of EC results in the construction of a complex network, and certain key cytokines, such as TGF-β and IFN-γ, play dual roles in tumor progression by promoting tumor growth and immune escape or exerting antitumor effects. Understanding the intricate interactions among these factors might provide insights into potential therapeutic targets for enhancing antitumor immunity in patients with EC. Further research is warranted to explore novel strategies for immune modulation and improving immunotherapeutic efficacy in EC patients.
Tumor metabolism is usually characterized by high heterogeneity and constant remodeling due to the evolution of cancer cells, and metabolic reprogramming is a distinctive feature of malignant tumors[124]. The dynamic interactions among tumor cells and various immune cells could lead to metabolic competition within the tumor ecosystem, limiting the availability of nutrients for immune cells and resulting in acidification of the TME, thereby impairing the functions of immune cells[125]. In a previous study, ESCC patients were divided into high- and low-risk subtypes based on three genes associated with tumor metabolism, namely, CD38, INPP5E and POLR3G, and the high-risk subgroup exhibited decreased CD38 and POLR3G expression and increased INPP5E expression[126]. Compared with patients in the low-risk subgroup, patients in the high-risk subgroup had increased Treg infiltration and decreased plasma cell infiltration in the TME, as well as significant metabolic differences in ESCC tissues[126]. Notably, ESCC was primarily associated with glycolysis, and EAC was strongly correlated with oxidative metabolism, glycolipid metabolism and the tricarboxylic acid cycle[127].
Under normoxic conditions, most tumors preferentially rely on glycolysis for energy, which is considered an advantage for survival and is known as the Warburg effect[128]. A recent study highlighted the inhibitory role of estrogen-related receptor gamma in the occurrence, proliferation and glycolytic activity of ESCC cells, and one of its specific agonists, DY131, could inhibit the proliferation and glycolytic activity of ESCC cells by modulating certain specific genes involved in the glycolytic pathway[128]. In addition, the combination of DY131 with PD-1 blockade may have a synergistic effect on the suppression of ESCC growth[128]. As a byproduct of glycolysis, lactate may play an important regulatory role in the development and progression of ESCC, and is closely correlated with immunosuppression in the TME[129]. Furthermore, intracellular hypoxia is also associated with the progression, treatment resistance and poor prognosis of various malignancies. Numerous genes associated with hypoxia, such as PGK1, PGM1 and
Certain metabolic pathways other than the glycolysis and hypoxia pathways are also involved in EC. Zhao et al[132] identified six genes associated with iron metabolism and iron death (PRNP, SLC3A2, SLC39A8, SLC39A14, ATP6V0A1, and LCN2) in ESCC, and these genes may be associated with the infiltration of immune cells, tumor mutational load and ESCC prognosis. In addition, lncRNAs such as LINC01068, TMEM92-AS1 and AC243967.2 have been reported to be correlated with iron metabolism and iron death, and be closely related to the infiltration of immune cells in ESCC[133]. Moreover, Zhang et al[134] reported that mitochondrial energy metabolism is associated with the TIME and poor prognosis in ESCC patients, and identified several fatty acid metabolism-related genes that are predictors of EC prognosis[135]. Additionally, tryptophan-derived metabolites have been shown to contribute to tumor immune escape, and been identified as biomarkers for EC metastasis and prognosis[136].
These insights emphasize the importance of metabolic alterations in the TME of patients with EC. Understanding the intricate metabolic interactions between tumor cells and immune cells could guide the development of targeted therapies for different subtypes of EC, and further research in these areas may open new avenues for the management of patients with EC.
In this review, we mainly described the potential mechanisms of immunosuppression in the TME of patients with EC, which opens up an interesting and promising field of future immunotherapies. The presence of decreased immune cells and increased immunosuppressive cells, including exhausted CD8+ T cells and NK cells, Tregs, M2 macrophages and MDSCs, in the TIME of EC is not rare, and these cells may contribute to tumor immune escape and tumor progression. Moreover, various other factors related to tumor cells also participate in the formation of an immunosuppressive micro-environment in EC, such as the downregulated expression of MHC molecules on tumor cells, the release of immunosuppressive cytokines by tumor cells and their surroundings, and altered tumor metabolism. With a deeper and more comprehensive understanding of the complexity and heterogeneity of the TME, such as tumor types, the distribution and function of infiltrating immune and nonimmune cell subsets, the expression of cytokines and the activation or inhibition of signaling pathways in the TME, we may better elucidate the mechanisms of the immunosuppressive microenvironment, better understand the differences in patient response to the same immunotherapeutic strategies, and accurately predict the efficacy of immunotherapeutic approaches; thus, personalized treatments can be developed to overcome the effects of immune suppressive factors, improve the efficacy of immunotherapy, and maximize the advantages of immunotherapy.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s classification
Scientific Quality: Grade A
Novelty: Grade A
Creativity or Innovation: Grade A
Scientific Significance: Grade A
P-Reviewer: Panaitescu C, Romania S-Editor: Wang JJ L-Editor: A P-Editor: Chen YX
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64722] [Article Influence: 16180.5] [Reference Citation Analysis (177)] |
| 2. | Siewert JR, Ott K. Are squamous and adenocarcinomas of the esophagus the same disease? Semin Radiat Oncol. 2007;17:38-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 238] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 3. | Rustgi AK, El-Serag HB. Esophageal carcinoma. N Engl J Med. 2014;371:2499-2509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 817] [Cited by in RCA: 998] [Article Influence: 90.7] [Reference Citation Analysis (0)] |
| 4. | Li M, Park JY, Sheikh M, Kayamba V, Rumgay H, Jenab M, Narh CT, Abedi-Ardekani B, Morgan E, de Martel C, McCormack V, Arnold M. Population-based investigation of common and deviating patterns of gastric cancer and oesophageal cancer incidence across populations and time. Gut. 2023;72:846-854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 19] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 5. | Lagergren J, Lagergren P. Recent developments in esophageal adenocarcinoma. CA Cancer J Clin. 2013;63:232-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 212] [Cited by in RCA: 234] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 6. | Joseph A, Raja S, Kamath S, Jang S, Allende D, McNamara M, Videtic G, Murthy S, Bhatt A. Esophageal adenocarcinoma: A dire need for early detection and treatment. Cleve Clin J Med. 2022;89:269-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 46] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 7. | Li X, Chen L, Luan S, Zhou J, Xiao X, Yang Y, Mao C, Fang P, Zeng X, Gao H, Yuan Y. The development and progress of nanomedicine for esophageal cancer diagnosis and treatment. Semin Cancer Biol. 2022;86:873-885. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 8. | Ajani JA, D'Amico TA, Bentrem DJ, Cooke D, Corvera C, Das P, Enzinger PC, Enzler T, Farjah F, Gerdes H, Gibson M, Grierson P, Hofstetter WL, Ilson DH, Jalal S, Keswani RN, Kim S, Kleinberg LR, Klempner S, Lacy J, Licciardi F, Ly QP, Matkowskyj KA, McNamara M, Miller A, Mukherjee S, Mulcahy MF, Outlaw D, Perry KA, Pimiento J, Poultsides GA, Reznik S, Roses RE, Strong VE, Su S, Wang HL, Wiesner G, Willett CG, Yakoub D, Yoon H, McMillian NR, Pluchino LA. Esophageal and Esophagogastric Junction Cancers, Version 2.2023, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2023;21:393-422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 234] [Reference Citation Analysis (1)] |
| 9. | Wang L, Han H, Wang Z, Shi L, Yang M, Qin Y. Targeting the Microenvironment in Esophageal Cancer. Front Cell Dev Biol. 2021;9:684966. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 10. | Zhu K, Liu Q, Zhou Y, Tao C, Zhao Z, Sun J, Xu H. Oncogenes and tumor suppressor genes: comparative genomics and network perspectives. BMC Genomics. 2015;16 Suppl 7:S8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 44] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 11. | Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423-1437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4060] [Cited by in RCA: 5770] [Article Influence: 524.5] [Reference Citation Analysis (0)] |
| 12. | Vesely MD, Kershaw MH, Schreiber RD, Smyth MJ. Natural innate and adaptive immunity to cancer. Annu Rev Immunol. 2011;29:235-271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1617] [Cited by in RCA: 1500] [Article Influence: 107.1] [Reference Citation Analysis (0)] |
| 13. | Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, Coussens LM, Gabrilovich DI, Ostrand-Rosenberg S, Hedrick CC, Vonderheide RH, Pittet MJ, Jain RK, Zou W, Howcroft TK, Woodhouse EC, Weinberg RA, Krummel MF. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24:541-550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2019] [Cited by in RCA: 3835] [Article Influence: 547.9] [Reference Citation Analysis (0)] |
| 14. | Lv B, Wang Y, Ma D, Cheng W, Liu J, Yong T, Chen H, Wang C. Immunotherapy: Reshape the Tumor Immune Microenvironment. Front Immunol. 2022;13:844142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 221] [Article Influence: 73.7] [Reference Citation Analysis (0)] |
| 15. | Xiao Y, Yu D. Tumor microenvironment as a therapeutic target in cancer. Pharmacol Ther. 2021;221:107753. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 1139] [Article Influence: 227.8] [Reference Citation Analysis (2)] |
| 16. | Chang RB, Beatty GL. The interplay between innate and adaptive immunity in cancer shapes the productivity of cancer immunosurveillance. J Leukoc Biol. 2020;108:363-376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 17. | Williams WV. Editorial: Targeted Immunotherapy for Cancer. Front Pharmacol. 2022;13:894681. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 18. | Cheng C, Zhuge L, Xiao X, Luan S, Yuan Y. Overcoming resistance to PD-1/PD-L1 inhibitors in esophageal cancer. Front Oncol. 2022;12:955163. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 19. | Dhupar R, Van Der Kraak L, Pennathur A, Schuchert MJ, Nason KS, Luketich JD, Lotze MT. Targeting Immune Checkpoints in Esophageal Cancer: A High Mutational Load Tumor. Ann Thorac Surg. 2017;103:1340-1349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 20. | Ohigashi Y, Sho M, Yamada Y, Tsurui Y, Hamada K, Ikeda N, Mizuno T, Yoriki R, Kashizuka H, Yane K, Tsushima F, Otsuki N, Yagita H, Azuma M, Nakajima Y. Clinical significance of programmed death-1 ligand-1 and programmed death-1 ligand-2 expression in human esophageal cancer. Clin Cancer Res. 2005;11:2947-2953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 566] [Cited by in RCA: 639] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 21. | Dolton G, Rius C, Wall A, Szomolay B, Bianchi V, Galloway SAE, Hasan MS, Morin T, Caillaud ME, Thomas HL, Theaker S, Tan LR, Fuller A, Topley K, Legut M, Attaf M, Hopkins JR, Behiry E, Zabkiewicz J, Alvares C, Lloyd A, Rogers A, Henley P, Fegan C, Ottmann O, Man S, Crowther MD, Donia M, Svane IM, Cole DK, Brown PE, Rizkallah P, Sewell AK. Targeting of multiple tumor-associated antigens by individual T cell receptors during successful cancer immunotherapy. Cell. 2023;186:3333-3349.e27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 70] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 22. | Zheng Y, Chen Z, Han Y, Han L, Zou X, Zhou B, Hu R, Hao J, Bai S, Xiao H, Li WV, Bueker A, Ma Y, Xie G, Yang J, Chen S, Li H, Cao J, Shen L. Immune suppressive landscape in the human esophageal squamous cell carcinoma microenvironment. Nat Commun. 2020;11:6268. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 292] [Article Influence: 58.4] [Reference Citation Analysis (0)] |
| 23. | Nakamura S, Ohuchida K, Ohtsubo Y, Yamada Y, Tsutsumi C, Okuda S, Hisano K, Mochida Y, Shinkawa T, Iwamoto C, Torata N, Mizuuchi Y, Shindo K, Nakata K, Moriyama T, Torisu T, Nagai E, Morisaki T, Kitazono T, Oda Y, Nakamura M. Single-cell transcriptome analysis reveals functional changes in tumour-infiltrating B lymphocytes after chemotherapy in oesophageal squamous cell carcinoma. Clin Transl Med. 2023;13:e1181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 24. | Almeida LG, Sakabe NJ, deOliveira AR, Silva MC, Mundstein AS, Cohen T, Chen YT, Chua R, Gurung S, Gnjatic S, Jungbluth AA, Caballero OL, Bairoch A, Kiesler E, White SL, Simpson AJ, Old LJ, Camargo AA, Vasconcelos AT. CTdatabase: a knowledge-base of high-throughput and curated data on cancer-testis antigens. Nucleic Acids Res. 2009;37:D816-D819. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 259] [Cited by in RCA: 320] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 25. | Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5:615-625. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1173] [Cited by in RCA: 1220] [Article Influence: 61.0] [Reference Citation Analysis (0)] |
| 26. | Caballero OL, Chen YT. Cancer/testis (CT) antigens: potential targets for immunotherapy. Cancer Sci. 2009;100:2014-2021. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 425] [Cited by in RCA: 423] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 27. | Chen X, Wang L, Li P, Song M, Qin G, Gao Q, Zhang Z, Yue D, Wang D, Nan S, Qi Y, Li F, Yang L, Huang L, Zhang M, Zhang B, Gao Y, Zhang Y. Dual TGF-β and PD-1 blockade synergistically enhances MAGE-A3-specific CD8(+) T cell response in esophageal squamous cell carcinoma. Int J Cancer. 2018;143:2561-2574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 73] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 28. | Kerkar SP, Wang ZF, Lasota J, Park T, Patel K, Groh E, Rosenberg SA, Miettinen MM. MAGE-A is More Highly Expressed Than NY-ESO-1 in a Systematic Immunohistochemical Analysis of 3668 Cases. J Immunother. 2016;39:181-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 54] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 29. | Oshima Y, Shimada H, Yajima S, Nanami T, Matsushita K, Nomura F, Kainuma O, Takiguchi N, Soda H, Ueda T, Iizasa T, Yamamoto N, Yamamoto H, Nagata M, Yokoi S, Tagawa M, Ohtsuka S, Kuwajima A, Murakami A, Kaneko H. NY-ESO-1 autoantibody as a tumor-specific biomarker for esophageal cancer: screening in 1969 patients with various cancers. J Gastroenterol. 2016;51:30-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 58] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 30. | Ishikawa T, Kageyama S, Miyahara Y, Okayama T, Kokura S, Wang L, Sato E, Yagita H, Itoh Y, Shiku H. Safety and antibody immune response of CHP-NY-ESO-1 vaccine combined with poly-ICLC in advanced or recurrent esophageal cancer patients. Cancer Immunol Immunother. 2021;70:3081-3091. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 31. | Li P, Chen X, Ping Y, Qin G, Huang L, Zhao Q, Zhang Z, Chen H, Wang L, Yang S, Zhang Y. Clinical Correlation of Function and TCR vβ Diversity of MAGE-C2-Specific CD8(+) T Cell Response in Esophageal Cancer. J Immunol. 2022;209:1039-1047. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 32. | Lonie JM, Brosda S, Bonazzi VF, Aoude LG, Patel K, Brown I, Sharma S, Lampe G, Addala V, Koufariotis LT, Wood S, Waddell N, Dolcetti R, Barbour AP. The oesophageal adenocarcinoma tumour immune microenvironment dictates outcomes with different modalities of neoadjuvant therapy - results from the AGITG DOCTOR trial and the cancer evolution biobank. Front Immunol. 2023;14:1220129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 33. | Ma Y, Yu J, Ma X, Li Q, Su Q, Cao B. Efficacy and adverse events of immune checkpoint inhibitors in esophageal cancer patients: Challenges and perspectives for immunotherapy. Asia Pac J Clin Oncol. 2024;20:180-187. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 34. | Vesely MD, Zhang T, Chen L. Resistance Mechanisms to Anti-PD Cancer Immunotherapy. Annu Rev Immunol. 2022;40:45-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 243] [Cited by in RCA: 260] [Article Influence: 86.7] [Reference Citation Analysis (6)] |
| 35. | Iglesias-Escudero M, Arias-González N, Martínez-Cáceres E. Regulatory cells and the effect of cancer immunotherapy. Mol Cancer. 2023;22:26. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 105] [Reference Citation Analysis (0)] |
| 36. | Oh DY, Fong L. Cytotoxic CD4(+) T cells in cancer: Expanding the immune effector toolbox. Immunity. 2021;54:2701-2711. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 264] [Article Influence: 66.0] [Reference Citation Analysis (0)] |
| 37. | Lei X, Khatri I, de Wit T, de Rink I, Nieuwland M, Kerkhoven R, van Eenennaam H, Sun C, Garg AD, Borst J, Xiao Y. CD4(+) helper T cells endow cDC1 with cancer-impeding functions in the human tumor micro-environment. Nat Commun. 2023;14:217. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 43] [Reference Citation Analysis (0)] |
| 38. | Whiteside TL. What are regulatory T cells (Treg) regulating in cancer and why? Semin Cancer Biol. 2012;22:327-334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 219] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 39. | Croft W, Evans RPT, Pearce H, Elshafie M, Griffiths EA, Moss P. The single cell transcriptional landscape of esophageal adenocarcinoma and its modulation by neoadjuvant chemotherapy. Mol Cancer. 2022;21:200. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 34] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 40. | Singh S, Bano A, Saraya A, Das P, Sharma R. Association of MARCH7 with tumor progression and T-cell infiltration in esophageal cancer. Med Oncol. 2022;40:67. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 41. | Liu Z, Zhang Y, Ma N, Yang Y, Ma Y, Wang F, Wang Y, Wei J, Chen H, Tartarone A, Velotta JB, Dayyani F, Gabriel E, Wakefield CJ, Kidane B, Carbonelli C, Long L, Liu Z, Su J, Li Z. Progenitor-like exhausted SPRY1(+)CD8(+) T cells potentiate responsiveness to neoadjuvant PD-1 blockade in esophageal squamous cell carcinoma. Cancer Cell. 2023;41:1852-1870.e9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 52] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 42. | Chen QY, Li YN, Wang XY, Zhang X, Hu Y, Li L, Suo DQ, Ni K, Li Z, Zhan JR, Zeng TT, Zhu YH, Li Y, Ma LJ, Guan XY. Tumor Fibroblast-Derived FGF2 Regulates Expression of SPRY1 in Esophageal Tumor-Infiltrating T Cells and Plays a Role in T-cell Exhaustion. Cancer Res. 2020;80:5583-5596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 43. | Sanmamed MF, Chen L. A Paradigm Shift in Cancer Immunotherapy: From Enhancement to Normalization. Cell. 2018;175:313-326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 589] [Cited by in RCA: 989] [Article Influence: 164.8] [Reference Citation Analysis (0)] |
| 44. | Mellman I, Chen DS, Powles T, Turley SJ. The cancer-immunity cycle: Indication, genotype, and immunotype. Immunity. 2023;56:2188-2205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 379] [Article Influence: 189.5] [Reference Citation Analysis (0)] |
| 45. | Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science. 2018;359:1350-1355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2848] [Cited by in RCA: 4622] [Article Influence: 660.3] [Reference Citation Analysis (0)] |
| 46. | Dong H, Zhu G, Tamada K, Chen L. B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat Med. 1999;5:1365-1369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1806] [Cited by in RCA: 1966] [Article Influence: 75.6] [Reference Citation Analysis (0)] |
| 47. | Kim JM, Chen DS. Immune escape to PD-L1/PD-1 blockade: seven steps to success (or failure). Ann Oncol. 2016;27:1492-1504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 303] [Cited by in RCA: 480] [Article Influence: 53.3] [Reference Citation Analysis (0)] |
| 48. | Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, Leming PD, Spigel DR, Antonia SJ, Horn L, Drake CG, Pardoll DM, Chen L, Sharfman WH, Anders RA, Taube JM, McMiller TL, Xu H, Korman AJ, Jure-Kunkel M, Agrawal S, McDonald D, Kollia GD, Gupta A, Wigginton JM, Sznol M. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443-2454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8900] [Cited by in RCA: 9914] [Article Influence: 762.6] [Reference Citation Analysis (0)] |
| 49. | Yagi T, Baba Y, Ishimoto T, Iwatsuki M, Miyamoto Y, Yoshida N, Watanabe M, Baba H. PD-L1 Expression, Tumor-infiltrating Lymphocytes, and Clinical Outcome in Patients With Surgically Resected Esophageal Cancer. Ann Surg. 2019;269:471-478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 141] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 50. | Noori M, Yousefi AM, Zali MR, Bashash D. Predictive value of PD-L1 expression in response to immune checkpoint inhibitors for esophageal cancer treatment: A systematic review and meta-analysis. Front Oncol. 2022;12:1021859. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 51. | Sharpe AH, Freeman GJ. The B7-CD28 superfamily. Nat Rev Immunol. 2002;2:116-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1234] [Cited by in RCA: 1281] [Article Influence: 55.7] [Reference Citation Analysis (0)] |
| 52. | Leach DR, Krummel MF, Allison JP. Enhancement of antitumor immunity by CTLA-4 blockade. Science. 1996;271:1734-1736. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2582] [Cited by in RCA: 2812] [Article Influence: 97.0] [Reference Citation Analysis (0)] |
| 53. | Perkins D, Wang Z, Donovan C, He H, Mark D, Guan G, Wang Y, Walunas T, Bluestone J, Listman J, Finn PW. Regulation of CTLA-4 expression during T cell activation. J Immunol. 1996;156:4154-4159. [PubMed] |
| 54. | Kelly RJ. Emerging Multimodality Approaches to Treat Localized Esophageal Cancer. J Natl Compr Canc Netw. 2019;17:1009-1014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 114] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 55. | Tian C, Wang X, Zhang S. CTLA-4 and its inhibitors in esophageal cancer: efficacy of therapy and potential mechanisms of adverse events. Am J Cancer Res. 2023;13:3140-3156. [PubMed] |
| 56. | Zheng Y, Li Y, Lian J, Yang H, Li F, Zhao S, Qi Y, Zhang Y, Huang L. TNF-α-induced Tim-3 expression marks the dysfunction of infiltrating natural killer cells in human esophageal cancer. J Transl Med. 2019;17:165. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 75] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 57. | Lu B, Chen L, Liu L, Zhu Y, Wu C, Jiang J, Zhang X. T-cell-mediated tumor immune surveillance and expression of B7 co-inhibitory molecules in cancers of the upper gastrointestinal tract. Immunol Res. 2011;50:269-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 56] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 58. | Iouzalen N, Andreae S, Hannier S, Triebel F. LAP, a lymphocyte activation gene-3 (LAG-3)-associated protein that binds to a repeated EP motif in the intracellular region of LAG-3, may participate in the down-regulation of the CD3/TCR activation pathway. Eur J Immunol. 2001;31:2885-2891. [PubMed] [DOI] [Full Text] |
| 59. | Gebauer F, Krämer M, Bruns C, Schlößer HA, Thelen M, Lohneis P, Schröder W, Zander T, Alakus H, Buettner R, Loeser H, Quaas A. Lymphocyte activation gene-3 (LAG3) mRNA and protein expression on tumour infiltrating lymphocytes (TILs) in oesophageal adenocarcinoma. J Cancer Res Clin Oncol. 2020;146:2319-2327. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 60. | Wang P, Chen Y, Long Q, Li Q, Tian J, Liu T, Wu Y, Ding Z. Increased coexpression of PD-L1 and TIM3/TIGIT is associated with poor overall survival of patients with esophageal squamous cell carcinoma. J Immunother Cancer. 2021;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 51] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 61. | Zhao K, Ma L, Feng L, Huang Z, Meng X, Yu J. CD155 Overexpression Correlates With Poor Prognosis in Primary Small Cell Carcinoma of the Esophagus. Front Mol Biosci. 2020;7:608404. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 62. | O'Donnell JS, Madore J, Li XY, Smyth MJ. Tumor intrinsic and extrinsic immune functions of CD155. Semin Cancer Biol. 2020;65:189-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 98] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 63. | Chen Z, Cao K, Zhang J, Liu Z, Lu L, Qi B, Shi L, Huang R, Zhao S. Concomitant expression of inhibitory molecules for T cell activation predicts poor survival in patients with esophageal squamous cell carcinoma. Curr Cancer Drug Targets. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 64. | Tang W, Chen S, Kang M, Liu J, Liu C. Investigation of BTLA tagging variants with risk of esophagogastric junction adenocarcinoma. Biosci Rep. 2019;39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 65. | Zhao N, Zhang Z, Wang Q, Li L, Wei Z, Chen H, Zhou M, Liu Z, Su J. DNA damage repair profiling of esophageal squamous cell carcinoma uncovers clinically relevant molecular subtypes with distinct prognoses and therapeutic vulnerabilities. EBioMedicine. 2023;96:104801. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 66. | Hou N, Ma J, Li W, Zhao L, Gao Q, Mai L. T-cell immunoglobulin and mucin domain-containing protein-3 and galectin-9 protein expression: Potential prognostic significance in esophageal squamous cell carcinoma for Chinese patients. Oncol Lett. 2017;14:8007-8013. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 67. | Tie Y, Tang F, Wei YQ, Wei XW. Immunosuppressive cells in cancer: mechanisms and potential therapeutic targets. J Hematol Oncol. 2022;15:61. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 299] [Article Influence: 99.7] [Reference Citation Analysis (0)] |
| 68. | Whiteside TL. FOXP3+ Treg as a therapeutic target for promoting anti-tumor immunity. Expert Opin Ther Targets. 2018;22:353-363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 132] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 69. | Nabeki B, Ishigami S, Uchikado Y, Sasaki K, Kita Y, Okumura H, Arigami T, Kijima Y, Kurahara H, Maemura K, Natsugoe S. Interleukin-32 expression and Treg infiltration in esophageal squamous cell carcinoma. Anticancer Res. 2015;35:2941-2947. [PubMed] |
| 70. | Xu T, Duan Q, Wang G, Hu B. CD4 + CD25high regulatory T cell numbers and FOXP3 mRNA expression in patients with advanced esophageal cancer before and after chemotherapy. Cell Biochem Biophys. 2011;61:389-392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 71. | Conroy MJ, Kennedy SA, Doyle SL, Hayes B, Kavanagh M, van der Stok EP, O'Sullivan K, Cathcart MC, Reynolds JV, Lysaght J. A study of the immune infiltrate and patient outcomes in esophageal cancer. Carcinogenesis. 2021;42:395-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 72. | Pan C, Wang Y, Liu Q, Hu Y, Fu J, Xie X, Zhang S, Xi M, Wen J. Phenotypic profiling and prognostic significance of immune infiltrates in esophageal squamous cell carcinoma. Oncoimmunology. 2021;10:1883890. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 73. | Lian J, Liu S, Yue Y, Yang Q, Zhang Z, Yang S, Zhang Y. Eomes promotes esophageal carcinoma progression by recruiting Treg cells through the CCL20-CCR6 pathway. Cancer Sci. 2021;112:144-154. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 74. | Han L, Chen S, Chen Z, Zhou B, Zheng Y, Shen L. Interleukin 32 Promotes Foxp3(+) Treg Cell Development and CD8(+) T Cell Function in Human Esophageal Squamous Cell Carcinoma Microenvironment. Front Cell Dev Biol. 2021;9:704853. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 75. | Liu J, Luo Y, Wang J, Xi C, Chen Y, Yang G, Ling Y. Key molecules involved in the Th17/Treg balance are associated with the pathogenesis of reflux esophagitis and Barrett's esophagus. Esophagus. 2021;18:388-397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 76. | Myers JA, Miller JS. Exploring the NK cell platform for cancer immunotherapy. Nat Rev Clin Oncol. 2021;18:85-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 830] [Article Influence: 166.0] [Reference Citation Analysis (0)] |
| 77. | Liu T, Dai X, Xu Y, Guan T, Hong L, Zaib T, Zhou Q, Cheng K, Zhou X, Ma C, Sun P. CD22 is a potential target of CAR-NK cell therapy for esophageal squamous cell carcinoma. J Transl Med. 2023;21:710. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 78. | Lim KS, Mimura K, Kua LF, Shiraishi K, Kono K. Implication of Highly Cytotoxic Natural Killer Cells for Esophageal Squamous Cell Carcinoma Treatment. J Immunother. 2018;41:261-273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 79. | Dhar P, Wu JD. NKG2D and its ligands in cancer. Curr Opin Immunol. 2018;51:55-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 162] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 80. | Ferrari de Andrade L, Tay RE, Pan D, Luoma AM, Ito Y, Badrinath S, Tsoucas D, Franz B, May KF Jr, Harvey CJ, Kobold S, Pyrdol JW, Yoon C, Yuan GC, Hodi FS, Dranoff G, Wucherpfennig KW. Antibody-mediated inhibition of MICA and MICB shedding promotes NK cell-driven tumor immunity. Science. 2018;359:1537-1542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 304] [Cited by in RCA: 343] [Article Influence: 49.0] [Reference Citation Analysis (0)] |
| 81. | Watanabe M, Kono K, Kawaguchi Y, Mizukami Y, Mimura K, Maruyama T, Izawa S, Fujii H. NK cell dysfunction with down-regulated CD16 and up-regulated CD56 molecules in patients with esophageal squamous cell carcinoma. Dis Esophagus. 2010;23:675-681. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 36] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 82. | Yang YM, Hong P, Xu WW, He QY, Li B. Advances in targeted therapy for esophageal cancer. Signal Transduct Target Ther. 2020;5:229. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 209] [Cited by in RCA: 315] [Article Influence: 63.0] [Reference Citation Analysis (0)] |
| 83. | Li J, Qiu G, Fang B, Dai X, Cai J. Deficiency of IL-18 Aggravates Esophageal Carcinoma Through Inhibiting IFN-γ Production by CD8(+)T Cells and NK Cells. Inflammation. 2018;41:667-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 84. | Zheng Y, Li Y, Tang B, Zhao Q, Wang D, Liu Y, Guo M, Zhao S, Qi Y, Zhang Y, Huang L. IL-6-induced CD39 expression on tumor-infiltrating NK cells predicts poor prognosis in esophageal squamous cell carcinoma. Cancer Immunol Immunother. 2020;69:2371-2380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 85. | Wu J, Gao FX, Wang C, Qin M, Han F, Xu T, Hu Z, Long Y, He XM, Deng X, Ren DL, Dai TY. IL-6 and IL-8 secreted by tumour cells impair the function of NK cells via the STAT3 pathway in oesophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2019;38:321. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 134] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 86. | Orecchioni M, Ghosheh Y, Pramod AB, Ley K. Macrophage Polarization: Different Gene Signatures in M1(LPS+) vs. Classically and M2(LPS-) vs. Alternatively Activated Macrophages. Front Immunol. 2019;10:1084. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 765] [Cited by in RCA: 1375] [Article Influence: 229.2] [Reference Citation Analysis (0)] |
| 87. | Uehara K, Iwashita H, Tanabe Y, Kurima K, Oshiro M, Kina S, Ota A, Iwashita A, Kinjo T. Esophageal Xanthoma: Presence of M2 Macrophages Suggests Association with Late Inflammatory and Reparative Processes. Open Med (Wars). 2017;12:335-339. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 88. | Jiang CH, Liang WH, Li FP, Xie YF, Yuan X, Zhang HJ, Li M, Li JF, Zhang AZ, Yang L, Liu CX, Pang LJ, Li F, Hu JM. Distribution and prognostic impact of M1 macrophage on esophageal squamous cell carcinoma. Carcinogenesis. 2021;42:537-545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 89. | Yang H, Zhang Q, Xu M, Wang L, Chen X, Feng Y, Li Y, Zhang X, Cui W, Jia X. CCL2-CCR2 axis recruits tumor associated macrophages to induce immune evasion through PD-1 signaling in esophageal carcinogenesis. Mol Cancer. 2020;19:41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 295] [Article Influence: 59.0] [Reference Citation Analysis (0)] |
| 90. | Lu Z, Zheng S, Liu C, Wang X, Zhang G, Wang F, Wang S, Huang J, Mao S, Lei Y, Wang Z, Sun N, He J. S100A7 as a potential diagnostic and prognostic biomarker of esophageal squamous cell carcinoma promotes M2 macrophage infiltration and angiogenesis. Clin Transl Med. 2021;11:e459. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 38] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 91. | Sun Y, Qian Y, Chen C, Wang H, Zhou X, Zhai W, Qiu L, Ning H, Zhao Y, Shi C, Han L, Qi Y, Wu Y, Gao Y. Extracellular vesicle IL-32 promotes the M2 macrophage polarization and metastasis of esophageal squamous cell carcinoma via FAK/STAT3 pathway. J Exp Clin Cancer Res. 2022;41:145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 31] [Article Influence: 10.3] [Reference Citation Analysis (1)] |
| 92. | Wang Y, Lyu Z, Qin Y, Wang X, Sun L, Zhang Y, Gong L, Wu S, Han S, Tang Y, Jia Y, Kwong DL, Kam N, Guan XY. FOXO1 promotes tumor progression by increased M2 macrophage infiltration in esophageal squamous cell carcinoma. Theranostics. 2020;10:11535-11548. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 113] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 93. | Sui X, Chen C, Zhou X, Wen X, Shi C, Chen G, Liu J, He Z, Yao Y, Li Y, Gao Y. Integrative analysis of bulk and single-cell gene expression profiles to identify tumor-associated macrophage-derived CCL18 as a therapeutic target of esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2023;42:51. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 24] [Reference Citation Analysis (0)] |
| 94. | Chen J, Zhao D, Zhang L, Zhang J, Xiao Y, Wu Q, Wang Y, Zhan Q. Tumor-associated macrophage (TAM)-derived CCL22 induces FAK addiction in esophageal squamous cell carcinoma (ESCC). Cell Mol Immunol. 2022;19:1054-1066. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 40] [Reference Citation Analysis (0)] |
| 95. | Mi X, Xu R, Hong S, Xu T, Zhang W, Liu M. M2 Macrophage-Derived Exosomal lncRNA AFAP1-AS1 and MicroRNA-26a Affect Cell Migration and Metastasis in Esophageal Cancer. Mol Ther Nucleic Acids. 2020;22:779-790. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 102] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 96. | Qiao X, Cheng Z, Xue K, Xiong C, Zheng Z, Jin X, Li J. Tumor-associated macrophage-derived exosomes LINC01592 induce the immune escape of esophageal cancer by decreasing MHC-I surface expression. J Exp Clin Cancer Res. 2023;42:289. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 30] [Reference Citation Analysis (0)] |
| 97. | Hegde S, Leader AM, Merad M. MDSC: Markers, development, states, and unaddressed complexity. Immunity. 2021;54:875-884. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 437] [Article Influence: 109.3] [Reference Citation Analysis (0)] |
| 98. | Gabitass RF, Annels NE, Stocken DD, Pandha HA, Middleton GW. Elevated myeloid-derived suppressor cells in pancreatic, esophageal and gastric cancer are an independent prognostic factor and are associated with significant elevation of the Th2 cytokine interleukin-13. Cancer Immunol Immunother. 2011;60:1419-1430. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 408] [Cited by in RCA: 481] [Article Influence: 34.4] [Reference Citation Analysis (0)] |
| 99. | Li P, Chen X, Qin G, Yue D, Zhang Z, Ping Y, Wang D, Zhao X, Song M, Zhao Q, Li J, Liu S, Zhang C, Lian J, Cao L, Li F, Huang L, Wang L, Yang L, Huang J, Li H, Zhang B, Zhang Y. Maelstrom Directs Myeloid-Derived Suppressor Cells to Promote Esophageal Squamous Cell Carcinoma Progression via Activation of the Akt1/RelA/IL8 Signaling Pathway. Cancer Immunol Res. 2018;6:1246-1259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 100. | Yue D, Liu S, Zhang T, Wang Y, Qin G, Chen X, Zhang H, Wang D, Huang D, Wang F, Wang L, Zhao S, Zhang Y. NEDD9 promotes cancer stemness by recruiting myeloid-derived suppressor cells via CXCL8 in esophageal squamous cell carcinoma. Cancer Biol Med. 2021;18:705-720. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 101. | Karakasheva TA, Waldron TJ, Eruslanov E, Kim SB, Lee JS, O'Brien S, Hicks PD, Basu D, Singhal S, Malavasi F, Rustgi AK. CD38-Expressing Myeloid-Derived Suppressor Cells Promote Tumor Growth in a Murine Model of Esophageal Cancer. Cancer Res. 2015;75:4074-4085. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 137] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 102. | Jiao ZJ, Gao JJ, Hua SH, Chen DY, Wang WH, Wang H, Wang XH, Xu HX. Correlation between circulating myeloid-derived suppressor cells and Th17 cells in esophageal cancer. World J Gastroenterol. 2012;18:5454-5461. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 21] [Cited by in RCA: 24] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 103. | Jianmin P, Qinchao H, Chunyang W, Jiayu Z, Siyu W, Li W, Juan X, Bin C. Depletion of Gr1+ myeloid cells attenuates high-fat-diet-aggravated esophageal squamous cell carcinoma in mice. Carcinogenesis. 2023;44:587-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 104. | Wen J, Xuan B, Gao Y, Liu Y, Wang L, He L, Meng X, Zhou T, Tao Y, Guo K, Wang Y. Lnc-17Rik promotes the immunosuppressive function of Myeloid-Derived suppressive cells in esophageal cancer. Cell Immunol. 2023;385:104676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 105. | Burr ML, Sparbier CE, Chan KL, Chan YC, Kersbergen A, Lam EYN, Azidis-Yates E, Vassiliadis D, Bell CC, Gilan O, Jackson S, Tan L, Wong SQ, Hollizeck S, Michalak EM, Siddle HV, McCabe MT, Prinjha RK, Guerra GR, Solomon BJ, Sandhu S, Dawson SJ, Beavis PA, Tothill RW, Cullinane C, Lehner PJ, Sutherland KD, Dawson MA. An Evolutionarily Conserved Function of Polycomb Silences the MHC Class I Antigen Presentation Pathway and Enables Immune Evasion in Cancer. Cancer Cell. 2019;36:385-401.e8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 335] [Cited by in RCA: 449] [Article Influence: 74.8] [Reference Citation Analysis (0)] |
| 106. | Rockett JC, Darnton SJ, Crocker J, Matthews HR, Morris AG. Expression of HLA-ABC, HLA-DR and intercellular adhesion molecule-1 in oesophageal carcinoma. J Clin Pathol. 1995;48:539-544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 27] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 107. | Liu Q, Hao C, Su P, Shi J. Down-regulation of HLA class I antigen-processing machinery components in esophageal squamous cell carcinomas: association with disease progression. Scand J Gastroenterol. 2009;44:960-969. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 108. | Yang Y, Zhang J, Miao F, Wei J, Shen C, Shen Y, Xie W. Loss of heterozygosity at 6p21 underlying [corrected] HLA class I downregulation in Chinese primary esophageal squamous cell carcinomas. Tissue Antigens. 2008;72:105-114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 109. | Zhao X, Sun Q, Tian H, Cong B, Jiang X, Peng C. Loss of heterozygosity at 6p21 and HLA class I expression in esophageal squamous cell carcinomas in China. Asian Pac J Cancer Prev. 2011;12:2741-2745. [PubMed] |
| 110. | Nie Y, Yang G, Song Y, Zhao X, So C, Liao J, Wang LD, Yang CS. DNA hypermethylation is a mechanism for loss of expression of the HLA class I genes in human esophageal squamous cell carcinomas. Carcinogenesis. 2001;22:1615-1623. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 153] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 111. | Mari L, Hoefnagel SJM, Zito D, van de Meent M, van Endert P, Calpe S, Sancho Serra MDC, Heemskerk MHM, van Laarhoven HWM, Hulshof MCCM, Gisbertz SS, Medema JP, van Berge Henegouwen MI, Meijer SL, Bergman JJGHM, Milano F, Krishnadath KK. microRNA 125a Regulates MHC-I Expression on Esophageal Adenocarcinoma Cells, Associated With Suppression of Antitumor Immune Response and Poor Outcomes of Patients. Gastroenterology. 2018;155:784-798. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 74] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 112. | Wu AA, Drake V, Huang HS, Chiu S, Zheng L. Reprogramming the tumor microenvironment: tumor-induced immunosuppressive factors paralyze T cells. Oncoimmunology. 2015;4:e1016700. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 210] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 113. | Veen LM, Skrabanja TLP, Derks S, de Gruijl TD, Bijlsma MF, van Laarhoven HWM. The role of transforming growth factor β in upper gastrointestinal cancers: A systematic review. Cancer Treat Rev. 2021;100:102285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 114. | Moreau JM, Velegraki M, Bolyard C, Rosenblum MD, Li Z. Transforming growth factor-β1 in regulatory T cell biology. Sci Immunol. 2022;7:eabi4613. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 136] [Article Influence: 45.3] [Reference Citation Analysis (0)] |
| 115. | Peng D, Fu M, Wang M, Wei Y, Wei X. Targeting TGF-β signal transduction for fibrosis and cancer therapy. Mol Cancer. 2022;21:104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 590] [Article Influence: 196.7] [Reference Citation Analysis (0)] |
| 116. | Tan B, Khattak A, Felip E, Kelly K, Rich P, Wang D, Helwig C, Dussault I, Ojalvo LS, Isambert N. Bintrafusp Alfa, a Bifunctional Fusion Protein Targeting TGF-β and PD-L1, in Patients with Esophageal Adenocarcinoma: Results from a Phase 1 Cohort. Target Oncol. 2021;16:435-446. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 117. | Tau GZ, Cowan SN, Weisburg J, Braunstein NS, Rothman PB. Regulation of IFN-gamma signaling is essential for the cytotoxic activity of CD8(+) T cells. J Immunol. 2001;167:5574-5582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 69] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 118. | Wu Q, Zhang W, Wang Y, Min Q, Zhang H, Dong D, Zhan Q. MAGE-C3 promotes cancer metastasis by inducing epithelial-mesenchymal transition and immunosuppression in esophageal squamous cell carcinoma. Cancer Commun (Lond). 2021;41:1354-1372. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 119. | Huang J, Li J, Li Y, Lu Z, Che Y, Mao S, Lei Y, Zang R, Zheng S, Liu C, Wang X, Li N, Sun N, He J. Interferon-inducible lncRNA IRF1-AS represses esophageal squamous cell carcinoma by promoting interferon response. Cancer Lett. 2019;459:86-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 120. | Wang Y, Liu D, Chen P, Koeffler HP, Tong X, Xie D. Negative feedback regulation of IFN-gamma pathway by IFN regulatory factor 2 in esophageal cancers. Cancer Res. 2008;68:1136-1143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 42] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 121. | Wang Y, Liu DP, Chen PP, Koeffler HP, Tong XJ, Xie D. Involvement of IFN regulatory factor (IRF)-1 and IRF-2 in the formation and progression of human esophageal cancers. Cancer Res. 2007;67:2535-2543. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 83] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 122. | Zhang C, Jiang F, Su C, Xie P, Xu L. Upregulation of long noncoding RNA SNHG20 promotes cell growth and metastasis in esophageal squamous cell carcinoma via modulating ATM-JAK-PD-L1 pathway. J Cell Biochem. 2019;120:11642-11650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 46] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 123. | Meraz MA, White JM, Sheehan KC, Bach EA, Rodig SJ, Dighe AS, Kaplan DH, Riley JK, Greenlund AC, Campbell D, Carver-Moore K, DuBois RN, Clark R, Aguet M, Schreiber RD. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431-442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1278] [Cited by in RCA: 1300] [Article Influence: 44.8] [Reference Citation Analysis (0)] |
| 124. | Faubert B, Solmonson A, DeBerardinis RJ. Metabolic reprogramming and cancer progression. Science. 2020;368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 501] [Cited by in RCA: 1501] [Article Influence: 300.2] [Reference Citation Analysis (0)] |
| 125. | Xia L, Oyang L, Lin J, Tan S, Han Y, Wu N, Yi P, Tang L, Pan Q, Rao S, Liang J, Tang Y, Su M, Luo X, Yang Y, Shi Y, Wang H, Zhou Y, Liao Q. The cancer metabolic reprogramming and immune response. Mol Cancer. 2021;20:28. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 721] [Article Influence: 180.3] [Reference Citation Analysis (0)] |
| 126. | Liu Y, Wang L, Fang L, Liu H, Tian H, Zheng Y, Fan T, Li C, He J. A Multi-Center Validated Subtyping Model of Esophageal Cancer Based on Three Metabolism-Related Genes. Front Oncol. 2021;11:772145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 127. | King RJ, Qiu F, Yu F, Singh PK. Metabolic and Immunological Subtypes of Esophageal Cancer Reveal Potential Therapeutic Opportunities. Front Cell Dev Biol. 2021;9:667852. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 128. | Wang T, Zhu Y, Chen L, Zhang W, Qi H, Shi X, Zhong M, Chen H, Li Q. ESRRG-PKM2 axis reprograms metabolism to suppress esophageal squamous carcinoma progression and enhance anti-PD-1 therapy efficacy. J Transl Med. 2023;21:605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 129. | Zhao F, Li Y, Dong Z, Zhang D, Guo P, Li Z, Li S. Identification of A Risk Signature Based on Lactic Acid Metabolism-Related LncRNAs in Patients With Esophageal Squamous Cell Carcinoma. Front Cell Dev Biol. 2022;10:845293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 10] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 130. | Tan L, Cheng D, Wen J, Huang K, Zhang Q. Identification of prognostic hypoxia-related genes signature on the tumor microenvironment in esophageal cancer. Math Biosci Eng. 2021;18:7743-7758. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 131. | He Z, Liu H, Guan H, Ji J, Jiang Y, Zhang N, Song Z, Wang X, Shen P, Wang H, Cui R. Construction of a Prognostic Model for Hypoxia-Related LncRNAs and Prediction of the Immune Landscape in the Digestive System Pan-Cancer. Front Oncol. 2022;12:812786. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 132. | Zhao M, Li M, Zheng Y, Hu Z, Liang J, Bi G, Bian Y, Sui Q, Zhan C, Lin M, Wang Q. Identification and analysis of a prognostic ferroptosis and iron-metabolism signature for esophageal squamous cell carcinoma. J Cancer. 2022;13:1611-1622. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 25] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 133. | Niu R, Zhao F, Dong Z, Li Z, Li S. A stratification system of ferroptosis and iron-metabolism related LncRNAs guides the prediction of the survival of patients with esophageal squamous cell carcinoma. Front Oncol. 2022;12:1010074. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 134. | Zhang Z, Jin G, Zhao J, Deng S, Chen F, Wuyun G, Zhao L, Li Q. Mitochondrial energy metabolism correlates with an immunosuppressive tumor microenvironment and poor prognosis in esophageal squamous cell carcinoma. Comput Struct Biotechnol J. 2023;21:4118-4133. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 135. | Guo Y, Pan S, Ke Y, Pan J, Li Y, Ma H. Seven Fatty Acid Metabolism-Related Genes as Potential Biomarkers for Predicting the Prognosis and Immunotherapy Responses in Patients with Esophageal Cancer. Vaccines (Basel). 2022;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 136. | Chen Y, Chen J, Guo D, Yang P, Chen S, Zhao C, Xu C, Zhang Q, Lin C, Zhong S, Zhang S. Tryptophan Metabolites as Biomarkers for Esophageal Cancer Susceptibility, Metastasis, and Prognosis. Front Oncol. 2022;12:800291. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |