Published online Apr 21, 2024. doi: 10.3748/wjg.v30.i15.2155
Peer-review started: January 5, 2024
First decision: January 22, 2024
Revised: February 2, 2024
Accepted: March 14, 2024
Article in press: March 14, 2024
Published online: April 21, 2024
Processing time: 105 Days and 0.9 Hours
Necroptosis has emerged as a novel molecular pathway that can be targeted by chemotherapy agents in the treatment of cancer. OSW-1, which is derived from the bulbs of Ornithogalum saundersiae Baker, exerts a wide range of pharmacological effects.
To explore whether OSW-1 can induce necroptosis in colorectal cancer (CRC) cells, thereby expanding its range of clinical applications.
We performed a sequence of functional experiments, including Cell Counting Kit-8 assays and flow cytometry analysis, to assess the inhibitory effect of OSW-1 on CRC cells. We utilized quantitative proteomics, employing tandem mass tag label
The results revealed that OSW-1 exerted a strong inhibitory effect on CRC cells, and this effect was accompanied by a necroptosis-like morphology that was observable via TEM. OSW-1 was shown to trigger necroptosis via activation of the RIPK1/RIPK3/MLKL pathway. Furthermore, the accumulation of p62/SQSTM1 was shown to mediate OSW-1-induced necroptosis through its interaction with RIPK1.
We propose that OSW-1 can induce necroptosis through the RIPK1/RIPK3/MLKL signaling pathway, and that this effect is mediated by the RIPK1-p62/SQSTM1 complex, in CRC cells. These results provide a theoretical foundation for the use of OSW-1 in the clinical treatment of CRC.
Core Tip: Colorectal cancer (CRC) is a significant health concern worldwide, and it has severe impacts on human lives. Identifying effective drugs for CRC treatment is very important. OSW-1, which is derived from the bulbs of Ornithogalum saundersiae, exhibits potent antitumor properties. This study confirmed the inhibitory effect of OSW-1 on CRC through both in vitro and in vivo experiments. Furthermore, tandem mass tag proteomic analysis was employed to predict differentially expressed proteins and potential underlying mechanisms. Our findings suggest that OSW-1 induces necroptosis via the RIPK1/RIPK3/MLKL signaling pathway, and this effect is mediated by the RIPK1-p62/SQSTM1 complex. These results provide a theoretical foundation for the use of OSW-1 in the clinical treatment of CRC.
- Citation: Wang N, Li CY, Yao TF, Kang XD, Guo HS. OSW-1 triggers necroptosis in colorectal cancer cells through the RIPK1/RIPK3/MLKL signaling pathway facilitated by the RIPK1-p62/SQSTM1 complex. World J Gastroenterol 2024; 30(15): 2155-2174
- URL: https://www.wjgnet.com/1007-9327/full/v30/i15/2155.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i15.2155
Colorectal cancer (CRC) is the most common type of gastrointestinal tumor, ranking third in incidence among gastr
Ornithogalum saundersiae Baker, which belongs to the Asparagaceae family, is a perennial plant that is native to South Africa and was introduced to China as an ornamental plant. OSW-1 [3β,16β,17α-trihydroxycholest-5-en-22-one-16-O-(2-O-4-methoxybenzoyl-β-D-xylopyranosyl)-(1→3)-(2-O-acetyl-α-L-arabinopyranoside)] (Figure 1A), which is extracted from the bulbs of Ornithogalum saundersiae, has been suggested to exert selective toxic effects against cancer cells. The anticancer properties of OSW-1 have been confirmed in diverse types of cancer, including triple-negative breast cancer, hepatocellular carcinomas, and lung cancer[6-9].
The application of quantitative proteomics to investigations of OSW-1-treated CRC cells provides a valuable approach for revealing clinically significant underlying mechanisms. Programmed cell death (PCD) includes various processes, such as necroptosis, apoptosis, pyroptosis, ferroptosis, and autophagy, and it has been shown to be a critical regulator of tissue development and to have significant implications for clinical outcomes. While much research has explored the mechanisms underlying apoptosis in cancer, the role of necroptosis in the progression of cancer has received less attention[10,11]. Necroptosis, which is a type of regulated cell death, is triggered by diverse stimuli, including pathogen recognition receptors, cell death receptor ligands, viral RNA sensors, DNA damage, and hypoxia sensors. The culmi
This research aimed to investigate the effect of OSW-1 on CRC cells and elucidate the mechanisms underlying necroptosis. We also conducted a tandem mass tag (TMT) proteomic analysis to predict differentially expressed proteins and possible underlying mechanisms. Our findings demonstrated that OSW-1 can indeed induce necroptosis in CRC cells. Furthermore, necroptosis triggered by OSW-1 is closely regulated by p62/SQSTM1, which is a critical regulatory molecule in autophagy. These findings provide novel insights into the molecular mechanisms underlying necroptosis, identifying OSW-1 as a potential therapeutic drug that should be further explored.
The human HT29 (iCell-h078) and HCT116 (Procell CL-0096) CRC cell lines were obtained from iCell Bioscience (Shanghai, China) and Procell Life Science & Technology Corporation (Wuhan, China). The cells were cultured in DMEM (Gibco BRL, Invitrogen, Carlsbad, CA, United States) and McCoy’s 5A medium (Gibco BRL, Invitrogen, Carlsbad, CA, United States), both supplemented with 10% fetal bovine serum (Newzerum, Christchurch, New Zealand) and 1% streptomycin and penicillin (Sigma, United States).
OSW-1 (C47H68O15, purity ≥ 98%) was obtained from GlpBio Technology Company (Montclair, CA, United States). OSW-1 was dissolved in dimethyl sulfoxide (DMSO, Sigma-Aldrich, St. Louis, MO, United States) to generate a stock solution and stored at -20 °C. To achieve the desired concentrations, OSW-1 was diluted in cell culture medium, ensuring that the DMSO concentration remained below 0.1% to minimize potential adverse effects.
Cells (8.0 × 103/well) were seeded in 96-well plates and treated with various concentrations of OSW-1 for 24 h. A Cell Counting Kit-8 (CCK-8) assay was used to assess cell proliferation. The absorbance was measured at 450 nm with a microplate reader (Thermo Fisher Scientific, MA, United States). Data analysis was conducted utilizing GraphPad Prism 9.0 software. (GraphPad Software, La Jolla, CA, United States). Each sample was analyzed in triplicate.
An Annexin V-FITC/Propidium iodide (PI) apoptosis detection kit (Elabscience Biotechnology, Wuhan, China) was used to detect cell apoptosis. After treating cells with OSW-1 for 24 h, the cells were digested to generate single-cell suspensions, which were stained according to the kit instructions. The experiment was replicated three times for each measurement.
Cells were seeded in a six-well plate and washed with PBS (Procell, Wuhan, China) after OSW-1 treatment for 24 h. JC-1 working solution (Elabscience Biotechnology, Wuhan, China) was added to the cells, which were then incubated at 37 °C for 30 minutes and washed with PBS. Each sample was analyzed in triplicate.
Cells were seeded in a six-well plate and washed with PBS after OSW-1 treatment for 24 h. Hoechst 33342/PI working solution was added, and the samples were incubated for 20 min in the dark. Each sample was analyzed in triplicate.
Lactate dehydrogenase (LDH) assay kit was used to measure LDH release and determine LDH activity (Beyotime Biotechnology, Shanghai, China). After treatment with OSW-1 for 24 h, 200 μL of LDH working solution was added to the cells and incubated for 30 min at room temperature according to the manufacturer's instructions. The absorbance at was measured 450 nm with a microplate reader. Each sample was analyzed in triplicate.
To assess changes in protein levels, a 6-plex TMT proteomic assay was employed. The proteomic experiments included two groups: The control group (treated with DMSO) and the OSW-1 treatment group. CRC cells were exposed to 1 μM OSW-1 for 24 h, and each group included three independent samples. Sangon Biotech (Shanghai, China) conducted the proteomic profiling, including enzymatic hydrolysis, labeling, mass spectrometry, and bioinformatics analysis, which included Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses.
Cells (5.0 × 105/well) were seeded in a six-well plate. The cells were exposed to OSW-1 for 24 h and then observed and photographed by optical microscopy (Nikon, Tokyo, Japan) to determine cell morphology.
CRC cells were plated on a sterile cover glass within a Petri dish. After treatment with OSW-1 for 24 h, the slides were gently washed with PBS, followed by incubation with an electron microscopy fixative (Servicebio, Wuhan, China) in the Petri dish. After the samples were fixed for 2 h at room temperature, the Petri dishes were transferred to 4 °C for storage. Specimens were affixed to cuprum grids, dehydrated with ethanol, and incubated with 2.6% lead citrate, after which exposure to CO2 was avoided for 8 min. Following desiccation with filter paper, the copper grids were placed on the grid board and allowed to air-dry overnight. Subsequently, the cell morphology of the samples was examined using transmission electron microscopy (TEM; HITACHI, Tokyo, Japan).
CRC cells were exposed to OSW-1 for 24 h, and PCR was carried out using an ABI 7500 PCR instrument and the SYBR Green Premix Pro Taq HS quantitative real-time PCR Kit (TaKaRa Biotechnology, Beijing, China). Relative mRNA expression levels were determined by normalization to the mRNA expression level of β-actin using the 2-ΔΔCt method (Table 1). The experiments were performed in triplicate. Downregulation of p62/SQSTM1 in CRC cells was achieved using the following siRNA duplex from GenePharma: CAUCCAGUAUUCAAAGCAUTT and AUGCUUUGAAUACUGGAUGTT. Cells were transfected using the GP-transfect-Mate Kit (Genepharma Company, Suzhou, China).
| Primer | Sequences (5’-3’) |
| p62-F | 5'-CTGGGACTGAGAAGGCTCAC-3' |
| p62-R | 5'-GCAGCTGATGGTTTGGAAAT-3' |
| LC3-F | 5'-AGCAGCATCCAACCAAAATC -3' |
| LC3-R | 5'-CTGTGTCCGTTCACCAACAG -3' |
| RIPK1-F | 5'-GGGAAGGTGTCTCTGTGTTTC-3' |
| RIPK1-R | 5'-CCTCGTTGTGCTCAATGCAG-3' |
| β-actin-F | 5'-AGTTGCGTTACACCCTTTCTTG-3' |
| β-actin-R | 5'-GCTGTCACCTTCACCGTTCC-3' |
After being treated with OSW-1 for 24 h, proteins were extracted from total cellular lysates using RIPA buffer (Proteintech, Wuhan, China), and the protein contents were quantified with a BCA protein assay kit (Elabscience, Wuhan, China). The protein samples were electrophoresed on a 10% SDS-PAGE gel (Severbio, Beijing, China) and electrotransferred to PVDF membranes (Millipore, MA, United States). The membranes were blocked at room temperature for 30 min and subsequently incubated with primary antibodies at 4 °C overnight. Then, secondary antibodies were added, and the immunoreactive bands were observed using enhanced chemiluminescence. The western blot bands were evaluated using ImageJ software.
According to the manufacturer's instructions, IP was performed with protein G magnetic beads after cells had been treated with OSW-1 for 24 h. After cells were lysed in RIPA buffer, the proteins were collected from the supernatants and incubated at 4 °C overnight with anti-RIPK1 (5 mg). To prepare a complex between the antibody and antigen, a mixture of protein G magnetic beads (60 μL) was added, and the mixture was incubated. The immunomagnetic bead-antibody-antigen complex was separated using a magnetic separator. After three washes of the beads with PBS, Western blotting was used to detect endogenous interactions between the specific antibody and other proteins.
Cells were cultured in confocal dishes (BIOFIL, Guangzhou, China) and stimulated with OSW-1 for 24 h. Subsequently, the cells were fixed with 4% paraformaldehyde for 30 min at room temperature. A permeabilization buffer of 0.5% Triton-X 100 was used, followed by blocking with a solution containing PBS, 5% goat serum, and 0.5% Triton-X 100 at room temperature for 30 min. Primary antibodies, including anti-RIPK1 and anti-p62 antibodies, were diluted in a primary antibody diluent (5% goat serum), added and incubated overnight at 4 °C. After washing with PBS, secondary antibodies (goat anti-rabbit Cy3 and goat anti-mouse FITC) were added, and the samples were incubated for 1 h at room temperature. Then, the samples were subjected to three consecutive washes with PBS. Finally, the nuclei were stained with DAPI. Observations were conducted at 40 × magnification with a 0.75 × zoom lens (Leica, Germany). Spectral Borealis lasers (green, 488 nm; red, 561 nm; blue, 405 nm) were used for excitation. A series of images were acquired with Leica SP8 software.
Images were captured to analyze the cellular autophagic flux using a confocal laser scanning microscope (Leica, Germany). For quantitative assessment of fluorescence intensity, ImageJ software was used to measure the integral optical density.
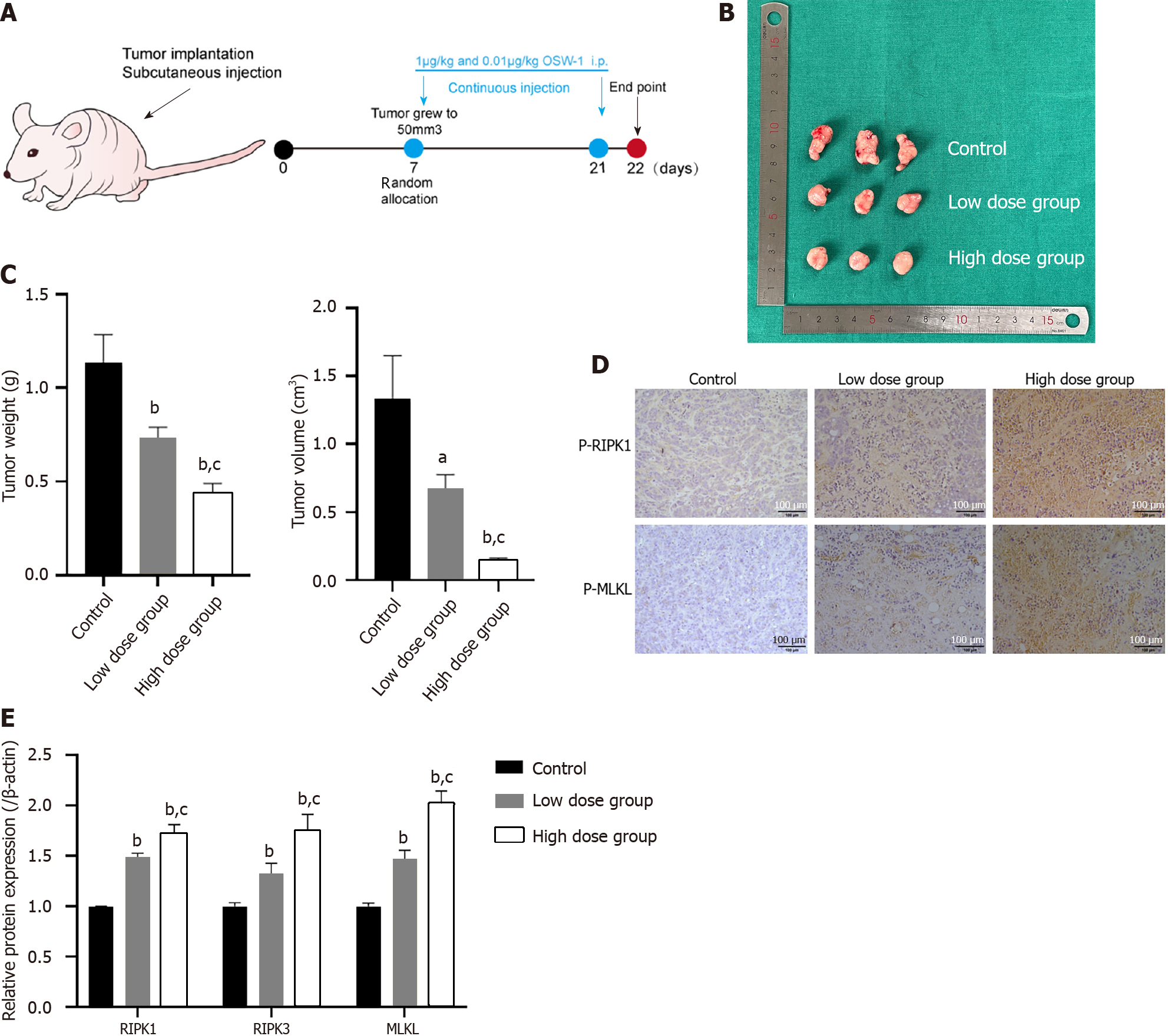
Four-week-old BALB/c-nude male mice were obtained from the Model Animal Research Institute of Dalian Medical University. A subcutaneous tumor model was established by injecting HT29 cells under the skin of each nude mouse. When the tumor volume reached 50 mm3, the nude mice were randomly divided into three groups: (1) The control group (treated with DMSO), (2) the low-dose OSW-1 group (0.01 µg/kg daily), and (3) the high-dose OSW-1 group (1 µg/kg daily). Tumor growth was monitored twice a week by measurements taken with a digital caliper. For further analysis, the mice were sacrificed humanely at 14 d, and the tumors were isolated. Tumor volumes were calculated with the following formula: V (mm3) = L (mm) × W2 (mm2) × 0.5. All the animal procedures and protocols were approved by the Committee on the Ethics of Animal Experiments of Dalian Medical University, No. AEE22108.
The paraffin sections were dewaxed using prewarmed xylene and dehydrated with an alcohol gradient. An appropriate amount of peroxidase blocking agent (Zsbio, Beijing, China) was added, followed by incubation at room temperature for 10 min. Subsequently, primary antibodies (Proteintech, Wuhan, China) were applied, and the samples were incubated overnight at 4 °C in a refrigerator. After overnight storage, a secondary antibody was added. The sections were then subjected to DAB staining and restained with hematoxylin. Finally, the slides were examined under a microscope.
All the graphs were generated using GraphPad Prism v.9 software. The data are presented as the mean ± SD and were analyzed using SPSS 23.0 software. One-way analysis of variance was used for data analysis, with statistical significance set at P < 0.05.
To investigate the ability of OSW-1 to suppress cell survival, we conducted a CCK-8 assay to assess the viability of CRC cells treated with different concentrations of OSW-1 for 24 h. The cytotoxicity results from the CCK-8 assay revealed a dose-dependent effect of OSW-1 on the viability of CRC cells, as shown in Figure 1B. Notably, there was a significant decrease in cell viability in the treated group compared to the control group. Furthermore, to determine the cell death rates, LDH release assays were conducted (Figure 1C). The results indicated a noticeable increase in LDH release with increasing OSW-1 concentration. OSW-1 (1 µM) significantly inhibited CRC cell survival, which was consistent with the CCK-8 results. Therefore, 1 µM OSW-1 was used as the concentration for the subsequent studies.
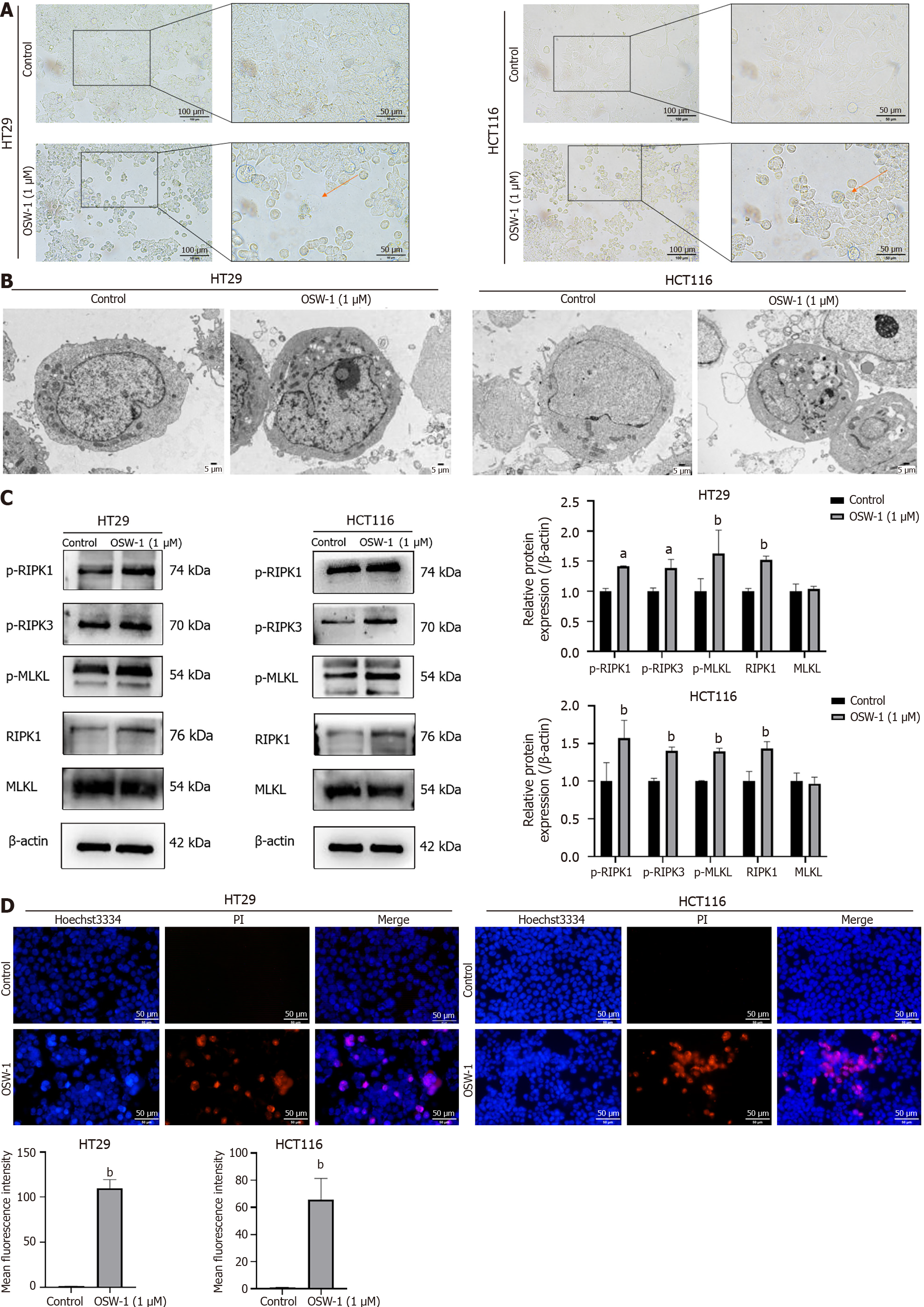
Moreover, a colony formation assay was performed to assess the effect of OSW-1 on colony formation ability, and the results revealed the significant inhibition of CRC cell colony formation (Figure 1D). As shown in Figure 1E, dual-fluorescence staining with Annexin V-FITC/PI demonstrated that CRC cells treated with OSW-1 underwent necrosis, as indicated by a notable increase in the percentage of necrotic and late apoptotic cells. To further investigate the potential mechanism underlying necrosis, the membrane-permeable fluorescent probe JC-1 was utilized. Under normal conditions, JC-1 aggregates in healthy cells. However, necroptosis can disrupt the mitochondrial membrane potential (Δψm), leading to the conversion of JC-1 aggregates (red) into monomers (green). We found that OSW-1 is a potent inducer of necroptosis that can cause JC-1 fluorescence to shift from red to green in CRC cells, which is indicative of a decrease in the Δψm (Figure 1F).
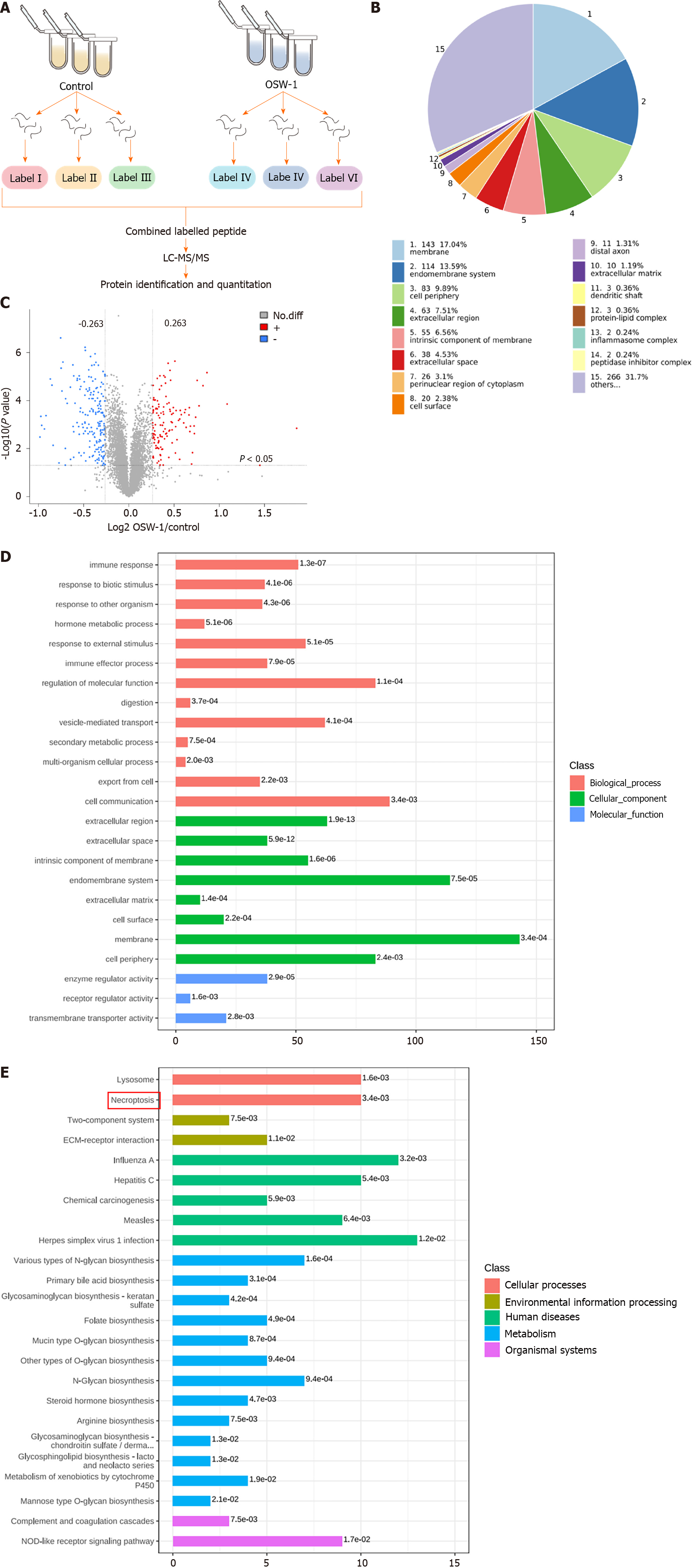
Our proteomic analysis revealed distinct patterns of protein expression between the OSW-1 treatment group and the control group (Figure 2A). After applying the criteria of a fold change > 1.2 and a significance level of P < 0.05 for filtering, our analysis identified 312 differentially expressed proteins in the OSW-1 treatment group. Among these, 143 were associated with the cell membrane, 114 with the endomembrane system, 83 with the cell periphery, 63 with the extracellular region, and 55 with the intrinsic component of the membrane (Figure 2B). A volcano plot was constructed, which shows 186 proteins with decreased expression and 126 proteins with increased expression in the OSW-1 treatment group (Figure 2C). To comprehensively understand the functional roles of proteins that were affected by OSW-1, we conducted enrichment analyses of GO terms, KEGG pathways, and protein domains. Our GO enrichment analysis (Figure 2D) revealed that the proteins that were affected by OSW-1 were predominantly associated with biological processes related to cell communication. Additionally, regarding cellular components, these proteins were enriched mainly in the cell membrane. Additionally, our analyses of KEGG pathway enrichment revealed that OSW-1 induced significant changes in the expression of proteins related to the necroptosis pathway (Figure 2E). Collectively, these findings suggest that OSW-1 may attenuate CRC development by modulating signaling pathways related to necroptosis.
Subsequently, we observed morphological changes in CRC cells following exposure to OSW-1 under an optical micro
Since RIPK1, RIPK3, and MLKL are important markers of necroptosis, our investigation examined the protein levels of these three necroptosis-associated molecules in OSW-1-treated CRC cells by western blotting analysis. As shown in Figure 3C, phosphorylated RIPK1, which serves as a marker of RIPK1 activation, was clearly increased in CRC cells following OSW-1 treatment. Additionally, in addition to phosphorylating RIPK3, which is the binding partner of RIPK1, OSW-1 also increased the phosphorylation of RIPK3 in CRC cells. Furthermore, OSW-1 equally enhanced the phosph
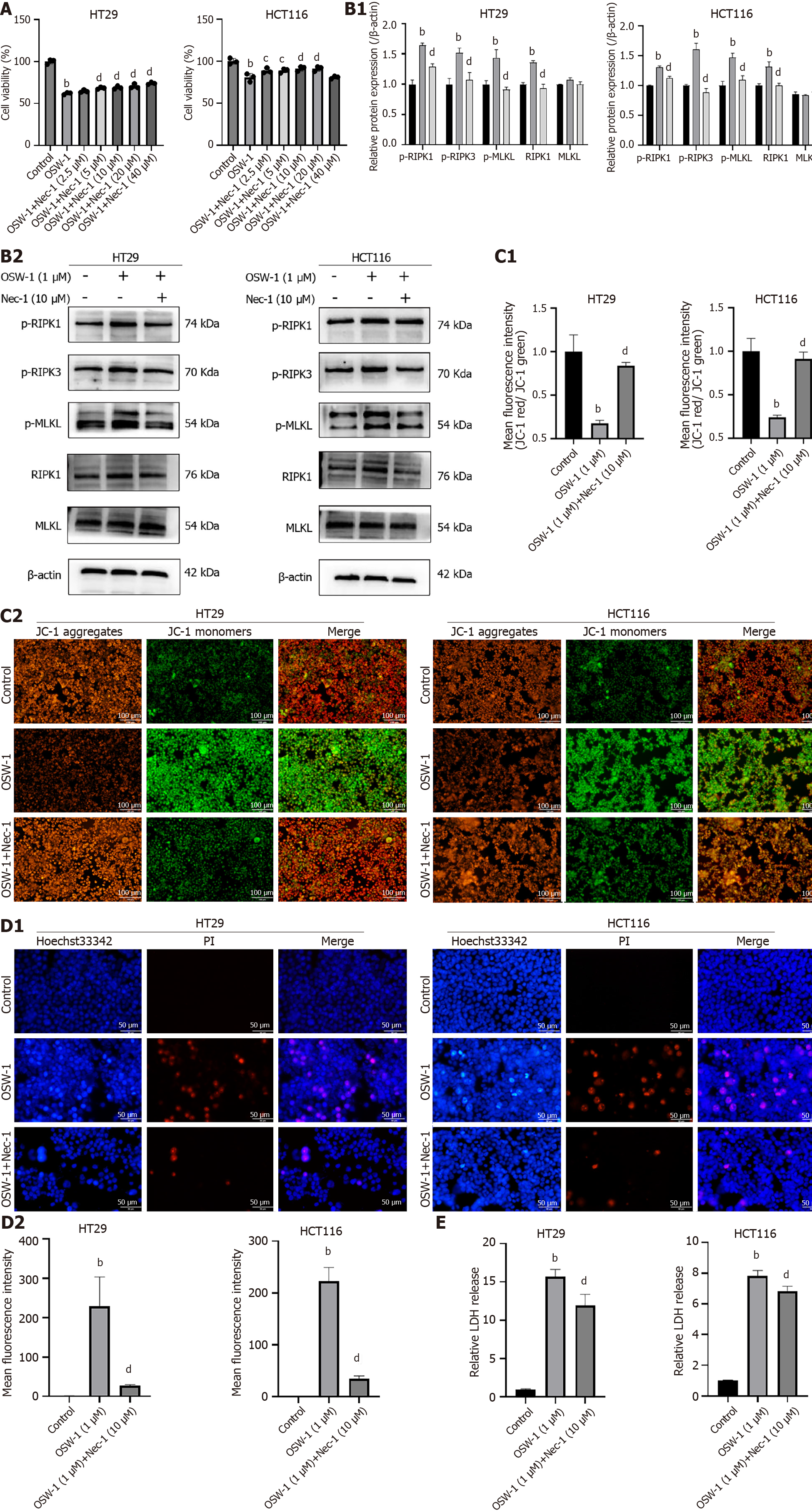
In this study, we focused on determining whether OSW-1-induced necroptosis requires the RIPK1/RIPK3/MLKL pathway, which is to be a critical pathway in necroptosis. The initial small-molecule inhibitor of RIPK1, necrostatin-1 (Nec-1), was shown to trigger necroptosis in cells. Since RIPK1 serves as a critical mediator of necroptosis, Nec-1 has been widely used to analyze this cellular process. To investigate the potential molecular mechanism underlying OSW-1-mediated necroptosis, we evaluated the impact of various Nec-1 concentrations on cell viability in cell culture experiments. The results revealed a noticeable increase in cell viability with increasing Nec-1 concentration (Figure 4A). Treatment with 10 µM Nec-1 significantly enhanced the survival of CRC cells, so this concentration was chosen for subsequent experiments.
To further explore the possible signaling pathways related to necroptosis, we investigated RIPK1, RIPK3, and MLKL in CRC cells treated with OSW-1 and Nec-1 via western blotting analysis. In combination with OSW-1, Nec-1 significantly inhibited both RIPK3 activation and MLKL activation as well as the subsequent necroptosis induced by OSW-1 (Figure 4B). Moreover, there was a distinct shift from JC-1 fluorescence from red to green when the cells were exposed to OSW-1. However, in combination with Nec-1, the intensity of green fluorescence significantly decreased (Figure 4C). In addition, Hoechst 33342/PI staining demonstrated a notable increase in the proportion of live cells after treatment with both OSW-1 and Nec-1 (Figure 4D). Moreover, the LDH release assay revealed a significant decrease in LDH release after treatment with both OSW-1 and Nec-1 (Figure 4E).
Based on our data, OSW-1 induces necroptosis in a manner that requires the activation of the RIPK1/RIPK3/MLKL pathway.
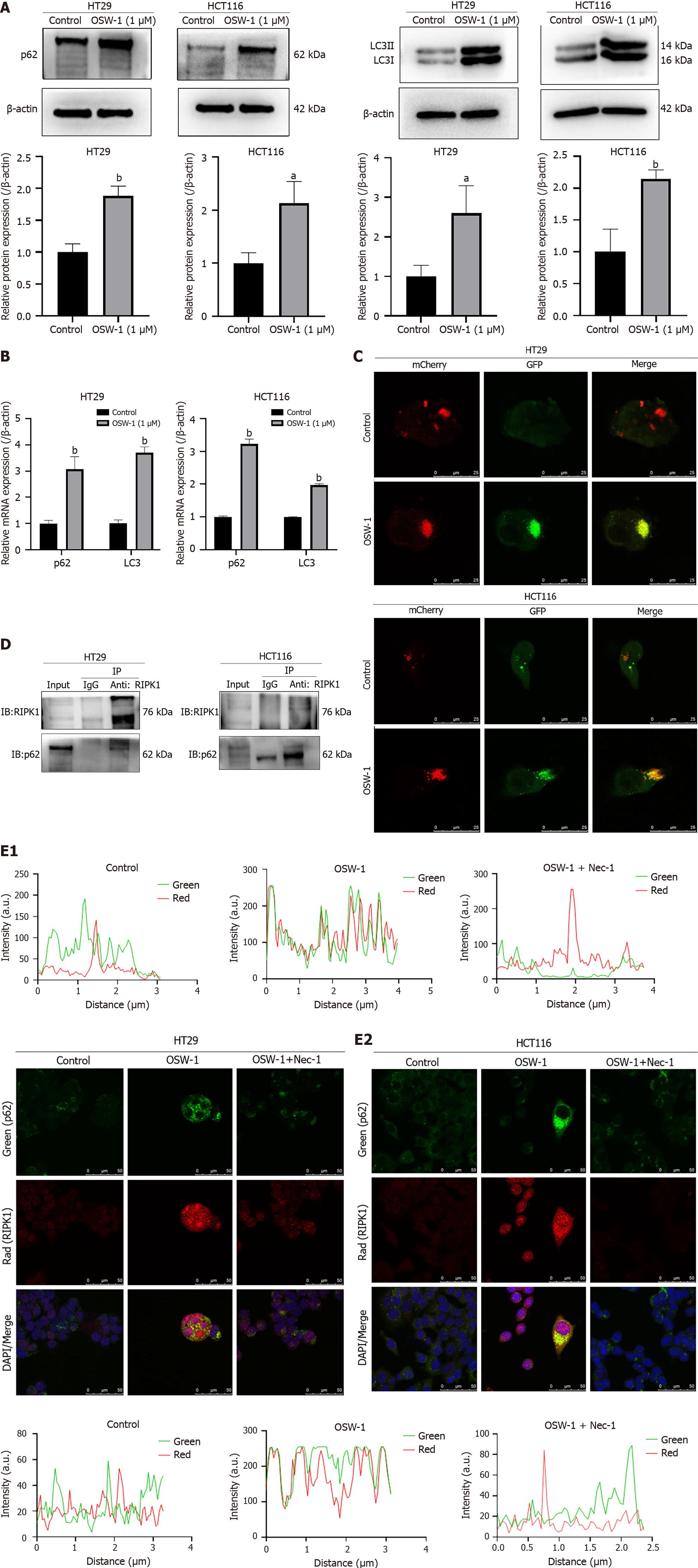
Based on the findings from the proteomic analysis, we focused on a notable protein, p62/SQSTM1, which is to be involved in the autophagic flux. According to previous studies, p62/SQSTM1 directly interacts with RIPK1. The region that is responsible for the binding of p62/SQSTM1 to RIPK1 is the ZZ structural domain (amino acids 122-167). We hypothesized that p62/SQSTM1 might function as a signaling platform and be involved in OSW-1-induced RIPK1-dependent necroptosis in CRC cells. Our primary objective was to test this hypothesis by assessing the protein expression of p62/SQSTM1 in CRC cells. Strikingly, the results showed significant upregulation of p62/SQSTM1 expression in the OSW-1 treatment group compared to the control group (Figure 5A). Our findings indicate that exposure of CRC cells to OSW-1 Leads to the accumulation of the p62/SQSTM1 proteins, which is generally associated with impaired autophagic degradation of p62/SQSTM1-bound substrates. We suspected that the upregulation of p62/SQSTM1 during OSW-1-induced necroptosis might be a result of impairment of the autophagic flux. Therefore, we assessed the protein levels of LC3-II, a crucial indicator of autophagy. These findings suggested that the level of LC3-II was increased after OSW-1 treatment in comparison to that in the control group (Figure 5A).
Additionally, the mRNA levels of p62/SQSTM1 and LC3 were elevated in CRC cells treated with OSW-1 (Figure 5B). These results suggested a connection between the aggregation of p62/SQSTM1 and the inhibition of autophagy-driven protein degradation. To verify this concept, we employed recombinant adenoviral vectors encoding GFP and mCherry-tagged LC3 (Ad-GFP & mCherry-LC3) to assess the autophagic flux. As shown in Figure 5C, OSW-1 progressively decreased the number of red dots (mCherry puncta lacking GFP fluorescence, indicative of autolysosomes) while increasing the number of yellow dots (overlapping mCherry with GFP fluorescence puncta, indicative of autophagosomes). These findings suggest that OSW-1 can impair the autophagic flux, contributing to the accumulation of p62/SQSTM1.
To explore the regulatory effects of p62/SQSTM1 on the RIPK1-related pathway, we conducted coimmunoprecipitation and immunofluorescence colocalization experiments. As shown in Figure 5D, the coimmunoprecipitation experiment further confirmed that OSW-1 promotes the interaction between RIPK1 and p62/SQSTM1, demonstrating the formation of a complex between p62/SQSTM1 and RIPK1. Moreover, immunocytochemical staining for p62/SQSTM1 (green) and RIPK1 (red) and analysis by confocal fluorescence microscopy revealed notable colocalization (yellow) of p62/SQSTM1 and RIPK1 compared to that in the control group. Following Nec-1 treatment, we observed a significant reduction in the colocalization of p62/SQSTM1 and RIPK1 compared to that in the OSW-1 group (Figure 5E).
To assess the role of p62/SQSTM1 in necroptosis, p62/SQSTM1 siRNA was transduced into CRC cells to reverse the effect of p62/SQSTM1 overexpression on necroptosis. When CRC cells that were transiently transfected with the control construct were exposed to OSW-1, they exhibited a reduction in p62/SQSTM1 expression (Figure 6A). Additionally, silencing p62/SQSTM1 Led to decreased levels of RIPK1 in CRC cells treated with OSW-1 (Figure 6A). Notably, RIPK1 expression was significantly decreased after specific siRNA-mediated knockdown of p62/SQSTM1 in CRC cells, suggesting that p62/SQSTM1 protects RIPK1 from degradation. To further elucidate the role of p62/SQSTM1 in OSW-1-induced necroptosis, western blotting experiments were conducted on CRC cells. The results indicated that the expression of phosphorylated RIPK1, phosphorylated RIPK3, and MLKL was decreased upon the silencing of p62/SQSTM1 (Figure 6B). Furthermore, Hoechst 33342/PI staining showed a notable increase in the proportion of live cells, indicating the restoration of necroptosis in p62/SQSTM1 siRNA-transfected cells (Figure 6C). These findings highlight the crucial role of p62/SQSTM1 in OSW-1-induced necroptosis in CRC cells.
In summary, our results demonstrated that impairment of the autophagic flux leads to the accumulation of p62/SQSTM1, which increases necroptosis in OSW-1-treated CRC cells via interaction with RIPK1 and phosphorylation of RIPK1/RIPK3/MLKL pathway-related proteins.
To assess the antitumor effects of OSW-1 on CRC in vivo, we established a mouse xenograft model. Following the development of palpable tumors, the mice were administered OSW-1 at doses of 0.01 µg/kg and 1 µg/kg for 14 d (Figure 7A). As expected, OSW-1 significantly suppressed the growth of tumors (Figure 7B). The tumor volumes in mice treated with OSW-1 were consistently smaller than those in control mice, with the high-dose group (1 µg/kg) displaying notably reduced tumor sizes compared to those in the low-dose group (0.01 µg/kg; Figure 7C). Moreover, the tumor weight exhibited a similar trend (Figure 7C). Furthermore, the influence of OSW-1 on CRC was assessed through immunohistochemistry (IHC). Compared with those in the control group, the expression levels of the necroptosis-related proteins p-RIPK1 and p-MLKL increased with increasing OSW-1 concentration (Figure 7D). These findings were consistent with the results of the in vitro experiments. Furthermore, the qRT-PCR results revealed a concentration-dependent increase in the expression of RIPK1 and MLKL in response to OSW-1 treatment (Figure 7E), which was consistent with the IHC results. In summary, our results suggest that OSW-1 suppresses tumor growth in a mouse xenograft model by activating the necroptosis pathway.
CRC is a heterogeneous malignant tumor that poses a significant threat to human survival[18-20]. Its pathogenic factors are complex and include gene mutation, age, family history, obesity, and physical inactivity[21,22]. With better understanding of CRC, the spectrum of treatment options has expanded, including endoscopic and surgical excision, radiotherapy, immunotherapy, and targeted therapy[23-25]. These treatments have led to a substantial reduction in cancer progression and an increased survival rate. However, despite these advancements, CRC remains the second most common cause of cancer-related death worldwide, primarily due to challenges encountered in clinical trials, such as resistance to radiotherapy and chemotherapy that aim to induce apoptosis[26,27]. To develop a novel strategy for preventing CRC progression, exploring alternative mechanisms of cell death in CRC by integrating high-throughput proteomics with conventional molecular techniques is imperative.
The natural product OSW-1, which is derived from plants, can selectively kill cancer cells[28]. In recent years, OSW-1 has attracted considerable attention due to its potent and selective cytotoxic effects against various types of cancer cell lines, suggesting a potentially novel mechanism of action. In the early stages of research, ovarian granulosa cell proliferation and the expression of the steroidal enzyme were thought to be inhibited by OSW-1[29]. In a previous study, OSW-1 was shown to exert cytotoxic effects against diverse cancer cells, including leukemia cells, and higher toxicity was observed against malignant cells than against normal cells[30]. However, the specific mechanism underlying the action of OSW-1 has not been elucidated. Our study revealed that OSW-1 effectively inhibited proliferation and suppressed survival in CRC cells.
Quantitative proteomic methods are widely utilized due to their ability to reveal the dynamics of protein expression and interactions on a global scale. This approach significantly contributes to comprehending gene functions and cellular processes. In this study, we employed a proteomic approach to identify proteins whose expression was altered in response to OSW-1 treatment. A total of 312 proteins exhibited differential expression. Considering the substantial number of differentially expressed proteins and the high enrichment factor, we focused our attention on the necroptosis pathway for further mechanistic exploration.
In recent years, necroptosis, which is a novel form of PCD, has emerged as an essential form of cell death that contributes to various diseases, and its role in tumors has received particular attention. Moreover, a large amount of evidence has shown that necroptosis can impact tumor development, rendering necroptosis as an important area of interest in cancer treatment[31-34]. Notably, the relationship between necroptosis and gastric cancer has been established[35]. Moreover, RIPK1, which is a pivotal signaling node in necroptosis, has been shown to cooperate with TRAF2 to suppress murine and human hepatocarcinogenesis[36]. Recent research has highlighted the ability of several chemotherapeutic drugs and natural products to induce necroptosis and inhibit tumor growth. For instance, jaceosidin has been identified as an inducer of necroptosis in human glioblastoma multiforme, suggesting its potential as a therapeutic agent for patients[37]. Additionally, the arborinane triterpene compound 3-O-acetylrubianol C, which is isolated from the Rubia Philippines, has been found to promote tumor necrosis factor-triggered necroptotic cell death[38].
Our findings indicate that OSW-1 effectively inhibited the proliferation of CRC cells. Moreover, the percentage of necrotic cells increased, as shown by Annexin V-FITC/PI dual-fluorescence staining, indicating the occurrence of necrosis. The change in the Δψm further confirmed the role of OSW-1 as an inducer of necroptosis. Our findings indicate that OSW-1 effectively triggered necroptosis in CRC cells. More strikingly, optical microscopy and TEM revealed morphological changes that are typical of necroptosis, such as membrane rupture and cytoplasmic vesiculation, in CRC cells. Additionally, a Hoechst 33342/PI double staining assay was used to characterize necroptosis. These data strongly suggested the occurrence of necroptosis in CRC cells following OSW-1 treatment.
The RIPK1/RIPK3/MLKL pathway Is widely recognized as the classic pathway that regulates necroptosis under diverse conditions[39]. In response to stimuli such as ischemia/reperfusion, inflammation, and certain medicines, RIPK1 binds to RIPK3 to form a necrosome, which triggers MLKL activation and translocation, ultimately resulting in cell lysis. This study utilized the RIPK1-specific inhibitor Nec-1 to explore whether OSW-1 induces necroptosis in CRC cells through the classical necroptotic pathway. Nec-1 demonstrated marked proliferative effects and inhibited cell death in CRC, promoting cell survival. Western blotting analysis revealed significant upregulation of p-RIPK1, p-RIPK3, and p-MLKL expression in CRC cells treated with OSW-1. However, Nec-1 effectively attenuated the increases in these protein levels. In addition, consistent with these findings, Hoechst 33342/PI staining indicated a substantial increase in the proportion of live cells after combined treatment with OSW-1 and Nec-1 compared to treatment with OSW-1 alone. Furthermore, the intensity of red fluorescence significantly decreased with the addition of Nec-1, suggesting that necroptosis was partially suppressed. Our results indicate that the RIPK1/RIPK3/MLKL pathway mediates necroptosis in CRC cells treated with OSW-1. Additionally, OSW-1-induced necroptosis can be partially reversed by Nec-1, demonstrating protective effects in CRC cells. However, while the classical pathway is a crucial regulator of necroptosis, other mechanisms also contribute to this process[40-42]. Further investigations are warranted to elucidate whether different pathways and molecules are involved in OSW-1-triggered necroptosis in CRC cells.
According to our study, OSW-1-induced necroptosis activates RIPK1 and related signaling pathways. However, the specific mechanism by which OSW-1 activates RIPK1 has not been determined. To further elucidate the mechanism underlying RIPK1-dependent necroptosis in CRC, we investigated p62/SQSTM1, which is binds to RIPK1 and regulates the ubiquitin-proteasome system and lysosomal autophagy in various diseases. Recent research has highlighted a connection between tumorigenesis and the upregulation or inefficient degradation of p62/SQSTM1[43]. High levels of p62/SQSTM1 have been shown to inhibit the activity of the E3 Ligase RNF168, increasing the sensitivity of cancer cells to radiotherapy[44,45]. Consistent with these findings, our data also indicated an accumulation of the p62/SQSTM1 protein levels after OSW-1 treatment, suggesting defective autophagic degradation of p62/SQSTM1-bound substrates, contributing to cancer therapy.
Notably, our results demonstrated that treatment of CRC cells with OSW-1 could lead to the formation of a complex between RIPK1 and p62/SQSTM1. We propose that the accumulation of p62/SQSTM1 induced by OSW-1 facilitates the formation of necrosomes, triggering the activation of the RIPK1/RIPK3 pathway and ultimately leading to necroptotic cell death. This hypothesis was confirmed by the data showing that OSW-1 treatment induced the interaction of p62/SQSTM1 with RIPK1 in CRC cells. Moreover, knockdown of p62/SQSTM1 resulted in decreased expression of p-RIPK, p-RIPK3 and p-MLKL. It would be worthwhile to explore whether other pathways and molecules regulate OSW-1-induced necroptosis in CRC cells.
OSW-1-treated cells and nude mice exhibited typical necroptosis-like characteristics, as shown by increased expression of p-RIPK1, p-RIPK3, and p-MLKL. Moreover, p62/SQSTM1 plays a crucial role in the regulation of necroptosis. When OSW-1 causes intracellular damage, the accumulation of p62/SQSTM1 may act as a signaling platform to activate necroptosis, possibly through the inhibition of the autophagic flux. From a therapeutic perspective, our findings suggest that OSW-1 might induce necroptosis in cancer cells under conditions of deficient or defective autophagy. This discovery provides a rationale for further investigating the use of OSW-1 as a promising antitumor drug, particularly in the context of individualized therapeutic approaches.
Our results demonstrated that OSW-1 could induce necroptosis through the RIPK1/RIPK3/MLKL pathway, and this effect was potentially mediated by the RIPK1-p62/SQSTM1 complex. This study provides a novel mechanism on the antitumor effects of OSW-1 and may provide a new therapeutic target from a new perspective of CRC cell death, which has important clinical significance.
Colorectal cancer (CRC) represents a significant health concern worldwide, and it has severe impacts on human lives. The search for effective drugs is crucial. OSW-1, which is derived from the bulbs of Ornithogalum saundersiae, exhibits potent antitumor properties. However, whether OSW-1 induces necroptosis in CRC cells to exert anticancer effects remains unclear.
We conducted a tandem mass tag proteomic analysis to elucidate the mechanisms underlying necroptosis. We explored the potential for the use of OSW-1 as a drug for the treatment of CRC.
This research aimed to investigate the influence of OSW-1 on CRC cells and elucidate the mechanisms underlying necroptosis.
We performed a sequence of functional experiments, including Cell Counting Kit-8 assays and flow cytometry analysis, to assess the inhibitory impact of OSW-1 on CRC cells. We utilized quantitative proteomics to analyze changes in protein expression. transmission electron microscopy (TEM) and immunofluorescence studies were also performed to examine the effects of OSW-1 on necroptosis. Additionally, western blotting, siRNA experiments, and immunoprecipitation were employed to evaluate protein interactions within CRC cells.
The results revealed a pronounced inhibitory effect of OSW-1 on CRC cells, which was accompanied by a necroptosis-like morphology that was observed via TEM. OSW-1 was shown to trigger necroptosis via activation of the RIPK1/RIPK3/MLKL pathway. Furthermore, the accumulation of p62/SQSTM1 was shown to mediate OSW-1-induced necroptosis through its interaction with RIPK1.
We propose that OSW-1 can induce necroptosis through the RIPK1/RIPK3/MLKL signaling pathway, which is facilitated by the RIPK1-p62/SQSTM1 complex in CRC cells.
This study provides a novel mechanism on the antitumor effects of OSW-1 and may provide a new therapeutic target from a new perspective of CRC cell death, which has important clinical significance.
The authors would like to acknowledge Jing-Yuan Zhao for statistical analysis assistance.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Nakaji K, Japan S-Editor: Li L L-Editor: A P-Editor: Yu HG
| 1. | Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11444] [Cited by in RCA: 13205] [Article Influence: 1467.2] [Reference Citation Analysis (3)] |
| 2. | Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683-691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3058] [Cited by in RCA: 3298] [Article Influence: 412.3] [Reference Citation Analysis (3)] |
| 3. | Holm M, Andersson E, Osterlund E, Ovissi A, Soveri LM, Anttonen AK, Kytölä S, Aittomäki K, Osterlund P, Ristimäki A. Detection of KRAS mutations in liquid biopsies from metastatic colorectal cancer patients using droplet digital PCR, Idylla, and next generation sequencing. PLoS One. 2020;15:e0239819. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 54] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 4. | Fakhr E, Zare F, Azadmanesh K, Teimoori-Toolabi L. LEF1 silencing sensitizes colorectal cancer cells to oxaliplatin, 5-FU, and irinotecan. Biomed Pharmacother. 2021;143:112091. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 5. | Ooft SN, Weeber F, Dijkstra KK, McLean CM, Kaing S, van Werkhoven E, Schipper L, Hoes L, Vis DJ, van de Haar J, Prevoo W, Snaebjornsson P, van der Velden D, Klein M, Chalabi M, Boot H, van Leerdam M, Bloemendal HJ, Beerepoot LV, Wessels L, Cuppen E, Clevers H, Voest EE. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci Transl Med. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 269] [Cited by in RCA: 488] [Article Influence: 97.6] [Reference Citation Analysis (0)] |
| 6. | Zhan Z, Liu Z, Lai J, Zhang C, Chen Y, Huang H. Anticancer Effects and Mechanisms of OSW-1 Isolated From Ornithogalum saundersiae: A Review. Front Oncol. 2021;11:747718. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 7. | Iguchi T, Kuroda M, Naito R, Watanabe T, Matsuo Y, Yokosuka A, Mimaki Y. Cholestane glycosides from Ornithogalum saundersiae bulbs and the induction of apoptosis in HL-60 cells by OSW-1 through a mitochondrial-independent signaling pathway. J Nat Med. 2019;73:131-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 8. | Ding X, Li Y, Li J, Yin Y. OSW-1 inhibits tumor growth and metastasis by NFATc2 on triple-negative breast cancer. Cancer Med. 2020;9:5558-5569. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 9. | Jin J, Jin X, Qian C, Ruan Y, Jiang H. Signaling network of OSW1induced apoptosis and necroptosis in hepatocellular carcinoma. Mol Med Rep. 2013;7:1646-1650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 10. | Liu L, Fan J, Ai G, Liu J, Luo N, Li C, Cheng Z. Berberine in combination with cisplatin induces necroptosis and apoptosis in ovarian cancer cells. Biol Res. 2019;52:37. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 143] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 11. | Youn MJ, So HS, Cho HJ, Kim HJ, Kim Y, Lee JH, Sohn JS, Kim YK, Chung SY, Park R. Berberine, a natural product, combined with cisplatin enhanced apoptosis through a mitochondria/caspase-mediated pathway in HeLa cells. Biol Pharm Bull. 2008;31:789-795. [PubMed] |
| 12. | Soliman M, Seo JY, Baek YB, Park JG, Kang MI, Cho KO, Park SI. Opposite Effects of Apoptotic and Necroptotic Cellular Pathways on Rotavirus Replication. J Virol. 2022;96:e0122221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 13. | Wu W, Wang X, Sun Y, Berleth N, Deitersen J, Schlütermann D, Stuhldreier F, Wallot-Hieke N, José Mendiburo M, Cox J, Peter C, Bergmann AK, Stork B. TNF-induced necroptosis initiates early autophagy events via RIPK3-dependent AMPK activation, but inhibits late autophagy. Autophagy. 2021;17:3992-4009. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 66] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 14. | Xu Y, Ma HB, Fang YL, Zhang ZR, Shao J, Hong M, Huang CJ, Liu J, Chen RQ. Cisplatin-induced necroptosis in TNFα dependent and independent pathways. Cell Signal. 2017;31:112-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 57] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 15. | Jang JY, Im E, Choi YH, Kim ND. Mechanism of Bile Acid-Induced Programmed Cell Death and Drug Discovery against Cancer: A Review. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 16. | Hannes S, Abhari BA, Fulda S. Smac mimetic triggers necroptosis in pancreatic carcinoma cells when caspase activation is blocked. Cancer Lett. 2016;380:31-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 61] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 17. | Sun W, Wu X, Gao H, Yu J, Zhao W, Lu JJ, Wang J, Du G, Chen X. Cytosolic calcium mediates RIP1/RIP3 complex-dependent necroptosis through JNK activation and mitochondrial ROS production in human colon cancer cells. Free Radic Biol Med. 2017;108:433-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 107] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 18. | Baidoun F, Elshiwy K, Elkeraie Y, Merjaneh Z, Khoudari G, Sarmini MT, Gad M, Al-Husseini M, Saad A. Colorectal Cancer Epidemiology: Recent Trends and Impact on Outcomes. Curr Drug Targets. 2021;22:998-1009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 154] [Article Influence: 38.5] [Reference Citation Analysis (2)] |
| 19. | Lamichhane P, Maiolini M, Alnafoosi O, Hussein S, Alnafoosi H, Umbela S, Richardson T, Alla N, Lamichhane N, Subhadra B, Deshmukh RR. Colorectal Cancer and Probiotics: Are Bugs Really Drugs? Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 20. | Shang A, Gu C, Wang W, Wang X, Sun J, Zeng B, Chen C, Chang W, Ping Y, Ji P, Wu J, Quan W, Yao Y, Zhou Y, Sun Z, Li D. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p- TGF-β1 axis. Mol Cancer. 2020;19:117. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 333] [Article Influence: 66.6] [Reference Citation Analysis (0)] |
| 21. | Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394:1467-1480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1570] [Cited by in RCA: 3008] [Article Influence: 501.3] [Reference Citation Analysis (3)] |
| 22. | Yiu AJ, Yiu CY. Biomarkers in Colorectal Cancer. Anticancer Res. 2016;36:1093-1102. [PubMed] |
| 23. | Xi Y, Xu P. Global colorectal cancer burden in 2020 and projections to 2040. Transl Oncol. 2021;14:101174. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 687] [Cited by in RCA: 1339] [Article Influence: 334.8] [Reference Citation Analysis (5)] |
| 24. | Guren MG. The global challenge of colorectal cancer. Lancet Gastroenterol Hepatol. 2019;4:894-895. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 74] [Article Influence: 12.3] [Reference Citation Analysis (1)] |
| 25. | Schreuders EH, Ruco A, Rabeneck L, Schoen RE, Sung JJ, Young GP, Kuipers EJ. Colorectal cancer screening: a global overview of existing programmes. Gut. 2015;64:1637-1649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 674] [Cited by in RCA: 913] [Article Influence: 91.3] [Reference Citation Analysis (0)] |
| 26. | Rabeneck L, Chiu HM, Senore C. International Perspective on the Burden of Colorectal Cancer and Public Health Effects. Gastroenterology. 2020;158:447-452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 45] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 27. | Kaminski MF, Robertson DJ, Senore C, Rex DK. Optimizing the Quality of Colorectal Cancer Screening Worldwide. Gastroenterology. 2020;158:404-417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 114] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 28. | Zhou Y, Garcia-Prieto C, Carney DA, Xu RH, Pelicano H, Kang Y, Yu W, Lou C, Kondo S, Liu J, Harris DM, Estrov Z, Keating MJ, Jin Z, Huang P. OSW-1: a natural compound with potent anticancer activity and a novel mechanism of action. J Natl Cancer Inst. 2005;97:1781-1785. [PubMed] |
| 29. | Tamura K, Honda H, Mimaki Y, Sashida Y, Kogo H. Inhibitory effect of a new steroidal saponin, OSW-1, on ovarian functions in rats. Br J Pharmacol. 1997;121:1796-1802. [PubMed] |
| 30. | Zhang Y, Fang F, Fan K, Zhang Y, Zhang J, Guo H, Yu P, Ma J. Effective cytotoxic activity of OSW-1 on colon cancer by inducing apoptosis in vitro and in vivo. Oncol Rep. 2017;37:3509-3519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 31. | Zhang DW, Shao J, Lin J, Zhang N, Lu BJ, Lin SC, Dong MQ, Han J. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science. 2009;325:332-336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1352] [Cited by in RCA: 1649] [Article Influence: 103.1] [Reference Citation Analysis (0)] |
| 32. | Maelfait J, Liverpool L, Bridgeman A, Ragan KB, Upton JW, Rehwinkel J. Sensing of viral and endogenous RNA by ZBP1/DAI induces necroptosis. EMBO J. 2017;36:2529-2543. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 105] [Cited by in RCA: 182] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 33. | Kaczmarek A, Vandenabeele P, Krysko DV. Necroptosis: the release of damage-associated molecular patterns and its physiological relevance. Immunity. 2013;38:209-223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 833] [Cited by in RCA: 1167] [Article Influence: 97.3] [Reference Citation Analysis (0)] |
| 34. | Lawlor KE, Khan N, Mildenhall A, Gerlic M, Croker BA, D'Cruz AA, Hall C, Kaur Spall S, Anderton H, Masters SL, Rashidi M, Wicks IP, Alexander WS, Mitsuuchi Y, Benetatos CA, Condon SM, Wong WW, Silke J, Vaux DL, Vince JE. RIPK3 promotes cell death and NLRP3 inflammasome activation in the absence of MLKL. Nat Commun. 2015;6:6282. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 397] [Cited by in RCA: 523] [Article Influence: 52.3] [Reference Citation Analysis (0)] |
| 35. | Xia Y, Zhang R, Wang M, Li J, Dong J, He K, Guo T, Ju X, Ru J, Zhang S, Sun Y. Development and validation of a necroptosis-related gene prognostic score to predict prognosis and efficiency of immunotherapy in gastric cancer. Front Immunol. 2022;13:977338. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 36. | Schneider AT, Gautheron J, Feoktistova M, Roderburg C, Loosen SH, Roy S, Benz F, Schemmer P, Büchler MW, Nachbur U, Neumann UP, Tolba R, Luedde M, Zucman-Rossi J, Panayotova-Dimitrova D, Leverkus M, Preisinger C, Tacke F, Trautwein C, Longerich T, Vucur M, Luedde T. RIPK1 Suppresses a TRAF2-Dependent Pathway to Liver Cancer. Cancer Cell. 2017;31:94-109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 126] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 37. | Park KR, Jeong Y, Lee J, Kwon IK, Yun HM. Anti-tumor effects of jaceosidin on apoptosis, autophagy, and necroptosis in human glioblastoma multiforme. Am J Cancer Res. 2021;11:4919-4930. [PubMed] |
| 38. | Kang K, Quan KT, Byun HS, Lee SR, Piao X, Ju E, Park KA, Sohn KC, Shen HM, Na M, Hur GM. 3-O-acetylrubianol C (3AR-C) induces RIPK1-dependent programmed cell death by selective inhibition of IKKβ. FASEB J. 2020;34:4369-4383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 39. | Zhang W, Fan W, Guo J, Wang X. Osmotic stress activates RIPK3/MLKL-mediated necroptosis by increasing cytosolic pH through a plasma membrane Na(+)/H(+) exchanger. Sci Signal. 2022;15:eabn5881. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 36] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 40. | Liu M, Li H, Yang R, Ji D, Xia X. GSK872 and necrostatin-1 protect retinal ganglion cells against necroptosis through inhibition of RIP1/RIP3/MLKL pathway in glutamate-induced retinal excitotoxic model of glaucoma. J Neuroinflammation. 2022;19:262. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 76] [Reference Citation Analysis (0)] |
| 41. | Vo DK, Urano Y, Takabe W, Saito Y, Noguchi N. 24(S)-Hydroxycholesterol induces RIPK1-dependent but MLKL-independent cell death in the absence of caspase-8. Steroids. 2015;99:230-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 30] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 42. | Saveljeva S, Mc Laughlin SL, Vandenabeele P, Samali A, Bertrand MJ. Endoplasmic reticulum stress induces ligand-independent TNFR1-mediated necroptosis in L929 cells. Cell Death Dis. 2015;6:e1587. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 119] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 43. | Cai-McRae X, Zhong H, Karantza V. Sequestosome 1/p62 facilitates HER2-induced mammary tumorigenesis through multiple signaling pathways. Oncogene. 2015;34:2968-2977. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 44. | Wang Y, Zhu WG, Zhao Y. Autophagy substrate SQSTM1/p62 regulates chromatin ubiquitination during the DNA damage response. Autophagy. 2017;13:212-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 44] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 45. | Wang Y, Zhang N, Zhang L, Li R, Fu W, Ma K, Li X, Wang L, Wang J, Zhang H, Gu W, Zhu WG, Zhao Y. Autophagy Regulates Chromatin Ubiquitination in DNA Damage Response through Elimination of SQSTM1/p62. Mol Cell. 2016;63:34-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 168] [Article Influence: 18.7] [Reference Citation Analysis (0)] |