Published online Apr 14, 2024. doi: 10.3748/wjg.v30.i14.1982
Peer-review started: December 27, 2023
First decision: January 27, 2024
Revised: February 19, 2024
Accepted: March 25, 2024
Article in press: March 25, 2024
Published online: April 14, 2024
Processing time: 107 Days and 15.7 Hours
Unmet needs exist in metabolic dysfunction-associated steatotic liver disease (MASLD) risk stratification. Our ability to identify patients with MASLD with advanced fibrosis and at higher risk for adverse outcomes is still limited. Incorporating novel biomarkers could represent a meaningful improvement to current risk predictors. With this aim, omics technologies have revolutionized the process of MASLD biomarker discovery over the past decades. While the research in this field is thriving, much of the publication has been haphazard, often using single-omics data and specimen sets of convenience, with many identified candidate biomarkers but lacking clinical validation and utility. If we incorporate these biomarkers to direct patients’ management, it should be considered that the roadmap for translating a newly discovered omics-based signature to an actual, analytically valid test useful in MASLD clinical practice is rigorous and, therefore, not easily accomplished. This article presents an overview of this area’s current state, the conceivable opportunities and challenges of omics-based laboratory diagnostics, and a roadmap for improving MASLD biomarker research.
Core Tip: Identifying patients with metabolic dysfunction-associated steatotic liver disease (MASLD) at higher risk for adverse outcomes is still a crucial clinical challenge. Novel and non-invasive screening, monitoring, and risk stratification methods are urgently needed. With this aim, omics technologies have revolutionized the process of MASLD biomarker discovery. Although many omics-based biomarkers were identified over the past decades, their translation into clinically useful tests that can guide management decisions has proven more difficult than expected. This review presents an overview of this area’s current state, the conceivable opportunities and challenges of omics-based laboratory diagnostics, and a roadmap for improving MASLD biomarker research.
- Citation: Trinks J, Mascardi MF, Gadano A, Marciano S. Omics-based biomarkers as useful tools in metabolic dysfunction-associated steatotic liver disease clinical practice: How far are we? World J Gastroenterol 2024; 30(14): 1982-1989
- URL: https://www.wjgnet.com/1007-9327/full/v30/i14/1982.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i14.1982
The exponential global increase in obesity and type 2 diabetes is to blame for the rising epidemic of metabolic dysfunction-associated steatotic liver disease (MASLD) in the last two decades. Globally, nearly a quarter of the world’s population is estimated to be affected by MASLD, with even higher rates in the Middle East, Northern Africa, and Central and South America. Without clear risk stratification and therapeutic options, MASLD has soon become the leading cause of chronic liver disease and liver-related morbidity and mortality worldwide[1].
Unfortunately, over half the adult population is expected to have MASLD by 2040, mainly affecting women, smokers, and those without metabolic syndrome[2]. This scenario will expand the number of patients with advanced liver fibrosis and end-stage liver disease, increasing the rates of liver transplantation and multiplying healthcare costs.
Current measures to overcome this situation are centered on the identification of people at the highest risk of pro
These limitations have driven the need for non-invasive MASLD screening, monitoring, and risk stratification meth
On the contrary, low-tech methods such as the anthropometric clinical indicators of visceral obesity have been pro
Since the Human Genome Project, the development of new technologies called “omics” has made it possible to measure a vast number of biological molecules within a tissue or cell in a high-throughput way. Many areas of research can be classified as omics, and the terms used to define them depend on the type of biological molecules globally ana
In MASLD research, new innovative omics technologies are extensively used in both preclinical (in vitro and in vivo) models and retrospective studies with archived samples, as they offer the possibility of in-depth screening for novel biomarkers and a better understanding of MASLD pathological processes[15]. However, translating their results into clinically useful tests that can guide management decisions has proven more difficult than expected. This article presents an overview of this area’s current state, the conceivable opportunities and challenges of omics-based laboratory diagnostics, and a roadmap for improving MASLD biomarker research.
We used PubMed (National Library of Medicine) and Reference Citation Analysis (RCA) databases to search and retrieve scientific articles to describe the current state of omics-derived biomarker development in MASLD. The “OR” and “AND” connectors were used to combine the descriptors: (“non-alcoholic fatty liver disease” or “non-alcoholic steatohepatitis”, “fatty liver” or “NAFLD” or “NASH” or “metabolic-associated fatty liver disease” or “MAFLD” “metabolic dysfunction–associated steatotic liver disease” or “MASLD”) and (“human”) and (“biomarker”) and (“omics” or “multi-omics” or “microbiome” or “genomics” or “proteomics” or “metabolomics” or “metagenomics” or “transcriptomics”).
Inclusion criteria were the availability of the full-text publication written in English up to December 2023. Review studies, publications present in more than one database, and out-of-scope studies were excluded. After reading the titles and abstracts of the studies, 24 of 163 studies found in the databases were excluded: 2 duplicate studies, 5 Chinese studies, 11 in vitro studies, and six evaluated individuals with other chronic liver diseases. The remaining studies were fully read according to the eligibility criteria. Thus, 139 articles were included in the analysis. The complete list of articles included in the analysis and their primary information (authorship and year of publication, type of sample, and omic technique/s used in the study) are shown in Supplementary Table 1.
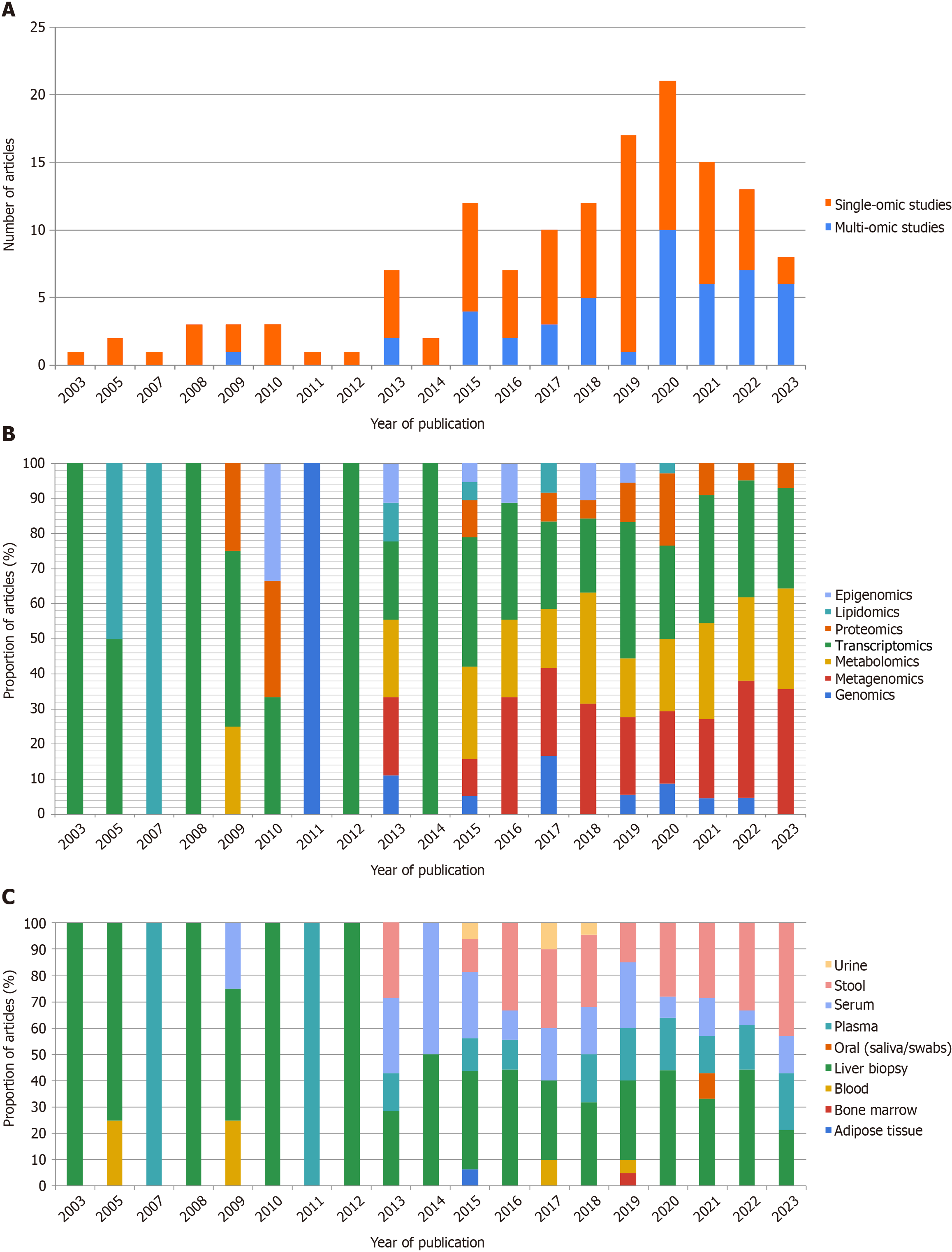
Primarily, omic papers on MASLD biomarker research were scarce 20 years ago. Since then, the number of omic articles has increased exponentially (Figure 1A). This systematic literature review showed that the recent and scanty publication of multi-omic studies focuses on three levels of omics data including transcripts, genes, and proteins. This is followed by other omics areas, comprising metabolites, epigenetic changes, and combinations thereof (Figure 1B).
Of the types of samples analyzed for MASLD biomarker research, the rise in the number of omic studies was followed by a boost of papers aimed at discovering MASLD biomarkers on a diverse array of biological samples (Figure 1C). However, liver tissue samples were the most frequently featured. Stool, plasma, and serum samples have dominated over the last few years (Figure 1C). These results indicate a current interest in the search for non-invasive biomarkers, primarily focusing on gut microbiota studies and its well-described relationship to MASLD development and progression.
The 139 studies were also analyzed according to a previously published set of criteria that evaluate the internal validity of individual studies[16,17]. The most common limitations observed in these studies were a small sample size (16.5%), followed by the presence of bias in the selection and stratification of patients due to the lack of histological diagnosis of MASLD using liver biopsies (12.2%). Moreover, disregard for potential confounding variables (such as age, gender, ethnicity, body mass index, or presence of comorbidities) and their appropriate adjustments in the data analysis was observed in 13 studies (9.35%).
The validity of a study can also be evaluated by its reproducibility. In the case of omics research, depositing raw data, complete protocols, and bioinformatics codes and workflows in a public repository is considered a first step to replicating a study’s findings[17]. Although many leading journals now demand to make data and protocols publicly available as a prerequisite for publication[18], this practice remains inconsistent across journals and omics studies. In fact, seven (5%) of the studies included in our literature search did not offer public access to their data nor indicate how others may obtain it in case specific legal or ethical restrictions prohibit public sharing of the data set.
However, these flaws aren’t recurrently detected in some fields of omics research. In the case of genomics, genome-wide association studies require high degree of certainty in the results (P values < 5 × 10-8), develop large, multi-centric replication studies, and have good compliance with data availability policies[19], as was observed in the two studies of this type (1.4%) analyzed in the literature search carried out in this review.
Despite this progress, the omics-derived biomarker research in MASLD has resulted in several candidate biomarkers that lack clinical validation and utility; that is, it is yet unknown if the identified biomarkers are accurate, reproducible, or reliable in terms of the analytical and clinical/biological validation, or even if there is enough evidence to consistently demonstrate that the use of the omics-based predictor results in a better outcome for patient care (utility).
There are many reasons for this disappointing output. First, the advent of omics-based biomarker studies has led to an excess of highly confounded reports by a superlative number of variables applied to a small sample size. Conditions for sample collection, transport, and storage can significantly affect the quantity and quality of the molecules (nucleic acid, proteins, metabolites) to be analyzed in the biological sample[20]. Moreover, using different NGS platforms or reagents with dissimilar lot numbers can be a source of technical bias that can alter results[21].
Second, the analysis of an outstanding number of data can be challenging. This is especially true if no consensus exists on the data processing pipeline or the software and packages to be used[22,23]. In the case of those omics studies focused on the search for MASLD biomarkers in the human microbiome, the need for complete and curated databases for the human microbiota’s less-studied viral and fungal components poses an obstacle to thorough data analysis[24].
Finally, the shift from a single-omic to a multi-omic approach to biomarker research is critical for increasing the chances of identifying an accurate biomarker for MASLD risk stratification. The number of multi-omics studies on MASLD biomarker research is still scarce but on the rise, as shown by the systematic literature review results. Multi-omic applications provide novel insights and a more holistic understanding of biological processes. Thus, this type of omic study could advance our ability to understand and treat the complex underlying biology of MASLD[25]. Unfortunately, the need for a rigorous study design, accurate sample size and statistical power calculation, and problematic data integration interfere with the expansion of these studies[26].
A lesson can be learned from cancer research. As a consequence of the premature use of omics-based tests developed to predict sensitivity to chemotherapeutic agents in lung and breast cancer clinical trials at Duke University, a committee of the Institute of Medicine generated a series of recommendations that are considered a roadmap of the best scientific practices for the development, validation and clinical translation of omics-based biomarkers and tests[27]. In 2013, the United States National Cancer Institute proposed a practical guideline[28,29] that lists 30 criteria for the development path of omics-based predictors from high-throughput technology to clinical trials (Figure 2). In brief, researchers should take into account: (1) Clinical specimen issues: Collection, processing, storage conditions, availability, quality, amount (mass or volume), and composition of appropriate clinical specimens; (2) Assay issues: Standardization of technical protocols, reagents, and scoring and reporting methods required for the analytical performance of the omics assay in the clinical setting; (3) Model development and evaluation: Avoidance of errors, inconsistencies, or bias in approachs for omics data pre-processing, preparation of the mathematical predictor model, and evaluation of its performance (validation); (4) Clinical trial design: Adherence to accepted standards for good clinical practice, including the development of a formal protocol, an informatics workflow for the analysis of clinical and omics data, a pre-defined study plan and statistical analysis strategy, and the pre-registration of the study and analysis plan in a public registry. The three possible study designs to evaluate the clinical utility are: Prospective-retrospective studies using stored samples; prospective clinical studies, where the biomarker is not involved in patient management decisions; or prospective clinical studies, where the biomarker guides patient management decisions; and (5) Ethical, legal, and regulatory issues: Commitment to protecting human subjects involved in the research, performance of certified laboratory tests if the results have significant clinical value and need to be communicated to the patient or the patient’s physician, documentation of intellectual property rights.
Although there are subtle differences in how this checklist is applied to a particular omics test and clinical setting, it is recognized that any field of omics-based biomarker research should take notice of the abovementioned criteria to determine when there is sufficient and reliable evidence to justify the clinical use of an omics-based biomarker, or even that it is ready for evaluation in a clinical trial.
After demonstrating its clinical usefulness, the new biomarker must prove its cost-effectiveness or cost-utility, mainly as its use will be widespread (up to 30% of the general population in some geographical regions). The benefit the application of the biomarker produces, measured by healthy life years’ indicator, must exceed the cost of the intervention. The application of the biomarker is considered cost-effective if it produces at least USD 50000 per quality-adjusted life year gained[30].
Regarding MASLD biomarker research, a significant step forward was recently taken by the non-invasive biomarkers for metabolic liver disease (NIMBLE) and liver investigation: testing marker utility in steatohepatitis (LITMUS) projects, as multiple circulating biomarkers made the first step in the biomarker qualification path[31,32].
The LITMUS project[32] evaluated the diagnostic accuracy of 5 single biomarkers (CK-18 M30, CK-18 M65, PRO-C3, PRO-C4, and PRO-C6), 9 multimarker scores (FIB-4, MACK-3, a scoring system proposed by Cao et al[33] in 2013, ADAPT, FIBC3, ABC3D, NFS, ELF, and SomaSignal), as well as liver stiffness measurement (LSM) and controlled attenuation parameter vibration-controlled transient elastography (VCTE) in detecting at-risk NASH and fibrosis severity in a European cohort of 966 biopsy-proven participants with MASLD (of which 335 patients had at-risk NASH and 271 advanced fibrosis).
None of the single markers or multimarker scores achieved the predefined acceptable area under the curve (AUC) of 0.8 to be considered a diagnostic marker of acceptable accuracy and for replacing biopsy in detecting people with both NASH and clinically significant fibrosis. However, the SomaSignal test, the ADAPT score, and the LSM VCTE could be used as prescreening methods in clinical trials. The SomaSignal test showed the best results, with AUC values higher for diagnosing advanced fibrosis than for detecting NASH and clinically significant fibrosis[32].
On the other hand, the NIMBLE project tested the performance metrics of 5 biomarker panels (NIS4, OWLiver, PROC3, ELF, and FibroMeter VCTE) for the diagnosis of NASH, at-risk NASH or fibrosis severity in an American cohort of 1073 biopsy-proven individuals with the full spectrum of MASLD[31].
In this case, panels with an area under the receiver operating characteristic of 0.7 or higher were considered a diag
However, despite several limitations (data not applicable to all ethnicities, the use of a curated patient population, limited quantity of sample material, lack of evaluation of omics-based biomarkers, etc.) added to the known inter-observer variability in the histology scoring of the reference method, these groundbreaking studies represent an advance towards having regulatory approved biomarkers for MASLD risk stratification[5,6]. Moreover, as many tested biomar
The expectation of omics technologies is that in the future, patients might be diagnosed and treated according to their personalized MASLD molecular signatures. However, we have a long and arduous path to reach our goal. A consensus regarding best practices on sampling conditions, technical issues, and data processing on discovery studies is mandatory before omics-based biomarkers for MASLD can be validated and their clinical utility tested.
The increasing burden of MASLD emphasizes the pressing need for a novel biomarker that can surpass the “imperfect” liver histology. Moreover, it should be simple, cheap, and easy to adapt to different situations (population screening or stratification in clinical trials). Due to the complex multisystemic pathophysiology of MASLD, it seems unlikely that a single biomarker featuring all these attributes will be identified in the near future. On the contrary, different algorithms integrating clinical data with an arrangement of previously reported omics-based, circulating, anthropometric, and/or imaging-based markers could be considered strong candidates for clinical evaluation.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Argentina
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: de la Monte SM, United States; Lee YM, Singapore S-Editor: Wang JJ L-Editor: A P-Editor: Zhao YQ
| 1. | Rinella ME, Lazarus JV, Ratziu V, Francque SM, Sanyal AJ, Kanwal F, Romero D, Abdelmalek MF, Anstee QM, Arab JP, Arrese M, Bataller R, Beuers U, Boursier J, Bugianesi E, Byrne CD, Castro Narro GE, Chowdhury A, Cortez-Pinto H, Cryer DR, Cusi K, El-Kassas M, Klein S, Eskridge W, Fan J, Gawrieh S, Guy CD, Harrison SA, Kim SU, Koot BG, Korenjak M, Kowdley KV, Lacaille F, Loomba R, Mitchell-Thain R, Morgan TR, Powell EE, Roden M, Romero-Gómez M, Silva M, Singh SP, Sookoian SC, Spearman CW, Tiniakos D, Valenti L, Vos MB, Wong VW, Xanthakos S, Yilmaz Y, Younossi Z, Hobbs A, Villota-Rivas M, Newsome PN; NAFLD Nomenclature consensus group. A multisociety Delphi consensus statement on new fatty liver disease nomenclature. Hepatology. 2023;78:1966-1986. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1212] [Cited by in RCA: 1304] [Article Influence: 652.0] [Reference Citation Analysis (0)] |
| 2. | Le MH, Yeo YH, Zou B, Barnet S, Henry L, Cheung R, Nguyen MH. Forecasted 2040 global prevalence of nonalcoholic fatty liver disease using hierarchical bayesian approach. Clin Mol Hepatol. 2022;28:841-850. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 159] [Article Influence: 53.0] [Reference Citation Analysis (0)] |
| 3. | Valery PC, Powell EE. Predicting clinical outcomes in people with NAFLD: no need for a crystal ball? Lancet Gastroenterol Hepatol. 2023;8:684-685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 4. | Sanyal AJ, Castera L, Wong VW. Noninvasive Assessment of Liver Fibrosis in NAFLD. Clin Gastroenterol Hepatol. 2023;21:2026-2039. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 73] [Article Influence: 36.5] [Reference Citation Analysis (0)] |
| 5. | Sebastiani G. The quest for the ideal NASH biomarker. Lancet Gastroenterol Hepatol. 2023;8:685-687. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 6. | Krag A, Rinella ME. Pioneering the path to NASH biomarker approval. Nat Med. 2023;29:2416-2417. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 7. | Taylor RS, Taylor RJ, Bayliss S, Hagström H, Nasr P, Schattenberg JM, Ishigami M, Toyoda H, Wai-Sun Wong V, Peleg N, Shlomai A, Sebastiani G, Seko Y, Bhala N, Younossi ZM, Anstee QM, McPherson S, Newsome PN. Association Between Fibrosis Stage and Outcomes of Patients With Nonalcoholic Fatty Liver Disease: A Systematic Review and Meta-Analysis. Gastroenterology. 2020;158:1611-1625.e12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 355] [Cited by in RCA: 735] [Article Influence: 147.0] [Reference Citation Analysis (0)] |
| 8. | Davison BA, Harrison SA, Cotter G, Alkhouri N, Sanyal A, Edwards C, Colca JR, Iwashita J, Koch GG, Dittrich HC. Suboptimal reliability of liver biopsy evaluation has implications for randomized clinical trials. J Hepatol. 2020;73:1322-1332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 293] [Article Influence: 58.6] [Reference Citation Analysis (0)] |
| 9. | Siddiqui MS, Yamada G, Vuppalanchi R, Van Natta M, Loomba R, Guy C, Brandman D, Tonascia J, Chalasani N, Neuschwander-Tetri B, Sanyal AJ; NASH Clinical Research Network. Diagnostic Accuracy of Noninvasive Fibrosis Models to Detect Change in Fibrosis Stage. Clin Gastroenterol Hepatol. 2019;17:1877-1885.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 182] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 10. | Zhou JH, Cai JJ, She ZG, Li HL. Noninvasive evaluation of nonalcoholic fatty liver disease: Current evidence and practice. World J Gastroenterol. 2019;25:1307-1326. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 173] [Cited by in RCA: 161] [Article Influence: 26.8] [Reference Citation Analysis (3)] |
| 11. | Miele L, Zocco MA, Pizzolante F, De Matthaeis N, Ainora ME, Liguori A, Gasbarrini A, Grieco A, Rapaccini G. Use of imaging techniques for non-invasive assessment in the diagnosis and staging of non-alcoholic fatty liver disease. Metabolism. 2020;112:154355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 12. | Almeida NS, Rocha R, Cotrim HP, Daltro C. Anthropometric indicators of visceral adiposity as predictors of non-alcoholic fatty liver disease: A review. World J Hepatol. 2018;10:695-701. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 13. | Razmpour F, Daryabeygi-Khotbehsara R, Soleimani D, Asgharnezhad H, Shamsi A, Bajestani GS, Nematy M, Pour MR, Maddison R, Islam SMS. Application of machine learning in predicting non-alcoholic fatty liver disease using anthropometric and body composition indices. Sci Rep. 2023;13:4942. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 14. | Dai X, Shen L. Advances and Trends in Omics Technology Development. Front Med (Lausanne). 2022;9:911861. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 167] [Article Influence: 55.7] [Reference Citation Analysis (0)] |
| 15. | Martinou E, Pericleous M, Stefanova I, Kaur V, Angelidi AM. Diagnostic Modalities of Non-Alcoholic Fatty Liver Disease: From Biochemical Biomarkers to Multi-Omics Non-Invasive Approaches. Diagnostics (Basel). 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 31] [Article Influence: 10.3] [Reference Citation Analysis (1)] |
| 16. | Ioannidis JP, Khoury MJ. Improving validation practices in "omics" research. Science. 2011;334:1230-1232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 166] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 17. | Perng W, Aslibekyan S. Find the Needle in the Haystack, Then Find It Again: Replication and Validation in the 'Omics Era. Metabolites. 2020;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 18. | Hamilton DG, Hong K, Fraser H, Rowhani-Farid A, Fidler F, Page MJ. Prevalence and predictors of data and code sharing in the medical and health sciences: systematic review with meta-analysis of individual participant data. BMJ. 2023;382:e075767. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 19. | Lin X. Learning Lessons on Reproducibility and Replicability in Large Scale Genome-Wide Association Studies. Harv Data Sci Rev. 2020;2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Reference Citation Analysis (0)] |
| 20. | Wu WK, Chen CC, Panyod S, Chen RA, Wu MS, Sheen LY, Chang SC. Optimization of fecal sample processing for microbiome study - The journey from bathroom to bench. J Formos Med Assoc. 2019;118:545-555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 111] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 21. | Zheng Y. Study Design Considerations for Cancer Biomarker Discoveries. J Appl Lab Med. 2018;3:282-289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 22. | Yamada R, Okada D, Wang J, Basak T, Koyama S. Interpretation of omics data analyses. J Hum Genet. 2021;66:93-102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 23. | Hu B, Canon S, Eloe-Fadrosh EA, Anubhav, Babinski M, Corilo Y, Davenport K, Duncan WD, Fagnan K, Flynn M, Foster B, Hays D, Huntemann M, Jackson EKP, Kelliher J, Li PE, Lo CC, Mans D, McCue LA, Mouncey N, Mungall CJ, Piehowski PD, Purvine SO, Smith M, Varghese NJ, Winston D, Xu Y, Chain PSG. Challenges in Bioinformatics Workflows for Processing Microbiome Omics Data at Scale. Front Bioinform. 2021;1:826370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 2] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 24. | Cobbin JC, Charon J, Harvey E, Holmes EC, Mahar JE. Current challenges to virus discovery by meta-transcriptomics. Curr Opin Virol. 2021;51:48-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 58] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 25. | Santiago-Rodriguez TM, Hollister EB. Multi 'omic data integration: A review of concepts, considerations, and approaches. Semin Perinatol. 2021;45:151456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 45] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 26. | Krassowski M, Das V, Sahu SK, Misra BB. State of the Field in Multi-Omics Research: From Computational Needs to Data Mining and Sharing. Front Genet. 2020;11:610798. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 143] [Cited by in RCA: 186] [Article Influence: 37.2] [Reference Citation Analysis (0)] |
| 27. | Evolution of Translational Omics: Lessons Learned and the Path Forward. Washington (DC): National Academies Press (US); 2012-Mar-23 . [PubMed] |
| 28. | McShane LM, Cavenagh MM, Lively TG, Eberhard DA, Bigbee WL, Williams PM, Mesirov JP, Polley MY, Kim KY, Tricoli JV, Taylor JM, Shuman DJ, Simon RM, Doroshow JH, Conley BA. Criteria for the use of omics-based predictors in clinical trials: explanation and elaboration. BMC Med. 2013;11:220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 97] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 29. | McShane LM, Cavenagh MM, Lively TG, Eberhard DA, Bigbee WL, Williams PM, Mesirov JP, Polley MY, Kim KY, Tricoli JV, Taylor JM, Shuman DJ, Simon RM, Doroshow JH, Conley BA. Criteria for the use of omics-based predictors in clinical trials. Nature. 2013;502:317-320. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 165] [Cited by in RCA: 168] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 30. | Scott MG. When do new biomarkers make economic sense? Scand J Clin Lab Invest Suppl. 2010;242:90-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 31. | Sanyal AJ, Shankar SS, Yates KP, Bolognese J, Daly E, Dehn CA, Neuschwander-Tetri B, Kowdley K, Vuppalanchi R, Behling C, Tonascia J, Samir A, Sirlin C, Sherlock SP, Fowler K, Heymann H, Kamphaus TN, Loomba R, Calle RA. Diagnostic performance of circulating biomarkers for non-alcoholic steatohepatitis. Nat Med. 2023;29:2656-2664. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 60] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 32. | Vali Y, Lee J, Boursier J, Petta S, Wonders K, Tiniakos D, Bedossa P, Geier A, Francque S, Allison M, Papatheodoridis G, Cortez-Pinto H, Pais R, Dufour JF, Leeming DJ, Harrison SA, Chen Y, Cobbold JF, Pavlides M, Holleboom AG, Yki-Jarvinen H, Crespo J, Karsdal M, Ostroff R, Zafarmand MH, Torstenson R, Duffin K, Yunis C, Brass C, Ekstedt M, Aithal GP, Schattenberg JM, Bugianesi E, Romero-Gomez M, Ratziu V, Anstee QM, Bossuyt PM; Liver Investigation: Testing Marker Utility in Steatohepatitis (LITMUS) consortium investigators. Biomarkers for staging fibrosis and non-alcoholic steatohepatitis in non-alcoholic fatty liver disease (the LITMUS project): a comparative diagnostic accuracy study. Lancet Gastroenterol Hepatol. 2023;8:714-725. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 101] [Article Influence: 50.5] [Reference Citation Analysis (0)] |
| 33. | Cao W, Zhao C, Shen C, Wang Y. Cytokeratin 18, alanine aminotransferase, platelets and triglycerides predict the presence of nonalcoholic steatohepatitis. PLoS One. 2013;8:e82092. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 56] [Article Influence: 4.7] [Reference Citation Analysis (0)] |