Published online Apr 14, 2024. doi: 10.3748/wjg.v30.i14.2018
Peer-review started: October 31, 2023
First decision: December 15, 2023
Revised: January 11, 2024
Accepted: February 29, 2024
Article in press: February 29, 2024
Published online: April 14, 2024
Processing time: 164 Days and 10.5 Hours
Colorectal cancer (CRC) ranks among the most prevalent malignant tumors globally. Recent reports suggest that Fusobacterium nucleatum (F. nucleatum) contributes to the initiation, progression, and prognosis of CRC. Butyrate, a short-chain fatty acid derived from the bacterial fermentation of soluble dietary fiber, is known to inhibit various cancers. This study is designed to explore whether F. nucleatum influences the onset and progression of CRC by impacting the intestinal metabolite butyric acid.
To investigate the mechanism by which F. nucleatum affects CRC occurrence and development.
Alterations in the gut microbiota of BALB/c mice were observed following the oral administration of F. nucleatum. Additionally, DLD-1 and HCT116 cell lines were exposed to sodium butyrate (NaB) and F. nucleatum in vitro to examine the effects on proliferative proteins and mitochondrial function.
Our research indicates that the prevalence of F. nucleatum in fecal samples from CRC patients is significantly greater than in healthy counterparts, while the prevalence of butyrate-producing bacteria is notably lower. In mice colonized with F. nucleatum, the population of butyrate-producing bacteria decreased, resulting in altered levels of butyric acid, a key intestinal metabolite of butyrate. Exposure to NaB can impair mitochondrial morphology and diminish mitochondrial membrane potential in DLD-1 and HCT116 CRC cells. Consequently, this leads to modulated production of adenosine triphosphate and reactive oxygen species, thereby inhibiting cancer cell proliferation. Additionally, NaB triggers the adenosine monophosphate-activated protein kinase (AMPK) signaling pathway, blocks the cell cycle in HCT116 and DLD-1 cells, and curtails the proliferation of CRC cells. The combined presence of F. nucleatum and NaB attenuated the effects of the latter. By employing small interfering RNA to suppress AMPK, it was demonstrated that AMPK is essential for NaB’s inhibition of CRC cell proliferation.
F. nucleatum can promote cancer progression through its inhibitory effect on butyric acid, via the AMPK signaling pathway.
Core Tip: In this study, we unravel the mechanism of Fusobacterium nucleatum (F. nucleatum) in the progression of colorectal cancer (CRC), focusing on the interplay between intestinal flora and metabolism. Our results reveal that F. nucleatum suppresses the production of the short-chain fatty acid butyric acid. We discovered that sodium butyrate impairs mitochondrial function and impedes the cell cycle in CRC cells. Furthermore, the study highlights that both sodium butyrate and F. nucleatum orchestrate CRC cell proliferation via the adenosine monophosphate-activated protein kinase signaling pathway. These insights may enhance our understanding of CRC pathogenesis and aid in the development of more effective treatment strategies.
- Citation: Wu QL, Fang XT, Wan XX, Ding QY, Zhang YJ, Ji L, Lou YL, Li X. Fusobacterium nucleatum-induced imbalance in microbiome-derived butyric acid levels promotes the occurrence and development of colorectal cancer. World J Gastroenterol 2024; 30(14): 2018-2037
- URL: https://www.wjgnet.com/1007-9327/full/v30/i14/2018.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i14.2018
Colorectal cancer (CRC) is one of the most common malignant tumors in the world and is the second and third most common cancer in women and men, respectively[1]. In recent years, CRC has ranked fifth in malignant tumor mortality in China, and the incidence rate is increasing at an annual rate of 4%[2]. At present, it is one of the deadliest cancers globally. The occurrence of CRC is influenced by multiple factors, including genetics, dietary habits, and environmental influences[3,4], and its pathogenesis is complex and not yet fully understood.
The intestinal microenvironment contains a complex microbial ecosystem. In recent years, the interaction between gut microbiota and the occurrence and development of CRC has been a hot research topic[5]. Research has found that the gut microbiota balance in patients with CRC is disrupted, with significant differences in composition and proportion of intestinal microbes compared to normal, healthy individuals[6,7]. As early as 1997, Dove et al[8] found that in Apcmin/+ mice who spontaneously develop multiple intestinal tumors and numerous intestinal polyps, the number of small intestinal tumors was significantly reduced under germ-free conditions compared to Apcmin/+ mice in a regular environment, establishing an early association between the gut microbiota and CRC occurrence and development. Preclinical and clinical evidence has also emphasized the role of gut microbiota in altering the therapeutic response of CRC patients to chemotherapy and immunotherapy[7].
In recent years, several studies have reported the association between various microorganisms and CRC development, including Fusobacterium nucleatum (F. nucleatum), Peptostreptococcus anaerobius, Parvimonas micra, Enterotoxigenic Bacteroides fragilis, Peptostreptococcus stomatis, and Escherichia coli[9-11]. Additionally, there have been studies applying high-abundance pathogenic bacteria and low-abundance probiotics to colorectal tumor diagnostic models[12]. Many studies have indicated that microbial imbalance and infection are major factors in CRC occurrence and development[13,14].
F. nucleatum has been reported to play a role in the occurrence, development, and prognosis of CRC. Castellarin et al[15] compared 99 cases of CRC patient tissues with corresponding normal mucosal tissues and found that the average abundance of F. nucleatum in cancerous tissues was 415 times higher than in normal specimens. The relative abundance of F. nucleatum increases from intramucosal carcinoma to advanced stages of CRC and with increasing tumor malignancy[16]. The abundance of F. nucleatum in tumor tissues and fecal samples of CRC patients is significantly higher than in the normal control group[17,18]. However, the exact mechanism by which F. nucleatum plays a role in the intestinal micro-ecology and its specific molecular mechanisms have not been fully clarified.
The gut microbiota can produce short-chain fatty acids (SCFAs), such as acetate, propionate, and butyrate, through the fermentation of dietary fiber[19]. Among the SCFA family, butyrate plays an extremely important role. It exerts anti-inflammatory and anti-tumor effects on the mucosa by regulating cell metabolism, maintaining microbial homeostasis, inhibiting cell proliferation, immunomodulation, and effecting genetic/epigenetic regulation. It also provides a good energy source for colonic cells[20]. Although butyrate acts through several signaling pathways, one of key interest is the adenosine monophosphate-activated protein kinase (AMPK) pathway, as butyrate can inhibit cell proliferation and promote autophagy through this pathway, two actions that are important in cancer progression.
AMPK is a serine/threonine kinase and serves as a major energy sensor in cells, playing a critical role in maintaining energy homeostasis[21]. Once activated, AMPK promotes catabolic processes to generate adenosine triphosphate (ATP), assisting cells in escaping death and leading to drug resistance and metastasis. On the other hand, AMPK has also been reported to be positively correlated with tumor suppressor genes such as p53 and LKB1. Therefore, AMPK activation results in cell cycle arrest and tumor growth inhibition, playing a key role in cancer prevention[22].
In this study, we elucidate the role of F. nucleatum in CRC development from the perspective of the intestinal flora and intestinal metabolites and investigate the role of the intestinal flora in CRC occurrence and development.
HCT116 and DLD-1 cell lines were purchased from American Type Culture Collection [(ATCC) Manassas, VA, United States]. The HCT116 and DLD-1 cells were cultured in Dulbecco’s modified Eagle medium in an incubator with 5% CO2 at 37° C. Penicillin G (100 U/mL), streptomycin (100 mg/mL) (Beyotime, Nanjing, China), and 10% foetal bovine serum (Invitrogen, Waltham, MA, United States) were added to the culture medium. Two colonic cell lines were used to demonstrate reproducibility.
F. nucleatum strain ATCC 25586 was purchased from the ATCC. F. nucleatum was cultured in Fastidious Anaerobe Broth under anaerobic conditions.
Adult 6-8 wk-old male BALB/c mice and a weight of 18-22 g (Beijing Vital River Laboratory Animal Technology Co., Ltd., Beijing, China) were housed at the experimental animal center of Wenzhou Medical University and maintained under specific pathogen-free conditions (12-h light/dark cycle, 21° C ± 2° C, humidity 50% ± 10%). All animal studies were conducted in compliance with the animal experiment guidelines of Wenzhou Medical University. Study protocols were approved by the Animal Experimental Ethics Committee (wydw2022-0217).
All mice have free access to standard feed and water. After mice were habituated to their surroundings for a week, they were randomly divided into two groups of 12 mice per group. A suspension of F. nucleatum bacteria (1 × 108) was administered by gavage once a day for 5 d to mice in the treatment group. The control group was gavaged with physiological saline. Intervention was ceased for 1 wk, during which time, normal drinking water and diet were consumed, after which mice were gavaged with F. nucleatum again for 5 d. After repeating three cycles, the mice were euthanized using 1% pentobarbital sodium (administered by intraperitoneal injection). Feces were collected for subsequent experiments.
Fluorescence in situ hybridization (FISH) was conducted to examine the abundance of F. nucleatum using a specific probe. Briefly, sections of formalin-fixed, paraffin-embedded colonic tissue were cut into 5 μm sections and hybridized in accordance with the manufacturer’s instructions (FOCOFISH, Guangzhou, China). The following universal bacterial probe (EUB338; Cy3-labeled) was used: 5’-GCTGCCTCCCGTAGGAGT-3’. The following probe specific to F. nucleatum (FUS664; FITC-labeled) was used: 5’-CTTGTAGTTCCGC(C/T)TACCTC-3’. The resulting slides were visualized and examined under a fluorescent microscope (BX53F; Olympus, Tokyo, Japan). Five random fields per sample (200 × magnification) were examined, and the average number of bacteria per field was calculated. A blinded reviewer examined five random fields per sample (200 × magnification), and the average number of bacteria per field was calculated. Demographics of patient tissues can be found in our previous study[23].
Cell cycle analysis was performed by flow cytometry using propidium iodide (PI). CRC cells were cultured on a six-well plate (2 × 105 cells per well) and incubated at 37° C and 5% CO2 overnight. Cells were treated with F. nucleatum culture supernatants and sodium butyrate (NaB) for 24 h, after which cells were collected and incubated in the dark for 30 min with 1 mL DNA staining solution and 10 μL permeabilization solution treatment. Flow cytometry and CytExpert were used for cell cycle analysis, including G0/G1, S, and G2/M phases, and the proportion of cells in each phase was calculated.
Cells were cultured as described above. After 24 h of treatment with NaB or F. nucleatum culture supernatants, cells were centrifuged and resuspended in DCFH-DA staining solution (10 μmol/L) for 20 min at 37° C. Every 3-5 min, cell suspensions were mixed by inversion to ensure full contact with the probe. Cells were washed three times with serum-free cell culture medium to remove the DCFH-DA that did not enter the cell. Flow cytometry was used to measure fluorescence intensity.
Cellular mitochondrial depolarization was measured using a JC-1 dye, according to the manufacturer’s instructions. Briefly, after 24 h of treatment with NaB or F. nucleatum culture supernatants, cells were collected, suspended in 0.5 mL of culture media, and thoroughly mixed with 0.5 mL of the JC-1 staining working solution and incubated for 20 min at 37° C. Supernatants were removed after incubation and cells were rinsed three times with JC-1 staining buffer. Cells were resuspended with the appropriate volume of JC-1 staining buffer, followed by flow cytometric analysis.
An ATP assay kit was used to measure ATP concentrations according to the manufacturer’s instructions (Beyotime, Shanghai, China). Briefly, after 24 h of treatment with NaB or F. nucleatum culture supernatants, cells were lysed with ATP lysis buffer and centrifuged at 12000 rpm for 5 min at 4°C. Supernatants were collected and stored on ice. In 1.5 mL EP tubes, 100 μL ATP working solution (ATP test solution: ATP test dilution = 1:9) was added before the ATP test, and the tubes were incubated for 3-5 min at room temperature. Next, 20 μL of the sample or standard were added to the tube, mixed with a pipette, and the RLU values were measured with an enzyme marker after at least two seconds. The luminescence values were normalized against sample protein concentration. The data and images were processed by GraphPad Prism 6 statistical software (La Jolla, CA, United States).
After treatments, supernatant was removed and cells were fixed overnight at 4° C using 1-2 mL of 2.5% glutaraldehyde. The following day, a cell scraper was used to remove the cell layer and collected into a 1.5 mL EP tube. Cells were rinsed three times for 10 min with phosphate buffered saline, after which 1% osmic acid was added and cells were fixed in the dark at room temperature for 1 h. Cells were rinsed 3 times for 10 min with double distilled water, then stained at room temperature for 1 h with 1-2 drops of uranium acetate solution. Samples were placed in successive volumes of 50%, 70%, 80%, 90%, 100%, and 100% acetone solutions for 10 min for progressive dehydration, after which a 1:1 volume ratio of acetone to epoxy resin embedding agent was added for penetration treatment. After incubation for 60 min at 37° C, a 1:4 volume ratio mixture of acetone and embedding agent were added, and samples were incubated overnight at 37 °C. The pure embedding solution was added the following day, and samples were allowed to dry for 1 h in a 37° C oven. Pure embedding solution was added and polymerized at 45° C for 3 h and 65° C for 48 h. Semi-thin slices were then cut, followed by ultra-thin sections, which were then stained and dried for observation with transmission electron microscopy.
Small interfering RNAs (siRNAs) specific for AMPKα were purchased from Tsingke Biotechnology Co., Ltd. (Beijing, China). Transfection of siRNA was performed using Lipo3000™ transfection reagent according to the manufacturer’s protocol. The specific siRNA sequences are as follows: AMPKα siRNA - 1 sense: 5’-GCAGAAGUAUGUAGAGC
Total protein concentrations were examined using the Bicinchoninic Acid Protein Assay. Samples were separated using sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes. Skim milk was used for incubation with specific primary antibodies (1:1000 dilution) at 4° C overnight. The antibodies included phosphorylated AMPK (p-AMPK) (Cell Signaling Technology, Danvers, MA, United States), cyclin B1 (Diagbio, Hangzhou, China), Cdk1 (Abways, Beijing, China), p21 (Diagbio), C-myc (Diagbio), and GAPDH (Abways). The samples were incubated with secondary antibodies (1:2000 dilution) (Biosharp, Harjumaa, Estonia) at room temperature for 1 h. Protein bands were visualized using a hypersensitive enhanced chemiluminescence kit (Beyotime). The Bio-Rad gel imaging system was used to photograph gels, and ImageJ software was used for analysis.
Total genomic DNA of mice was obtained from their cecal contents. 16S/18S rRNA genes were amplified using specific primers. All polymerase chain reaction (PCR) steps were conducted in the reaction media (30 μL) containing 15 μL of High-Fidelity PCR Master Mix (New England Biolabs, Ipswich, MA, United States). The following thermocycling conditions were used: Predegeneration at 98° C for 1 min, denaturation at 98° C for 10 s, annealing at 50° C for 30 s, and extension at 7° C for 30 s (30 cycles); the final elongation step was carried out at 72° C for 5 min. PCR products were mixed and purified for quantification and identification. Sequencing libraries were generated by a TIANSeq Fast DNA Library Prep Kit (Illumina; Tiangen Biotech, Beijing, China). The library quality was evaluated using the Qubit@ 2.0 Fluorometer (Thermo Scientific, Waltham, MA, United States) and Agilent Bioanalyzer 2100 system (Santa Clara, CA, United States). Finally, the sequencing of the constructed library was performed on the Illumina platform using the 2 × 250 bp paired-end protocol and data analysis was conducted.
The above experiments were performed at least thrice. GraphPad Prism 6.0 software was used for graphing and statistical analysis. The BD FACS Aria II flow cytometer was used to measure the mitochondrial membrane potential and detect reactive oxygen species (ROS), and Treestar Flowjo 10.0 software (Eugene, OR, United States) was used to analyze the results. Cell cycle detection and data analysis were conducted by a CytExpert 2.3 flow cytometer (Beckman Coulter, Brea, CA, United States). All variables were presented as mean ± standard deviation. Other statistics were analyzed using Student’s t-test. P < 0.05 was considered statistically significant.
To study the difference in the flora in CRC tissues, 39 fresh clinical tissue samples (including 24 CRC samples, 10 normal tissue samples, and 5 paracancerous tissue samples) were collected for 16S rDNA sequencing. The results showed that there were significant differences between normal and CRC tissues at the family level (P < 0.05, Figure 1A). Fusobacterium was found at a higher abundance in CRC tissues, but in lower abundance in normal and adjacent tissues. Clostridium butyrate-producing bacteria were also found to drop in abundance as CRC spread, mostly because their abundance was substantially higher in normal tissue samples than in CRC and adjacent tissues.
Ten tissue samples were chosen from the aforementioned samples, including both normal and CRC tissue. FISH was used to determine the quantity of F. nucleatum in colorectal tissue. As shown in Figure 1B, the abundance of F. nucleatum in CRC tissue is significantly higher than in normal colorectal tissue, indicating that F. nucleatum is strongly associated with CRC occurrence and progression.
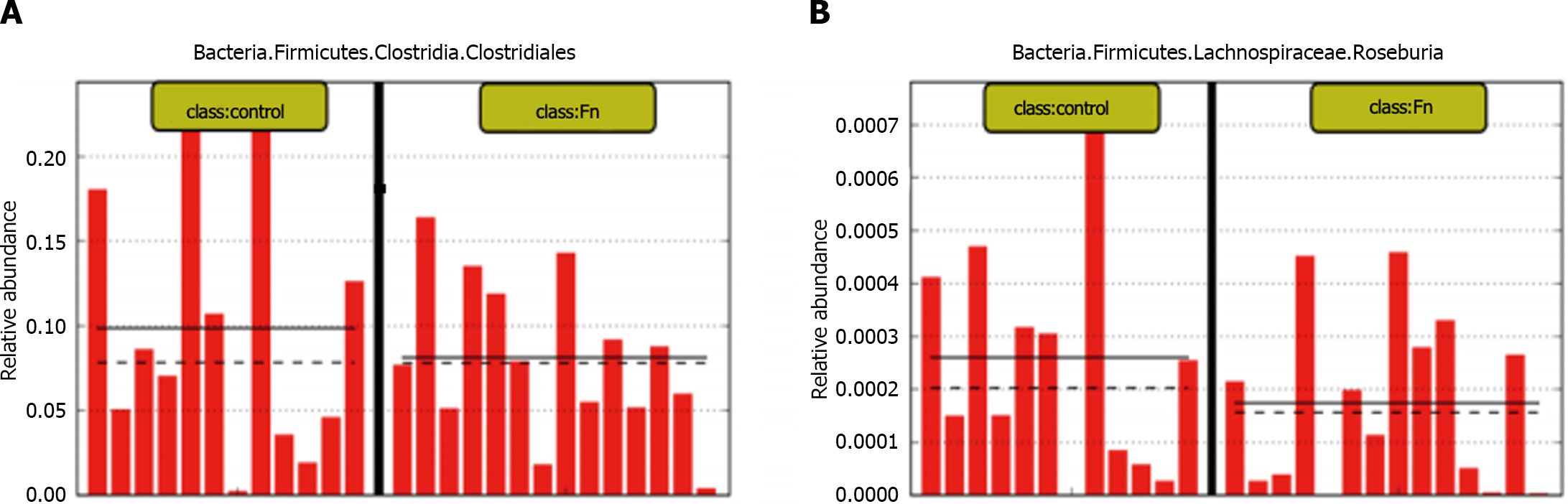
To further study the effect of intestinal flora on CRC development, we sequenced 16S rDNA from 105 stool samples, including 44 CRC patients and 61 healthy people. The results of LefSe analysis showed that there are significant differences in bacteria between CRC and normal/adjacent tissues (Figure 2A and B). Compared to CRC patients, the abundance of Lac_Lachnospira, Lac_Roseburia and Rum_Faecalibacterium significantly increased in healthy fecal samples (P < 0.05), which is the characteristic bacteria in fecal samples of healthy people (Figure 2C). The abundance of Trichospira and Rochella is much higher in fecal samples from healthy people compared to patients with stage I-IV CRC. There was no significant difference between CRC stages. The abundance of Rum_Faecalibacterium in healthy people and patients with stage I CRC fecal samples were the same (Figure 2D), demonstrating that although there is a significant difference in the abundance of butyrate-producing bacteria between CRC patients and healthy people, its role in CRC development needs to be further explored.
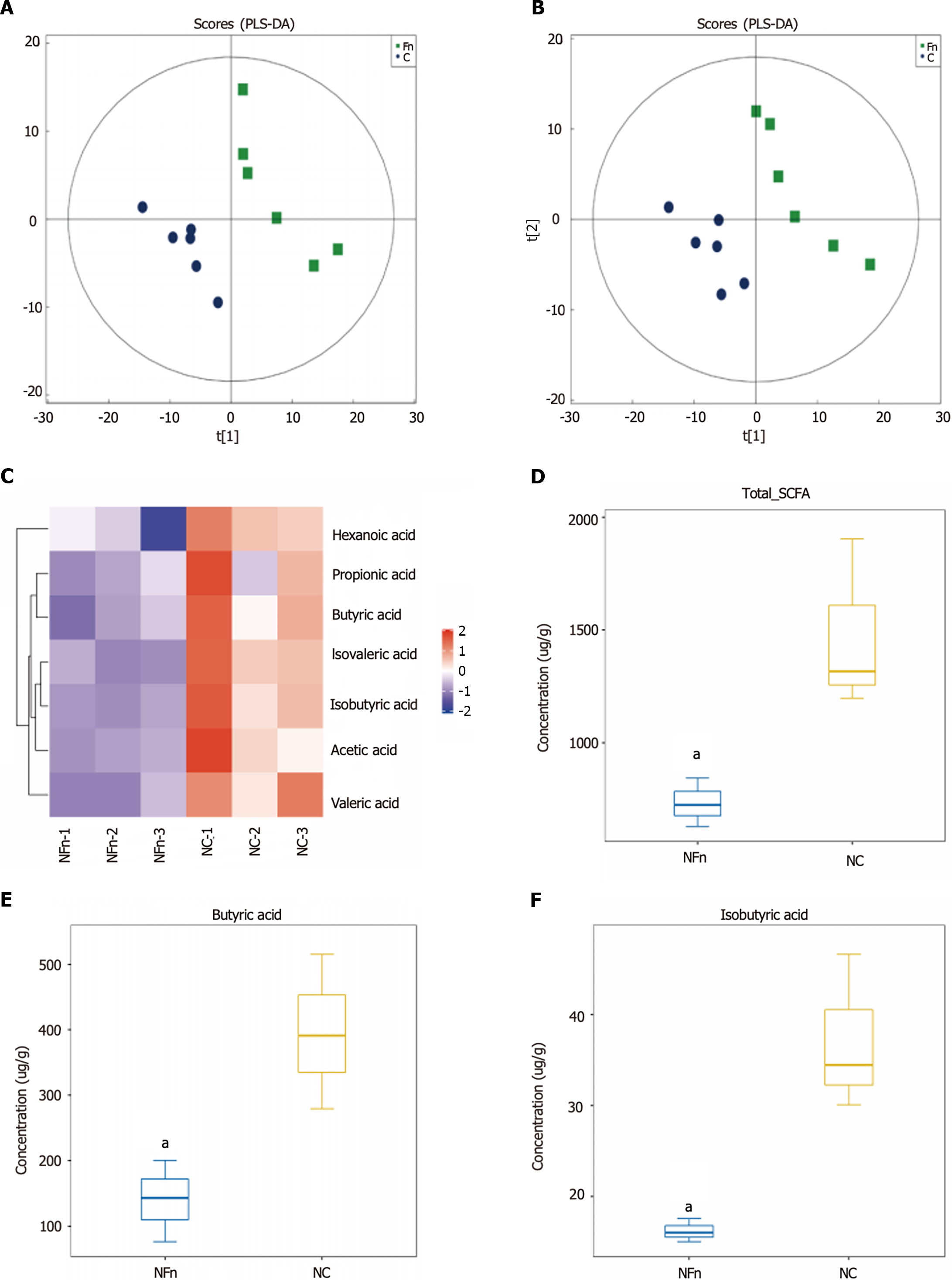
To further clarify the role and relationship between F. nucleatum and butyric acid bacteria in CRC occurrence and development, BALB/c mice were gavaged with an F. nucleatum suspension (1 × 108 CFU/d). 16S rDNA sequencing was used to detect changes in bacterial abundance in fecal samples. As shown in Figure 3A and B, intragastric administration of F. nucleatum resulted in significantly fewer butyric acid-producing bacteria in fecal samples compared to the control group.
Fecal samples from mice treated with F. nucleatum were collected, and GC-MS targeted metabolomics were used to discover and evaluate the quantities of short chain fatty acids (SCFAs). The GC-MS data shown in Figure 4A and B shows a significant difference in the PC1 direction between the F. nucleatum-treated group and the control group, indicating a significant divergence in metabolic pathways between the two groups. According to targeted SCFA detection, the amount of SCFAs in the F. nucleatum-treated group was much lower than in the untreated group (Figure 4C). Butyric acid/isobutyric acid levels in fecal samples significantly decreased following F. nucleatum treatment (P < 0.05) (Figure 4D-F), which is one of the most prominent components of SCFAs. This suggests that in mice, F. nucleatum treatment changes intestinal metabolic patterns, due to a significant decrease in the proportion of butyric acid-producing bacteria. As a result, F. nucleatum may affect the gut microbiota in addition to influencing metabolites and metabolic rate.
We treated human CRC cells DLD-1 and HCT116 cells with 2 mmol/L NaB and used flow cytometry to examine the effects on the cell cycle of HCT116 and DLD-1 cells. The proportion of G2/M phase cells dramatically increased after 24 h of NaB administration, as shown in Figure 5A and B. This suggests that NaB can prevent HCT116 and DLD-1 cells from entering the G1 phase. There were no discernible alterations in the cell cycle when we exposed cells to F. nucleatum supernatant (FAB). However, these effects were reduced when cells were treated with NaB and F. nucleatum supernatant, showing that F. nucleatum can suppress the efficacy of NaB.
An aberrant drop in mitochondrial membrane potential and a rise in ROS levels indicate mitochondrial malfunction. According to research, NaB can cause mitochondrial damage as part of its anti-tumor effect. We employed the JC-1 dye to measure mitochondrial membrane potential. HCT116 and DLD-1 cells were treated with 2 mmol/L NaB for 24 h and found that the mitochondrial membrane potential significantly decreased in both cell lines, whereas treatment with F. nucleatum supernatant increased the membrane potential of DLD-1 cells (P < 0.001) but did not significantly alter the mitochondrial membrane potential of HCT116 cells. The effect of NaB on mitochondrial membrane potential is eliminated when cells are treated with NaB and FAB, indicating that F. nucleatum metabolites can inhibit the damage caused to the mitochondrial membrane by NaB (Figure 6A and B).
ROS levels are a significant indicator of cell damage induced by normal physiological functions and environmental influences. When HCT116 and DLD-1 cells were treated with NaB, ROS levels significantly increased (P < 0.001) and the peak level shifted to the right. When cells were treated with FAB, there was a small reduction in ROS levels, but no significant difference when compared to the control group. When cells were co-treated with FAB and NaB, the effect of NaB was reversed (Figure 6C and D). This suggests that F. nucleatum metabolites have a protective impact on CRC cells, possibly by reducing the damage caused by NaB to mitochondria.
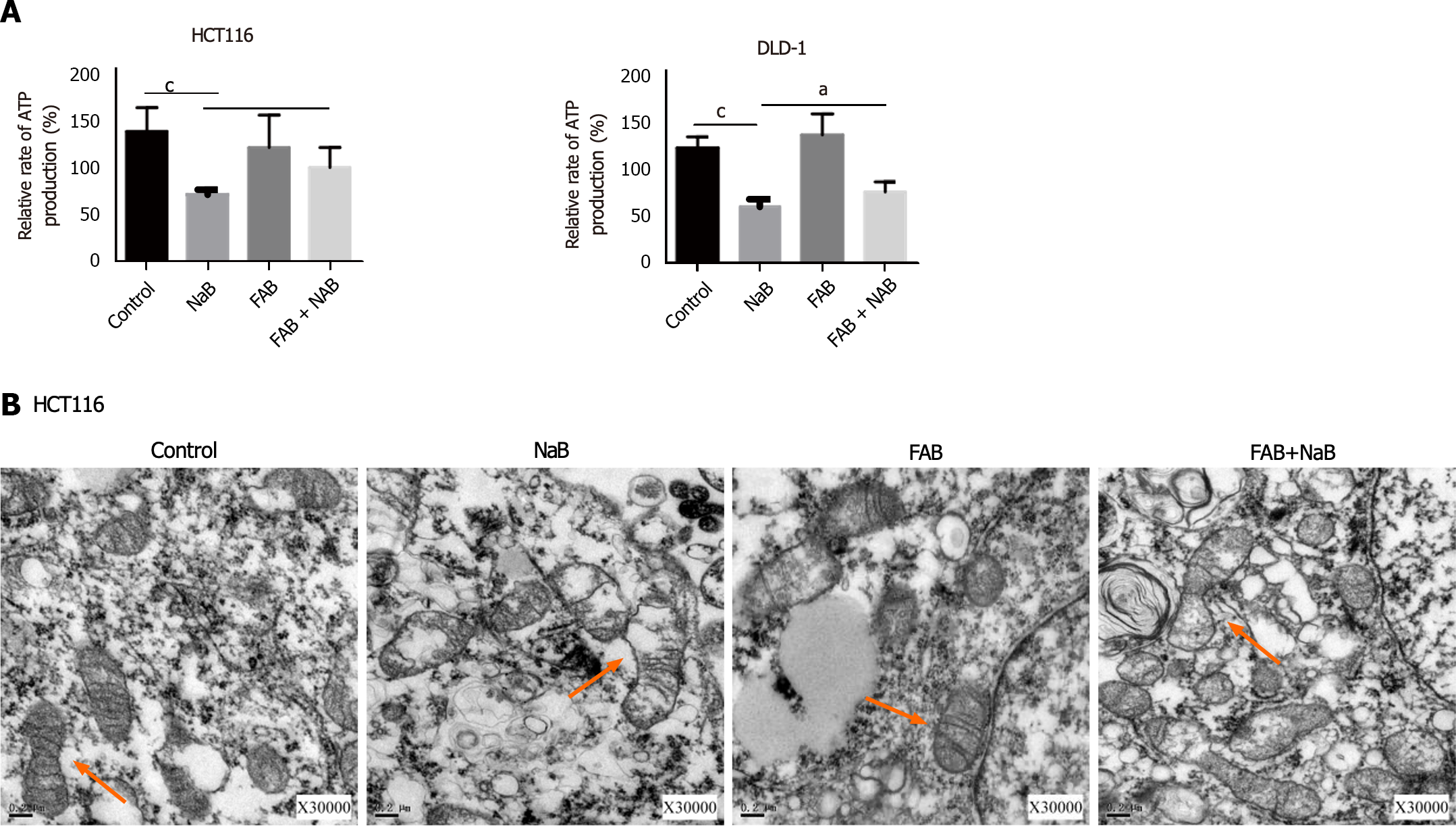
We assessed the amount of ATP present in HCT116 and DLD-1 cells to further test whether NaB had an impact on energy metabolism in CRC cells. Cells treated with NaB showed a substantial drop in ATP levels compared to the control group, but this impact was mitigated by FAB. Transmission electron microscopy showed that the mitochondria of the control group were largely undamaged, with distinct cristae and no visible damage. However, following NaB treatment, the mitochondrial matrix and cristae start to vanish, and in extreme situations, matrix overflow and mitochondrial membrane damage occurred. After the addition of FAB, the shape of the mitochondria was improved and the damage caused by NaB was diminished (Figure 7).
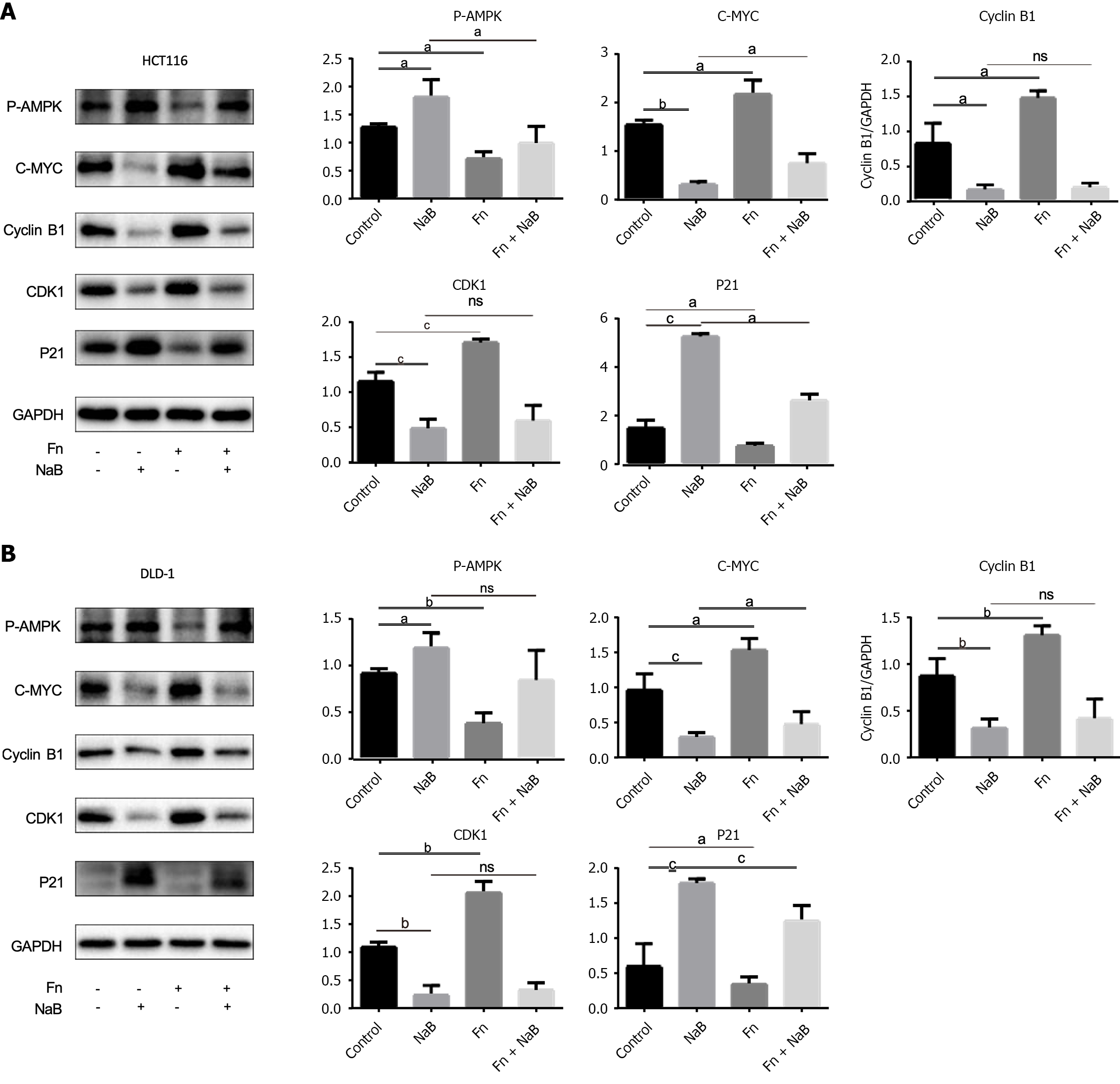
According to our findings, NaB affects energy metabolism of CRC cell lines. The serine/threonine kinase AMPK is a significant energy sensor in cells and is essential for preserving energy balance. AMPK activation results in cell cycle arrest and inhibition of tumor development and will therefore be essential to cancer prevention. We investigated the expression levels of cycle-related proteins in the AMPK signaling pathway. Protein analysis revealed that AMPK was activated, p-AMPK expression increased, proliferation proteins such as CDK1, C-myc, and cyclin B1 were decreased, and cycle arrest protein P21 expression increased after NaB treatment of DLD-1 and HCT116 cells. In contrast, F. nucleatum treatment reduced AMPK phosphorylation and promoted the expression of cyclic-related proteins CDK1 and C-myc, indicating that F. nucleatum treatment promoted cell cycle progression in CRC cells. However, F. nucleatum and NaB were administered together, the effect of F. nucleatum was significantly attenuated (Figure 8), which is consistent with the findings that early NaB blocked the CRC cell cycle.
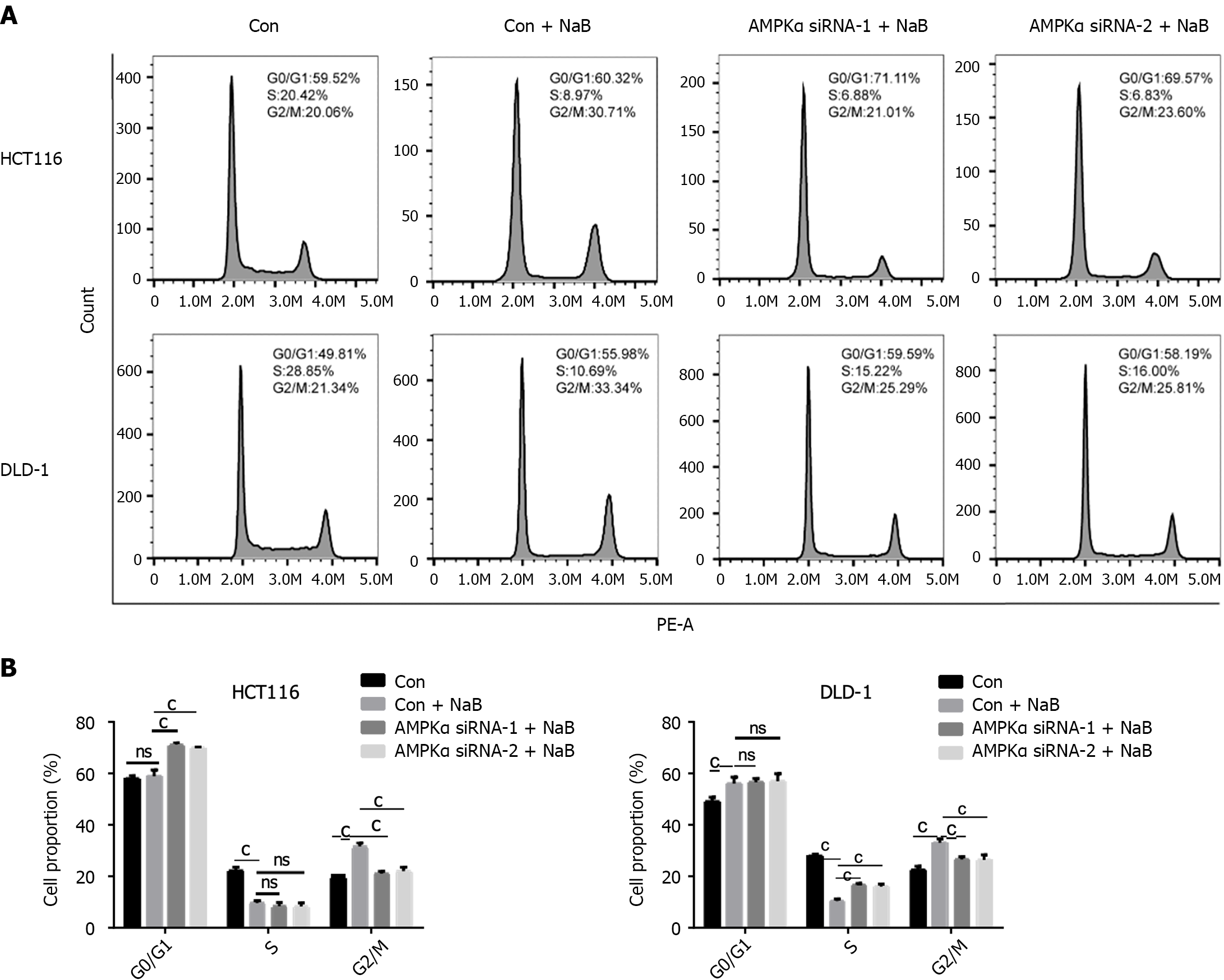
To establish the role of AMPK in NaB-induced cell inhibition, we used AMPK-specific siRNA to knock down AMPK expression. In HCT116 and DLD-1 cells (Figure 9), AMPK-specific siRNA prevented AMPK phosphorylation and increased the expression of downstream associated proliferative proteins following NaB treatment (Figure 10). Flow cytometry analysis also revealed that AMPK-specific siRNA prevented the cycle arrest caused by NaB (Figure 11). These findings indicate that AMPK is required for NaB to inhibit CRC cell proliferation.
There is a close relationship between the intestinal microecology and the occurrence and development of CRC. Dysregulation of microbial balance is often observed in CRC patients. According to some studies, F. nucleatum is abundant in the tumor microenvironment and fecal samples of CRC patients, thus its presence is regarded as one of the risk factors for the incidence and progression of CRC[24]. Yu et al[25] discovered that F. nucleatum content is prevalent in CRC tissues of patients whose cancer recurs after chemotherapy. 16S rDNA sequencing was employed in this experiment to detect bacterial diversity between CRC tissues and normal/paracancerous tissues. Fusobacteriaceae was discovered to be primarily concentrated in CRC tissues, whereas Clostrididae was shown to be abundant in normal tissues (Figure 1A). Using FISH technology to detect F. nucleatum content in CRC tissue and normal tissue samples, we found that the abundance of F. nucleatum in CRC tissue was significantly higher than that in normal tissue (Figure 1B), consistent with previous sequencing results. This indicates that F. nucleatum plays an important role in the occurrence and development of CRC.
This experiment used 16S rDNA sequencing on fecal samples from CRC patients and healthy individuals to further investigate the relationship between intestinal flora and CRC and found significant differences between the flora of CRC tissue and normal/paracancerous tissue (Figure 2A and B). Among these bacteria, butyric acid generating bacteria such as Lac_Lachnospira, Lac_Roseburia, and Rum_Faecalibacterium are common in feces of healthy patients (Figure 2C). Our research also found that the abundance of Lac_Lachnospira, Lac_Roseburia, and Rum faecalibacterium decreased significantly as CRC progressed from stage I to later stages (Figure 2D), indicating that butyric acid-producing bacteria play an important role in the progression of CRC.
After administering F. nucleatum intragastrically to BALB/c mice, their feces were analyzed to assess variations in bacterial populations and metabolic processes. The feces of mice treated with F. nucleatum contained much lower levels of butyric acid generating bacteria (Lac_Rosebaria and Clo_Closteriales) (Figure 3A and B). Interestingly, changes in the intestinal flora caused changes in intestinal metabolic patterns (Figure 4A and B), as well as changes in the production of SCFAs, such as butyric acid (Figure 4C-F). This finding implies that F. nucleatum may promote CRC progression by changing intestinal metabolites in vitro and in vivo.
Furthermore, studies have shown that F. nucleatum competes with the beneficial butyrate-producing Clostridium butyricum units, and that an abundance of F. nucleatum leads to a decrease in Clostridium butyricum abundance[26]. Therefore, it is vital to understand the association between F. nucleatum and butyric acid. We have previously treated DLD-1 cells with F. nucleatum and collected metabolites for nuclear magnetic resonance analysis. The results showed that after treating CRC cells with F. nucleatum, the extracellular concentration of butyrate significantly decreased[27]. Therefore, we speculate that F. nucleatum can actively consume butyric acid, promoting CRC development. In the future, we will further explore the relationship between F. nucleatum and Clostridium butyricum.
Butyrate is the preferred energy source for colonic cells and has been shown to inhibit tumor development through a variety of mechanisms[28], including anti-inflammatory and immunomodulatory effects, down-regulation of the Wnt signaling pathway[29], inhibition of tumor cell proliferation and migration[21], limitation of tumor angiogenesis[22], induction of apoptosis[30], and promotion of tumor cell differentiation[31]. In this study, we used flow cytometry to investigate the effect of NaB on the cell cycle in DLD-1 and HCT116 CRC cell lines. Our findings showed that NaB arrested the cell cycle at the G2/M phase. When CRC cells were co-treated with F. nucleatum supernatant and NaB, there was a significant drop in the number of cells in the G2/M phase, demonstrating that F. nucleatum can mitigate the effect of NaB on the cell cycle (Figure 5A and B).
ATP is the most direct source of energy in organisms, acting as the driving force for numerous biological operations. The mitochondrion is at the heart of cellular energy metabolism, producing the majority of ATP[32]. ROS are byproducts of aerobic respiration as well as signaling molecules that affect numerous cellular activities[33], and they play an important role in regulating various physiological functions in animals. In this study, we discovered that NaB treatment reduced ATP generation while increasing ROS production in CRC cells. The effect of NaB was partially inhibited when treated with F. nucleatum (Figures 6C, D, and 7A).
Furthermore, we used transmission electron microscopy to assess alterations in mitochondrial membrane potential and examined mitochondrial morphology in CRC cell lines. The mitochondrial membrane potential was shown to decrease after NaB treatment. In addition, the mitochondrial membrane was disrupted, the matrix began to melt, and the crista began to blur. When compared to the NaB group, the mitochondrial membrane potential increased in cells co-treated with NaB and F. nucleatum supernatants (FAB), as did the mitochondrial morphology. These findings reveal that NaB can disrupt energy metabolism by altering mitochondrial structure, whereas F. nucleatum metabolites have a protective impact and can mitigate NaB damage (Figure 7B).
AMPK is a highly conserved serine/threonine protein kinase that regulates cell cycle checkpoints in cancer cells in response to energy stress to coordinate proliferation and energy availability[34]. Because our experimental results show that NaB and F. nucleatum can impact ATP generation in CRC cells, we hypothesize that both affect CRC development via the AMPK pathway. After 24 h of treatment with F. nucleatum, the expression of p-AMPK was reduced, showing a relationship between F. nucleatum and the AMPK pathway. We evaluated the expression levels of cell cycle-related proteins in the AMPK pathway to further analyze the relationship between F. nucleatum, NaB, and the AMPK pathway. Western blotting revealed that NaB increased the expression of Thr172 p-AMPK and activated AMPK in both CRC cell lines (HCT116 and DLD-1) (Figure 8A and B). Furthermore, the AMPK-related proliferative proteins c-Myc, CDK1 and cyclin B1 were downregulated, whereas the cycle inhibitory protein P21 was upregulated. F. nucleatum treatment increased the expression of cyclin-related proteins c-Myc, CDK1 and cyclin B1, while inhibiting the production of phosphorylated AMPK and P21. These findings suggest that F. nucleatum and NaB can both regulate the proliferation of CRC cells via AMPK.
We employed AMPK-specific siRNA to knock down AMPK expression and then co-treated HCT116 and DLD-1 cells with NaB to explore the role of AMPK in CRC cell proliferation. AMPK-specific siRNA substantially decreases AMPK phosphorylation and the expression of its downstream proteins c-Myc, cyclinB1, and CDK1 (Figure 9A and B). When cells were co-treated with AMPK-specific siRNA and NaB, AMPK-specific siRNA prevented the induction of cell cycle arrest observed with NaB alone (Figures 10 and 11). These data suggest that NaB requires AMPK to inhibit CRC cell proliferation.
In conclusion, F. nucleatum can regulate the intestinal metabolite butyrate, and NaB can regulate energy metabolism and prevent CRC proliferation via the AMPK pathway. The findings of this study may provide a better understanding of CRC pathogenesis , thereby facilitating the development of more effective therapeutic approaches.
Colorectal cancer (CRC) ranks among the most prevalent malignant neoplasms globally. Fusobacterium nucleatum (F. nucleatum) has been implicated in the initiation, progression, and prognostic outcomes of CRC. Butyrate, a short-chain fatty acid (SCFA) derived from the bacterial fermentation of soluble dietary fiber, exhibits inhibitory effects on several types of cancers.
Recent research has demonstrated that the SCFA butyrate can suppress the proliferation, enrichment, and adherence of F. nucleatum in CRC tissues. This suppression is achieved by the downregulation of adhesion-associated outer membrane proteins, including RadD, FomA, and FadA. Consequently, this leads to a decrease in the colonization and invasion of F. nucleatum and mitigates its contribution to chemoresistance. Therefore, this study aims to investigate whether F. nucleatum can influence the synthesis of the intestinal metabolite butyric acid, thereby facilitating the development of CRC.
Exploring whether F. nucleatum affects the production of intestinal metabolite butyrate to promote CRC development.
Fecal samples were collected from mice in the treatment group following oral administration of F. nucleatum for the analysis of SCFAs and 16S rDNA. Concurrently, CRC cells underwent co-treatment with F. nucleatum and sodium butyrate (NaB) in vitro to assess alterations in the cell cycle, mitochondrial functionality, and the expression of pertinent proteins.
The abundance of F. nucleatum is markedly elevated in fecal specimens and CRC tissues from patients with CRC. F. nucleatum suppresses the synthesis of the SCFA butyric acid. NaB impairs mitochondrial functionality and impedes the cell cycle in CRC cells. Both NaB and F. nucleatum modulate the growth of CRC cells via the adenosine monophosphate-activated protein kinase (AMPK) signaling pathway. The presence of AMPK is essential for NaB’s effectiveness in inhibiting CRC cell proliferation.
Our findings showed that the abundance of F. nucleatum is significantly high in fecal samples and CRC tissues from CRC patients. F. nucleatum impedes the synthesis of the SCFA butyric acid. NaB compromises mitochondrial functionality and obstructs the cell cycle in CRC cells. The growth of CRC cells is modulated by both NaB and F. nucleatum via the AMPK signaling pathway. The presence of AMPK is critical for the ability of NaB to curb CRC cell proliferation.
The outcomes of this research could enhance our comprehension of CRC pathogenesis, potentially leading to the formulation of more efficacious therapeutic strategies.
The author thanks Dr. Xu Chang from the First Affiliated Hospital of Wenzhou Medical University for providing cancer pathological sections.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Jordan P, Portugal S-Editor: Wang JJ L-Editor: Filipodia P-Editor: Chen YX
| 1. | Wong MCS, Huang J, Huang JLW, Pang TWY, Choi P, Wang J, Chiang JI, Jiang JY. Global Prevalence of Colorectal Neoplasia: A Systematic Review and Meta-Analysis. Clin Gastroenterol Hepatol. 2020;18:553-561.e10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 57] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 2. | Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11444] [Cited by in RCA: 13213] [Article Influence: 1468.1] [Reference Citation Analysis (3)] |
| 3. | Seol JE, Kim J, Lee BH, Hwang DY, Jeong J, Lee HJ, Ahn YO, Lee JE, Kim DH. Folate, alcohol, ADH1B and ALDH2 and colorectal cancer risk. Public Health Nutr. 2020;1-8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 4. | Carr PR, Weigl K, Edelmann D, Jansen L, Chang-Claude J, Brenner H, Hoffmeister M. Estimation of Absolute Risk of Colorectal Cancer Based on Healthy Lifestyle, Genetic Risk, and Colonoscopy Status in a Population-Based Study. Gastroenterology. 2020;159:129-138.e9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 70] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 5. | Park EM, Chelvanambi M, Bhutiani N, Kroemer G, Zitvogel L, Wargo JA. Targeting the gut and tumor microbiota in cancer. Nat Med. 2022;28:690-703. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 298] [Article Influence: 99.3] [Reference Citation Analysis (0)] |
| 6. | Kim J, Lee HK. Potential Role of the Gut Microbiome In Colorectal Cancer Progression. Front Immunol. 2021;12:807648. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 111] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 7. | Wong CC, Yu J. Gut microbiota in colorectal cancer development and therapy. Nat Rev Clin Oncol. 2023;20:429-452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 277] [Reference Citation Analysis (0)] |
| 8. | Dove WF, Clipson L, Gould KA, Luongo C, Marshall DJ, Moser AR, Newton MA, Jacoby RF. Intestinal neoplasia in the ApcMin mouse: independence from the microbial and natural killer (beige locus) status. Cancer Res. 1997;57:812-814. [PubMed] |
| 9. | Yu J, Feng Q, Wong SH, Zhang D, Liang QY, Qin Y, Tang L, Zhao H, Stenvang J, Li Y, Wang X, Xu X, Chen N, Wu WK, Al-Aama J, Nielsen HJ, Kiilerich P, Jensen BA, Yau TO, Lan Z, Jia H, Li J, Xiao L, Lam TY, Ng SC, Cheng AS, Wong VW, Chan FK, Yang H, Madsen L, Datz C, Tilg H, Wang J, Brünner N, Kristiansen K, Arumugam M, Sung JJ. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. 2017;66:70-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 917] [Cited by in RCA: 783] [Article Influence: 97.9] [Reference Citation Analysis (0)] |
| 10. | Pleguezuelos-Manzano C, Puschhof J, Rosendahl Huber A, van Hoeck A, Wood HM, Nomburg J, Gurjao C, Manders F, Dalmasso G, Stege PB, Paganelli FL, Geurts MH, Beumer J, Mizutani T, Miao Y, van der Linden R, van der Elst S; Genomics England Research Consortium, Garcia KC, Top J, Willems RJL, Giannakis M, Bonnet R, Quirke P, Meyerson M, Cuppen E, van Boxtel R, Clevers H. Mutational signature in colorectal cancer caused by genotoxic pks(+) E. coli. Nature. 2020;580:269-273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 564] [Cited by in RCA: 721] [Article Influence: 144.2] [Reference Citation Analysis (0)] |
| 11. | Motamedi H, Ari MM, Shahlaei M, Moradi S, Farhadikia P, Alvandi A, Abiri R. Designing multi-epitope vaccine against important colorectal cancer (CRC) associated pathogens based on immunoinformatics approach. BMC Bioinformatics. 2023;24:65. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 12. | Kong C, Liang L, Liu G, Du L, Yang Y, Liu J, Shi D, Li X, Ma Y. Integrated metagenomic and metabolomic analysis reveals distinct gut-microbiome-derived phenotypes in early-onset colorectal cancer. Gut. 2023;72:1129-1142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 128] [Article Influence: 64.0] [Reference Citation Analysis (0)] |
| 13. | Lee MH. Harness the functions of gut microbiome in tumorigenesis for cancer treatment. Cancer Commun (Lond). 2021;41:937-967. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 33] [Reference Citation Analysis (0)] |
| 14. | Seely KD, Morgan AD, Hagenstein LD, Florey GM, Small JM. Bacterial Involvement in Progression and Metastasis of Colorectal Neoplasia. Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 15. | Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, Holt RA. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1164] [Cited by in RCA: 1497] [Article Influence: 106.9] [Reference Citation Analysis (0)] |
| 16. | Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, Watanabe H, Masuda K, Nishimoto Y, Kubo M, Hosoda F, Rokutan H, Matsumoto M, Takamaru H, Yamada M, Matsuda T, Iwasaki M, Yamaji T, Yachida T, Soga T, Kurokawa K, Toyoda A, Ogura Y, Hayashi T, Hatakeyama M, Nakagama H, Saito Y, Fukuda S, Shibata T, Yamada T. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med. 2019;25:968-976. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 404] [Cited by in RCA: 878] [Article Influence: 146.3] [Reference Citation Analysis (0)] |
| 17. | Tunsjø HS, Gundersen G, Rangnes F, Noone JC, Endres A, Bemanian V. Detection of Fusobacterium nucleatum in stool and colonic tissues from Norwegian colorectal cancer patients. Eur J Clin Microbiol Infect Dis. 2019;38:1367-1376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 18. | Wang N, Fang JY. Fusobacterium nucleatum, a key pathogenic factor and microbial biomarker for colorectal cancer. Trends Microbiol. 2023;31:159-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 164] [Article Influence: 82.0] [Reference Citation Analysis (0)] |
| 19. | Kim SM, Vetrivel P, Ha SE, Kim HH, Kim JA, Kim GS. Apigetrin induces extrinsic apoptosis, autophagy and G2/M phase cell cycle arrest through PI3K/AKT/mTOR pathway in AGS human gastric cancer cell. J Nutr Biochem. 2020;83:108427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 65] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 20. | Chattopadhyay I, Gundamaraju R, Jha NK, Gupta PK, Dey A, Mandal CC, Ford BM. Interplay between Dysbiosis of Gut Microbiome, Lipid Metabolism, and Tumorigenesis: Can Gut Dysbiosis Stand as a Prognostic Marker in Cancer? Dis Markers. 2022;2022:2941248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 33] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 21. | Zeng H, Briske-Anderson M. Prolonged butyrate treatment inhibits the migration and invasion potential of HT1080 tumor cells. J Nutr. 2005;135:291-295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 51] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 22. | Zgouras D, Wächtershäuser A, Frings D, Stein J. Butyrate impairs intestinal tumor cell-induced angiogenesis by inhibiting HIF-1alpha nuclear translocation. Biochem Biophys Res Commun. 2003;300:832-838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 65] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 23. | Yu T, Ji L, Lou L, Ye S, Fang X, Li C, Jiang F, Gao H, Lou Y, Li X. Fusobacterium nucleatum Affects Cell Apoptosis by Regulating Intestinal Flora and Metabolites to Promote the Development of Colorectal Cancer. Front Microbiol. 2022;13:841157. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Reference Citation Analysis (0)] |
| 24. | Hashemi Goradel N, Heidarzadeh S, Jahangiri S, Farhood B, Mortezaee K, Khanlarkhani N, Negahdari B. Fusobacterium nucleatum and colorectal cancer: A mechanistic overview. J Cell Physiol. 2019;234:2337-2344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 124] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 25. | Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, Qian Y, Kryczek I, Sun D, Nagarsheth N, Chen Y, Chen H, Hong J, Zou W, Fang JY. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell. 2017;170:548-563.e16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 814] [Cited by in RCA: 1477] [Article Influence: 184.6] [Reference Citation Analysis (0)] |
| 26. | Zheng DW, Dong X, Pan P, Chen KW, Fan JX, Cheng SX, Zhang XZ. Phage-guided modulation of the gut microbiota of mouse models of colorectal cancer augments their responses to chemotherapy. Nat Biomed Eng. 2019;3:717-728. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 303] [Article Influence: 50.5] [Reference Citation Analysis (0)] |
| 27. | Huang JP, Yang LN, Fang XT, Yu TT, Li X, Lou YL. [Fusobacterium nucleatum and Cdk5 promote colorectal cancer cell migration]. Chinese J Microecol. 2020;32:889-892,896. [DOI] [Full Text] |
| 28. | O'Keefe SJ. Diet, microorganisms and their metabolites, and colon cancer. Nat Rev Gastroenterol Hepatol. 2016;13:691-706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 521] [Cited by in RCA: 778] [Article Influence: 86.4] [Reference Citation Analysis (0)] |
| 29. | Bordonaro M, Lazarova DL, Sartorelli AC. Butyrate and Wnt signaling: a possible solution to the puzzle of dietary fiber and colon cancer risk? Cell Cycle. 2008;7:1178-1183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 104] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 30. | Chirakkal H, Leech SH, Brookes KE, Prais AL, Waby JS, Corfe BM. Upregulation of BAK by butyrate in the colon is associated with increased Sp3 binding. Oncogene. 2006;25:7192-7200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 59] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 31. | Comalada M, Bailón E, de Haro O, Lara-Villoslada F, Xaus J, Zarzuelo A, Gálvez J. The effects of short-chain fatty acids on colon epithelial proliferation and survival depend on the cellular phenotype. J Cancer Res Clin Oncol. 2006;132:487-497. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 162] [Cited by in RCA: 155] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 32. | Zhang J, Qiao W, Luo Y. Mitochondrial quality control proteases and their modulation for cancer therapy. Med Res Rev. 2023;43:399-436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 24] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 33. | Kasai S, Shimizu S, Tatara Y, Mimura J, Itoh K. Regulation of Nrf2 by Mitochondrial Reactive Oxygen Species in Physiology and Pathology. Biomolecules. 2020;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 128] [Cited by in RCA: 363] [Article Influence: 72.6] [Reference Citation Analysis (0)] |
| 34. | Steinberg GR, Hardie DG. New insights into activation and function of the AMPK. Nat Rev Mol Cell Biol. 2023;24:255-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 436] [Article Influence: 218.0] [Reference Citation Analysis (0)] |