Published online Apr 28, 2024. doi: 10.3748/wjg.v30.i16.2184
Peer-review started: January 20, 2024
First decision: January 31, 2024
Revised: February 9, 2024
Accepted: March 19, 2024
Article in press: March 19, 2024
Published online: April 28, 2024
Processing time: 96 Days and 13.4 Hours
MicroRNAs (miRNAs), small non-coding RNAs composed of 18–24 nucleotides, are potent regulators of gene expression, contributing to the regulation of more than 30% of protein-coding genes. Considering that miRNAs are regulators of inflammatory pathways and the differentiation of intestinal epithelial cells, there is an interest in exploring their importance in inflammatory bowel disease (IBD). IBD is a chronic and multifactorial disease of the gastrointestinal tract; the main forms are Crohn's disease and ulcerative colitis. Several studies have investigated the dysregulated expression of miRNAs in IBD, demonstrating their important roles as regulators and potential biomarkers of this disease. This editorial presents what is known and what is expected regarding miRNAs in IBD. Although the important regulatory roles of miRNAs in IBD are clearly established, biomarkers for IBD that can be applied in clinical practice are lacking, emphasizing the importance of further studies. Discoveries regarding the influence of miRNAs on the inflammatory process and the exploration of their role in gene regulation are expected to provide a basis for the use of miRNAs not only as potent biomarkers in IBD but also as therapeutic targets for the control of inflammatory processes in personalized medicine.
Core Tip: MicroRNAs (miRNAs) function in the regulation of inflammatory pathways and the differentiation of intestinal epithelial cells. There is substantial evidence for the important regulatory roles of miRNAs in inflammatory bowel disease (IBD), suggesting that they may serve as biomarkers. Therefore, this editorial aims to present what is already known and what the expectations are regarding the role of miRNAs in IBD.
- Citation: Oliveira ECS, Quaglio AEV, Grillo TG, Di Stasi LC, Sassaki LY. MicroRNAs in inflammatory bowel disease: What do we know and what can we expect? World J Gastroenterol 2024; 30(16): 2184-2190
- URL: https://www.wjgnet.com/1007-9327/full/v30/i16/2184.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i16.2184
MicroRNAs (miRNAs) are small non-coding RNAs composed of 18 to 24 nucleotides that are recognized as potent downregulators of gene expression or messenger RNA translation[1-3]. They regulate more than 30% of protein-coding genes and play important roles in cell survival, differentiation, proliferation, apoptosis, cell cycle control, and homeostasis[1,2,4].
Because miRNAs regulate inflammatory cellular signaling pathways and the intestinal epithelial cell differentiation, with an important role in the homeostasis of the intestinal mucosa[1,5,6], we explored the importance of miRNAs in inflammatory bowel diseases (IBD). IBD is a chronic disease of the gastrointestinal tract with a multifactorial and imbricated etiology, involving genetic, immune, and environmental factors. The main representatives of IBD are Crohn's disease (CD) and ulcerative colitis (UC), which differ both clinically and pathophysiologically[7].
In the last decade, several clinical and experimental studies of IBD have improved our understanding of miRNAs and contributed to the search for new and more accurate diagnostic markers and targets for treatment. Based on this, this editorial aims to present what is already known and what the expectations are regarding the role of miRNAs in IBD.
miRNAs play an important role as cellular and homeostasis regulators and may interfere with important inflammatory signaling pathways, such as the nuclear transcription factor kappa B (NF-κB), interleukin 23 (IL23)/IL23R, and IL-6/STAT3 pathways[8-11]. Therefore, alterations in the expression of certain miRNAs may be related to various immune diseases, including IBD. To evaluate their expression profiles in diseases, miRNAs can be quantified using samples of body fluids (circulating miRNAs), such as blood and feces, as well as through homogenized tissue biopsies (tissue miRNAs) using microarray profiling, quantitative real-time PCR, and next-generation sequencing techniques[12-15].
One of the first studies focusing on miRNAs in IBD identified three under expressed miRNAs (miR-192, miR-375, and miR-422b) and eight overexpressed miRNAs (miR-16, miR-21, miR-23a, miR-24, miR-29a, miR-126, miR-195, and Let-7f) in tissues from patients with active UC compared to tissues from healthy individuals[5]. Another study conducted by the same research group evaluated colonic tissues from patients with CD and identified three upregulated miRNAs (miR-23b, miR-106a, and miR-191) and two downregulated miRNAs (miR-19b and miR-629) when compared with levels in colonic tissues from healthy individuals[6]. Neither upregulated nor downregulated miRNAs in CD patients with were altered in UC patients[6], indicating that the miRNA expression profile differs between CD and UC.
These studies prompted researchers to investigate the role of miRNA dysregulation in IBD, both as regulators of inflammatory processes and as IBD biomarkers. Several studies have focused on the detection of miRNA biomarkers for IBD, revealing miR-223, miR-155, and miRNA-320a as key candidates.
A study using serum samples from IBD patients suggested that miR-223 is a potential biomarker, as levels of this miRNA were higher in both CD and UC samples than in healthy individuals[16]. Additionally, miR-223 expression is associated with the active phase of the disease[16]. Another study using the same sample type corroborated the increase in miR-223 expression in patients when compared with that in healthy individuals[17]. However, when active patients were compared with those in remission, miR-223 expression showed no significant differences[17]. Similarly, miR-155 expression differed between patients and healthy individuals[17], and this was corroborated by another study that demonstrated higher miR-155 expression levels in the colon tissues of patients than in samples from healthy individuals[18], suggesting that miR-155 is a potential biomarker of IBD activity. Similarly, a recent study on IBD demonstrated that miR-320a expression is higher in blood samples of IBD patients than in samples from healthy individuals and is significantly higher in patients with active IBD than in patients in remission[19], highlighting that miR-320a is another promising biomarker of disease activity in IBD patients.
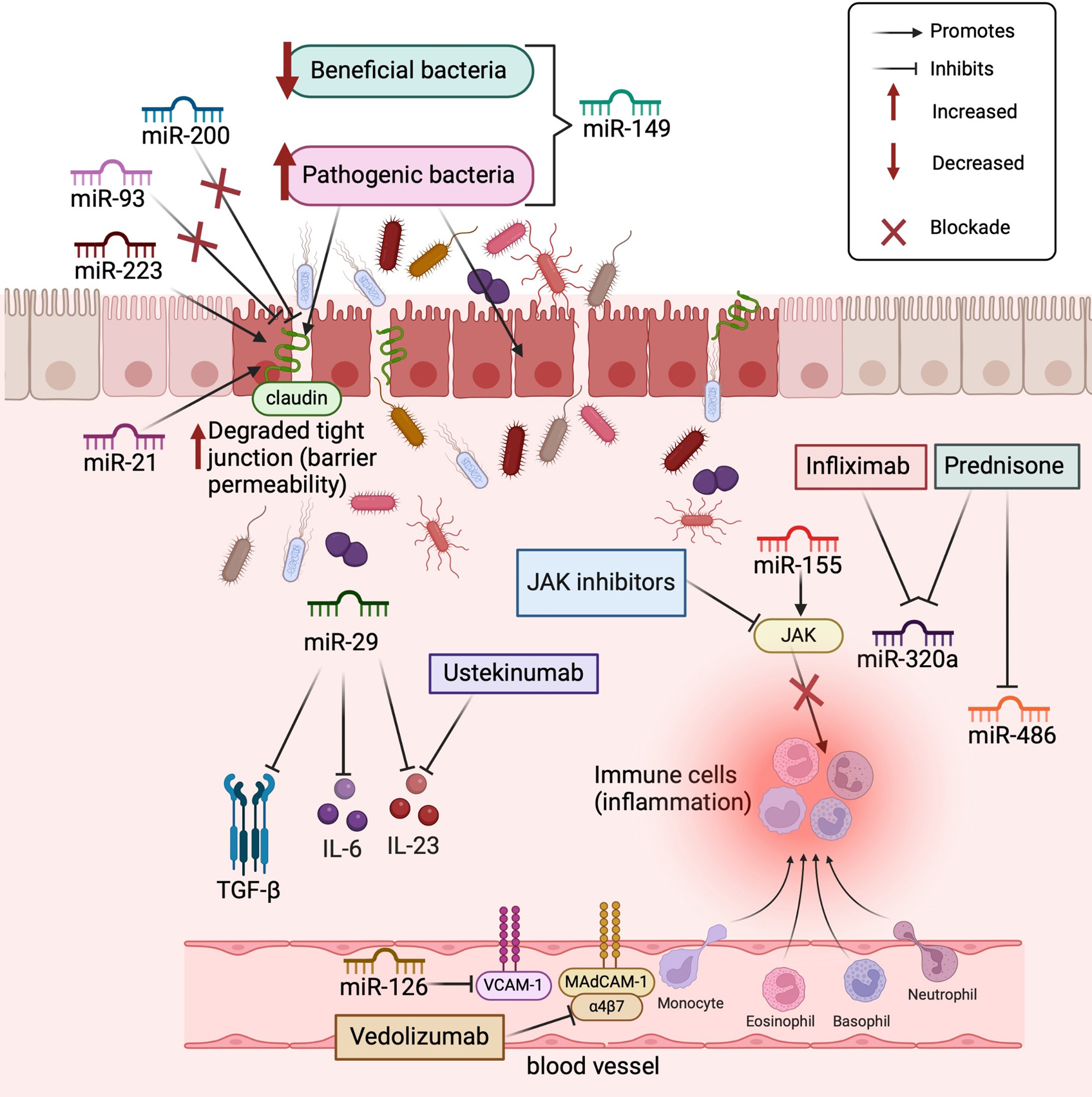
The search for miRNA biomarkers of IBD has also led to the identification of the roles of miRNAs in inflammatory signaling pathways modulated by various drugs, such as prednisone, Janus kinase (JAK) inhibitors, and monoclonal antibodies, including infliximab, ustekinumab, and vedolizumab (Figure 1). Furthermore, some miRNAs can be regulated when exposed to certain drugs, as demonstrated in the pediatric population, where it was observed that miR-146a, miR-146b, and miR-320a were reduced with the use of infliximab and prednisone, whereas miR-486 expression was reduced only with the use of prednisone[20].
In a study of dextran sodium sulfate (DSS)-induced intestinal inflammation in mice, tail vein injection of miR-29 linked to a nanoparticle significantly inhibited the intestinal anti-inflammatory process, which was related to reductions in transforming growth factor-β, IL-6, and IL-23 expression[21]. Notably, the cytokine IL-23 is the target of the monoclonal antibody ustekinumab, which is currently used to treat CD, indicating that miR-29 is a potential therapeutic target for IBD. In contrast, miR-155 targets regulatory proteins in the JAK signaling pathway, which controls immune cells and, consequently, inflammatory processes[22]. This mechanism is the same as that of JAK inhibitors, such as tofacitinib and upadacitinib, which are oral drugs used to treat UC.
In vitro and in vivo analyses have shown that miR-126 inhibits leukocyte adhesion to endothelial cells through the regulation of vascular cell adhesion molecule-1 (VCAM-1)[23]. Vedolizumab, indicated for the treatment of UC and CD, also inhibits the migration of leukocytes into inflamed intestinal tissue, blocking their interaction with mucosal addressin-cell adhesion molecule 1 (MAdCAM-1) in the intestinal vasculature[24], a mechanism similar to that described for miR-126. The development of new drugs capable of modulating the expression of these miRNAs is a promising approach for controlling the inflammatory response.
Other studies have also reported an important relationship between miRNAs and intestinal permeability, considering that a loss of the intestinal barrier is one of the processes that triggers intestinal inflammation in IBD patients[25,26]. In this context, miR-21 plays an important role in regulating the intestinal epithelial barrier, as it blocks the production of the RhoB (Ras homolog gene family member B) protein[25]. A reduction in RhoB protein levels results in the loss of tight junctions, increasing intestinal permeability with subsequent increased exposure to antigens, which serves as a trigger for the intestinal inflammatory process[25].
Besides miR-21, other miRNAs destroy tight junctions and consequently weaken the intestinal epithelial barrier, including miR-191, miR-212, miR-675, miR-874, miR-122a, miR-34c, miR-150, and miR-01a[26]. In contrast, miR-200 and miR-93 assist in the protection of tight junctions, maintaining intestinal barrier function[26]. These recent findings indicate that several miRNAs are important targets for the development of treatments aimed at maintaining the integrity of the intestinal barrier, thereby preventing overexposure to antigens as a trigger in the inflammatory processes involved in IBD pathophysiology.
A recent review published by our research group[27] reported that some miRNAs modulate the intestinal microbiota and induce dysbiosis in IBD patients, whereas the intestinal microbiota can also regulate the expression of miRNAs, establishing a complex relationship between these taxa and their host[27]. For example, in a DSS-induced intestinal inflammation model, miR-149 deletion induces changes in the microbiota and promotes intestinal inflammatory processes[28]. Additionally, the use of probiotics in mice, in addition to improving dysbiosis, reduces the expression of miR-155, miR-223, miR-150, and miR-143, which act on both intestinal permeability and the pro-inflammatory response, improving intestinal inflammation[29].
From the first studies on miRNAs to recent research, there has been a great evolution in our understanding of their functions in the immune system and inflammatory processes. Despite this, there is a lack of data to support the use of miRNAs as biomarkers for chronic diseases, such as IBD, limiting the application of scientific knowledge to clinical practice. To date, specific miRNAs have not been described as biomarkers for differentiating UC from CD, as markers of inflammatory activity, or as predictors of response to clinical treatment. However, research work has yielded new discoveries and insights into the roles of miRNAs in the pathogenesis and maintenance of the inflammatory process in IBD. In the near future, these findings are expected to lead to novel applications of these biomarkers in clinical practice.
Regarding IBD monitoring, few biomarkers for disease activity, such as fecal calprotectin, have been validated[30,31] and therefore colonoscopy is needed to visualize the intestinal mucosa. Considering that colonoscopy is invasive with inherent risks, new markers with high accuracy are required. miRNAs participate in various processes, including inflammation. Therefore, miRNAs can serve as appropriate biomarkers for the diagnosis and therapeutic monitoring of IBD patients. In addition to contributing to a more specific diagnosis and treatment, the identification of miRNAs as blood or fecal markers of IBD provides a less invasive[8], faster, and more accurate alternative to colonoscopy. The disadvantage of miRNA markers is the high cost of tests, which hinders their applicability in clinical practice.
Regarding the role of miRNAs in differentiating disease activity from remission, miR-223 expression is higher in the serum, tissue from the terminal ileum, and fecal samples of active CD compared to inactive CD[32]. Furthermore, miR-223 levels in serum, intestinal tissue, and fecal samples were correlated with Crohn´s Disease Activity Index, and fecal miR-223 was correlated with fecal calprotectin. These findings indicated that fecal miR-223 may be a novel, noninvasive biomarker for estimating disease activity in CD patients[32].
The Selecting Therapeutic Targets in IBD (Stride) II and IBD consensus established clinical response and remission as well as normalization of C-reactive protein as immediate and short-term targets, and endoscopic healing, restoration of quality of life, and absence of disability as long-term targets in IBD treatment, including mucosal healing as therapeutic goals[33,34]. The expectations include histological healing in UC and transmural healing in CD[33]. The identification of specific miRNAs correlated with mucosal healing, histological healing, or even transmural healing would be a revolutionary milestone in IBD, facilitating patient monitoring and the development of treat-to-target strategies using a simple blood marker.
Several miRNAs act on the same signaling pathways that are targets of drugs used to treat IBD[13,20-24], suggesting the potential use of miRNAs as IBD targets. The modulation of these miRNAs may positively interfere with patient responses to treatment, which is a promising strategy for drug development, as previously reported for miR-29[21], miR-155[22], and miR-126[23].
Regarding the modulation of miRNAs as a therapeutic strategy, a clinical trial is evaluating the efficacy of the small molecule drug candidate obefazimod for the treatment of moderate to severe active UC[35]. Obefazimod is the only known molecule that modulates miRNAs, as it enhances miR-124 expression, which is responsible for modulating inflammation and the innate immune response activated in IBD[35]. Results from this clinical trial are expected soon. Considering this mechanism of action, future studies should include other miRNAs as therapeutic targets; for example, the activation of miRNAs that help maintain the intestinal barrier, including miR-200 and miR-93, or inhibition of miRNAs that negatively regulate inflammatory processes, including miR-223 and miR-320a, should be evaluated.
In addition to their potential use as diagnostic markers and therapeutic targets, miRNAs have been studied as predictors of clinical and endoscopic responses in IBD patients. Reduced serum expression levels of let-7e at week 14 and miR-126 at week 54 were associated with clinical remission at weeks 14 and 54 and endoscopic remission at week 54 in 37 patients with CD treated with anti-tumor necrosis factor α therapy[36]. Another study found that increased let-7d and let-7e expression were associated with clinical remission at week 14 after infliximab induction therapy in CD, suggesting that these miRNAs are possible therapeutic biomarkers in CD patients treated with infliximab[37].
In severe acute colitis, a study evaluated tissue miRNAs associated with the response to intravenous (IV) steroids and in response to infliximab or cyclosporine in steroid-refractory patients[13]. Initially, 15 miRNAs associated with the response to IV steroids were identified (hp_hsa-mir-3934, hp_hsamir-3667, hp_hsa-mir-100, hsa-miR-603, hsa-miR-718, hsa-miR-4259, hp_hsa-mir-193b, hsa-miR-3150a-5p, hp_hsa-mir-1260b, hsa-miR-938, hsa-miR-3128, hsa-miR-4423-3p, hsa-miR-518b, hsa-miR-1468, and hsa-miR-3152-3p), in addition to six miRNAs associated with the response to infliximab (hsa-miR-4423-3p, hsa-miR-3128, hsa-miR-3152-3p, hp_hsa-miR-193b, hsa-mi-R938, and hp_hsa-miR-100) and four miRNA associated with the ciclosporin response (hsa-mi-R4423-3p, hsa-mi-R938, hsa-mi-R518b, and hp_hsa-miR-100). In a validation cohort study, among the miRNAs initially identified, only two were significantly differentially expressed between responders and non-responders: miR-3934 for IV steroids and miR-938 for second-line treatment (infliximab or cyclosporine)[13].
In the pediatric population, five serum miRNAs (miR-126, let-7c, miR-146a, miR-146b, and miR-320a) were associated with the clinical response; further studies are needed to validate these miRNAs as biomarkers of infliximab and glucocorticoid treatment response within this specific population[38].
In the future, it will be important to consider the modulation of miRNAs by drugs or probiotics and the use of miRNAs as treatment itself (Table 1). A recent review has highlighted promising results in preclinical cancer studies when using a single miRNA to target multiple genes[39]. Despite these advances, several limitations and challenges must be overcome to enable the use of miRNAs in IBD in clinical practice, as described in Table 2.
| What is already known about miRNAs in IBD? | What can we expect from miRNAs in IBD? | Ref. |
| Regulate cellular processes and homeostasis | Discover new functions in the pathophysiology of IBD | [8-11] |
| Differentially expressed between patients with IBD and healthy controls | Can be used as diagnostic markers for IBD | [5,6,16-19] |
| Differentially expressed between UC and CD | Can be used to differentiate UC from CD | [6] |
| Differentially expressed with respect to disease activity | Can be used as biomarkers of inflammatory activity | [16,17,19,32] |
| Regulated in response to drug exposure | Can be used as markers of drug responses | [20] |
| Regulate the expression of inflammatory cytokines | Potential therapeutic targets in IBD | [21,22,23] |
| Act on the intestinal barrier | Potential therapeutic targets in IBD | [25,26] |
| Modulate the intestinal microbiota | Potential therapeutic targets in IBD | [27-29] |
| High exam cost |
| Better definition of miRNAs as diagnostic markers in IBD (differentiation between UC and CD and diagnosis of disease activity) |
| Few validation studies of miRNAs as blood, fecal, and endoscopic biomarkers |
| Better definition of the differences between the expression of fecal, blood, and tissue miRNAs |
| Lack of evidence validating miRNAs as a tool for evaluating mucosal healing |
| Lack of evidence validating miRNAs as a tool for evaluating histological remission |
| Lack of evidence validating miRNAs predictors of clinical and endoscopic responses |
| Lack of evidence validating miRNAs in treatment monitoring |
| Lack of evidence validating miRNAs as predictors of severe disease |
| Insufficient data on the role of miRNAs in modulating the inflammatory response |
| Insufficient data on the effects of miRNAs on the intestinal barrier, intestinal microbiota, and the response to probiotics |
Previous studies have demonstrated that miRNAs are important mediators of the inflammatory process in IBD patients and represent potential therapeutic targets for the development of new drugs. Experimental and clinical studies have focused on the modulation of miRNA expression, either by stimulating miRNA expression with anti-inflammatory functions or inhibiting miRNA expression with pro-inflammatory functions. In this editorial, we present the main findings and future perspectives regarding miRNAs and their roles in IBD patients. Currently, there is no specific miRNA biomarker for IBD, nor is there a specific marker for UC or CD. It is expected that in the future, miRNAs will be developed as sensitive and specific diagnostic, therapeutic, and prognostic biomarkers in IBD, providing a non-invasive and accessible tool for effective monitoring.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Brazil
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Moldovan CA, Romania; Zheng L, China S-Editor: Li L L-Editor: Filipodia P-Editor: Chen YX
| 1. | Archanioti P, Gazouli M, Theodoropoulos G, Vaiopoulou A, Nikiteas N. Micro-RNAs as regulators and possible diagnostic bio-markers in inflammatory bowel disease. J Crohns Colitis. 2011;5:520-524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 50] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 2. | Coskun M, Bjerrum JT, Seidelin JB, Nielsen OH. MicroRNAs in inflammatory bowel disease--pathogenesis, diagnostics and therapeutics. World J Gastroenterol. 2012;18:4629-4634. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 69] [Cited by in RCA: 81] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 3. | Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther. 2016;1:15004. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1765] [Cited by in RCA: 1703] [Article Influence: 189.2] [Reference Citation Analysis (0)] |
| 4. | Dalal SR, Kwon JH. The Role of MicroRNA in Inflammatory Bowel Disease. Gastroenterol Hepatol (N Y). 2010;6:714-722. [PubMed] |
| 5. | Wu F, Zikusoka M, Trindade A, Dassopoulos T, Harris ML, Bayless TM, Brant SR, Chakravarti S, Kwon JH. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology. 2008;135:1624-1635.e24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 365] [Cited by in RCA: 405] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 6. | Wu F, Zhang S, Dassopoulos T, Harris ML, Bayless TM, Meltzer SJ, Brant SR, Kwon JH. Identification of microRNAs associated with ileal and colonic Crohn's disease. Inflamm Bowel Dis. 2010;16:1729-1738. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 236] [Cited by in RCA: 239] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 7. | Oligschlaeger Y, Yadati T, Houben T, Condello Oliván CM, Shiri-Sverdlov R. Inflammatory Bowel Disease: A Stressed "Gut/Feeling". Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 74] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 8. | James JP, Riis LB, Malham M, Høgdall E, Langholz E, Nielsen BS. MicroRNA Biomarkers in IBD-Differential Diagnosis and Prediction of Colitis-Associated Cancer. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 71] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 9. | Mikami Y, Philips RL, Sciumè G, Petermann F, Meylan F, Nagashima H, Yao C, Davis FP, Brooks SR, Sun HW, Takahashi H, Poholek AC, Shih HY, Afzali B, Muljo SA, Hafner M, Kanno Y, O'Shea JJ. MicroRNA-221 and -222 modulate intestinal inflammatory Th17 cell response as negative feedback regulators downstream of interleukin-23. Immunity. 2021;54:514-525.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 39] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 10. | Ren W, Zhang X, Li Q, Pu C, Zhang D. Activating IL-6/STAT3 Enhances Protein Stability of Proteasome 20S α+β in Colorectal Cancer by miR-1254. Biomed Res Int. 2022;2022:4250013. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 11. | Zhang LZ, Xue H, Qiao CX, You WL, Di AT, Zhao G. MiR-223 promotes pyroptosis of enteritis cells through activating NF-κB signalling pathway by targeting SNIP1 in inflammatory bowel disease. Autoimmunity. 2021;54:362-372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 12. | Liu D, Saikam V, Skrada KA, Merlin D, Iyer SS. Inflammatory bowel disease biomarkers. Med Res Rev. 2022;42:1856-1887. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 80] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 13. | Morilla I, Uzzan M, Laharie D, Cazals-Hatem D, Denost Q, Daniel F, Belleannee G, Bouhnik Y, Wainrib G, Panis Y, Ogier-Denis E, Treton X. Colonic MicroRNA Profiles, Identified by a Deep Learning Algorithm, That Predict Responses to Therapy of Patients With Acute Severe Ulcerative Colitis. Clin Gastroenterol Hepatol. 2019;17:905-913. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 45] [Article Influence: 7.5] [Reference Citation Analysis (1)] |
| 14. | Mohammadi A, Kelly OB, Filice M, Kabakchiev B, Smith MI, Silverberg MS. Differential Expression of microRNAs in Peripheral Blood Mononuclear Cells Identifies Autophagy and TGF-Beta-Related Signatures Aberrantly Expressed in Inflammatory Bowel Disease. J Crohns Colitis. 2018;12:568-581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 15. | Rashid H, Hossain B, Siddiqua T, Kabir M, Noor Z, Ahmed M, Haque R. Fecal MicroRNAs as Potential Biomarkers for Screening and Diagnosis of Intestinal Diseases. Front Mol Biosci. 2020;7:181. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 16. | Wang H, Zhang S, Yu Q, Yang G, Guo J, Li M, Zeng Z, He Y, Chen B, Chen M. Circulating MicroRNA223 is a New Biomarker for Inflammatory Bowel Disease. Medicine (Baltimore). 2016;95:e2703. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 50] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 17. | Schönauen K, Le N, von Arnim U, Schulz C, Malfertheiner P, Link A. Circulating and Fecal microRNAs as Biomarkers for Inflammatory Bowel Diseases. Inflamm Bowel Dis. 2018;24:1547-1557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 102] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 18. | Shao X, Li J, Xu F, Chen D, Liu K. MiR-155-Mediated Deregulation of GPER1 Plays an Important Role in the Gender Differences Related to Inflammatory Bowel Disease. Can J Infect Dis Med Microbiol. 2020;2020:8811477. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 19. | Cordes F, Demmig C, Bokemeyer A, Brückner M, Lenze F, Lenz P, Nowacki T, Tepasse P, Schmidt HH, Schmidt MA, Cichon C, Bettenworth D. MicroRNA-320a Monitors Intestinal Disease Activity in Patients With Inflammatory Bowel Disease. Clin Transl Gastroenterol. 2020;11:e00134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 20. | Heier CR, Fiorillo AA, Chaisson E, Gordish-Dressman H, Hathout Y, Damsker JM, Hoffman EP, Conklin LS. Identification of Pathway-Specific Serum Biomarkers of Response to Glucocorticoid and Infliximab Treatment in Children with Inflammatory Bowel Disease. Clin Transl Gastroenterol. 2016;7:e192. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 52] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 21. | Fukata T, Mizushima T, Nishimura J, Okuzaki D, Wu X, Hirose H, Yokoyama Y, Kubota Y, Nagata K, Tsujimura N, Inoue A, Miyoshi N, Haraguchi N, Takahashi H, Hata T, Matsuda C, Kayama H, Takeda K, Doki Y, Mori M, Yamamoto H. The Supercarbonate Apatite-MicroRNA Complex Inhibits Dextran Sodium Sulfate-Induced Colitis. Mol Ther Nucleic Acids. 2018;12:658-671. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 22. | Pathak S, Grillo AR, Scarpa M, Brun P, D'Incà R, Nai L, Banerjee A, Cavallo D, Barzon L, Palù G, Sturniolo GC, Buda A, Castagliuolo I. MiR-155 modulates the inflammatory phenotype of intestinal myofibroblasts by targeting SOCS1 in ulcerative colitis. Exp Mol Med. 2015;47:e164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 84] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 23. | Tang Y, Chen Y, Guo Q, Zhang L, Liu H, Wang S, Wu X, Shen X, Tao L. MiR-126-Loaded Immunoliposomes against Vascular Endothelial Inflammation In Vitro and Vivo Evaluation. Pharmaceutics. 2023;15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 2] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 24. | Feagan BG, Rutgeerts P, Sands BE, Hanauer S, Colombel JF, Sandborn WJ, Van Assche G, Axler J, Kim HJ, Danese S, Fox I, Milch C, Sankoh S, Wyant T, Xu J, Parikh A; GEMINI 1 Study Group. Vedolizumab as induction and maintenance therapy for ulcerative colitis. N Engl J Med. 2013;369:699-710. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1576] [Cited by in RCA: 1866] [Article Influence: 155.5] [Reference Citation Analysis (1)] |
| 25. | Yang Y, Ma Y, Shi C, Chen H, Zhang H, Chen N, Zhang P, Wang F, Yang J, Zhu Q, Liang Y, Wu W, Gao R, Yang Z, Zou Y, Qin H. Overexpression of miR-21 in patients with ulcerative colitis impairs intestinal epithelial barrier function through targeting the Rho GTPase RhoB. Biochem Biophys Res Commun. 2013;434:746-752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 118] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 26. | Moein S, Vaghari-Tabari M, Qujeq D, Majidinia M, Nabavi SM, Yousefi B. MiRNAs and inflammatory bowel disease: An interesting new story. J Cell Physiol. 2019;234:3277-3293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 56] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 27. | Oliveira ECS, Quaglio AEV, Magro DO, Di Stasi LC, Sassaki LY. Intestinal Microbiota and miRNA in IBD: A Narrative Review about Discoveries and Perspectives for the Future. Int J Mol Sci. 2023;24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 22] [Reference Citation Analysis (0)] |
| 28. | Feng Q, Li Y, Zhang H, Wang Z, Nie X, Yao D, Han L, Chen WD, Wang YD. Deficiency of miRNA-149-3p shaped gut microbiota and enhanced dextran sulfate sodium-induced colitis. Mol Ther Nucleic Acids. 2022;30:208-225. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 29. | Rodríguez-Nogales A, Algieri F, Garrido-Mesa J, Vezza T, Utrilla MP, Chueca N, García F, Rodríguez-Cabezas ME, Gálvez J. Intestinal anti-inflammatory effect of the probiotic Saccharomyces boulardii in DSS-induced colitis in mice: Impact on microRNAs expression and gut microbiota composition. J Nutr Biochem. 2018;61:129-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 102] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 30. | State M, Negreanu L, Voiosu T, Voiosu A, Balanescu P, Mateescu RB. Surrogate markers of mucosal healing in inflammatory bowel disease: A systematic review. World J Gastroenterol. 2021;27:1828-1840. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 21] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (1)] |
| 31. | Sakurai T, Saruta M. Positioning and Usefulness of Biomarkers in Inflammatory Bowel Disease. Digestion. 2023;104:30-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 64] [Reference Citation Analysis (0)] |
| 32. | Zhang J, Guo Z, Wang Z, Zhu W, Li Q. Fecal miR-223 is a noninvasive biomarker for estimating Crohn's disease activity. Immun Inflamm Dis. 2023;11:e1131. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 33. | Turner D, Ricciuto A, Lewis A, D'Amico F, Dhaliwal J, Griffiths AM, Bettenworth D, Sandborn WJ, Sands BE, Reinisch W, Schölmerich J, Bemelman W, Danese S, Mary JY, Rubin D, Colombel JF, Peyrin-Biroulet L, Dotan I, Abreu MT, Dignass A; International Organization for the Study of IBD. STRIDE-II: An Update on the Selecting Therapeutic Targets in Inflammatory Bowel Disease (STRIDE) Initiative of the International Organization for the Study of IBD (IOIBD): Determining Therapeutic Goals for Treat-to-Target strategies in IBD. Gastroenterology. 2021;160:1570-1583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 473] [Cited by in RCA: 1645] [Article Influence: 411.3] [Reference Citation Analysis (1)] |
| 34. | Raine T, Bonovas S, Burisch J, Kucharzik T, Adamina M, Annese V, Bachmann O, Bettenworth D, Chaparro M, Czuber-Dochan W, Eder P, Ellul P, Fidalgo C, Fiorino G, Gionchetti P, Gisbert JP, Gordon H, Hedin C, Holubar S, Iacucci M, Karmiris K, Katsanos K, Kopylov U, Lakatos PL, Lytras T, Lyutakov I, Noor N, Pellino G, Piovani D, Savarino E, Selvaggi F, Verstockt B, Spinelli A, Panis Y, Doherty G. ECCO Guidelines on Therapeutics in Ulcerative Colitis: Medical Treatment. J Crohns Colitis. 2022;16:2-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 584] [Cited by in RCA: 558] [Article Influence: 186.0] [Reference Citation Analysis (0)] |
| 35. | Vermeire S, Sands BE, Tilg H, Tulassay Z, Kempinski R, Danese S, Bunganič I, Nitcheu J, Santo J, Scherrer D, Biguenet S, Ehrlich HJ, Steens JM, Gineste P, Sandborn WJ. ABX464 (obefazimod) for moderate-to-severe, active ulcerative colitis: a phase 2b, double-blind, randomised, placebo-controlled induction trial and 48 wk, open-label extension. Lancet Gastroenterol Hepatol. 2022;7:1024-1035. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 34] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 36. | Guglielmi G, Crucitta S, Bertani L, Ruglioni M, Baiano Svizzero G, Ceccarelli L, Del Re M, Danesi R, Costa F, Fogli S. Expression of Circulating let-7e and miR-126 May Predict Clinical Remission in Patients With Crohn's Disease Treated With Anti-TNF-α Biologics. Inflamm Bowel Dis. 2024;30:441-446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 1] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 37. | Fujioka S, Nakamichi I, Esaki M, Asano K, Matsumoto T, Kitazono T. Serum microRNA levels in patients with Crohn's disease during induction therapy by infliximab. J Gastroenterol Hepatol. 2014;29:1207-1214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 39] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 38. | Batra SK, Heier CR, Diaz-Calderon L, Tully CB, Fiorillo AA, van den Anker J, Conklin LS. Serum miRNAs Are Pharmacodynamic Biomarkers Associated With Therapeutic Response in Pediatric Inflammatory Bowel Disease. Inflamm Bowel Dis. 2020;26:1597-1606. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 39. | Liang L, He X. A narrative review of microRNA therapeutics: understanding the future of microRNA research. Precis Cancer Med. 2021;4:1-7. [DOI] [Full Text] |