Published online Mar 28, 2024. doi: 10.3748/wjg.v30.i12.1764
Peer-review started: November 11, 2023
First decision: January 24, 2024
Revised: February 3, 2024
Accepted: March 6, 2024
Article in press: March 6, 2024
Published online: March 28, 2024
Processing time: 138 Days and 6.2 Hours
Increasing evidence has demonstrated that N6-methyladenosine (m6A) RNA modification plays an essential role in a wide range of pathological conditions. Impaired autophagy is a critical hallmark of acute pancreatitis (AP).
To explore the role of the m6A modification of ZKSCAN3 in the regulation of autophagy in AP.
The AP mouse cell model was established by cerulein-treated mouse pancreatic acinar cells (MPC-83), and the results were confirmed by the levels of amylase and inflammatory factors. Autophagy activity was evaluated by specific identification of the autophagy-related microstructure and the expression of autophagy-related genes. ZKSCAN3 and ALKBH5 were knocked down to study the function in AP. A m6A RNA binding protein immunoprecipitation assay was used to study how the m6A modification of ZKSCAN3 mRNA is regulated by ALKBH.
The increased expression of amylase and inflammatory factors in the supernatant and the accumulation of autophagic vacuoles verified that the AP mouse cell model was established. The downregulation of LAMP2 and upregulation of LC3-II/I and SQSTM1 demonstrated that autophagy was impaired in AP. The expression of ZKSCAN3 was upregulated in AP. Inhibition of ZKSCAN3 increased the expression of LAMP2 and decreased the expression of the inflammatory factors, LC3-II/I and SQSTM1. Furthermore, ALKBH5 was upregulated in AP. Knockdown of ALKBH5 downregulated ZKSCAN3 expression and restored decreased autophagic flux in AP. Notably, the bioinformatic analysis revealed 23 potential m6A modification sites on ZKSCAN3 mRNA. The m6A modification of ZKSCAN3 mRNA was significantly decreased in AP. Knockdown of ALKBH5 increased the modification of ZKSCAN3 mRNA, which confirmed that ALKBH5 upregulated ZKSCAN3 expression in a m6A-dependent manner.
ALKBH5 inhibits autophagic flux through m6A demethylation of ZKSCAN3 mRNA in AP, thereby aggravating the severity of the disease.
Core Tip: Acute pancreatitis (AP) is a common emergency in digestive system. Impaired autophagy is one of important pathogenic mechanisms of AP, however, its regulatory mechanism remains unclear. N6-methyladenosine modification and ZKSCAN3 are crucial regulatory factors of autophagy, but their roles in AP are not well-defined. This study confirmed that the demethylase ALKBH5 can inhibit autophagy flux by upregulating ZKSCAN3, thereby exacerbating the inflammatory severity of AP. The findings of this study provided new insights into the autophagy regulation mechanism and offered a novel direction for early intervention in AP.
- Citation: Zhang T, Zhu S, Huang GW. ALKBH5 suppresses autophagic flux via N6-methyladenosine demethylation of ZKSCAN3 mRNA in acute pancreatitis. World J Gastroenterol 2024; 30(12): 1764-1776
- URL: https://www.wjgnet.com/1007-9327/full/v30/i12/1764.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i12.1764
Acute pancreatitis (AP) is one of the most common digestive emergencies. The global prevalence and incidence of AP are approximately 76/100000 and 34/100000, respectively, and the number of new cases is increasing at an annual rate of 3%[1-3]. With the progression of therapeutic concepts and interventions, the prognosis of AP has significantly improved. However, due to the unclear pathogenesis of AP, clinicians are still unable to effectively intervene specifically in local or systemic inflammation. The pathogenesis of AP is complex and multifactorial and induces significant and sustained pathological disruption[4,5]. Therefore, further study of the mechanism underlying the progression of AP will provide insight into the development of future therapeutic strategies.
Autophagy is a highly conserved catabolic process in which abnormal biomolecules and organelles are degraded and degradation products are recycled. Autophagy plays an important role in maintaining cellular homeostasis. The entire autophagy process is defined as autophagic flux, and disrupted integrity of the process is called impaired autophagy[6]. Studies have shown that impaired autophagy plays an important role in the development of various diseases, such as neurodegeneration, inflammation, infection, tumors, and metabolic disorders[7-9]. In recent years, the important role of autophagy in AP has been gradually recognized. The basal level of autophagy in the mouse exocrine pancreas is significantly greater than that in the endocrine pancreas and other organs[10]. In experimental pancreatitis, interfering with the expression of upstream regulatory molecules or autophagy-related genes can induce inflammatory changes in exocrine pancreatic cells[11]. Impaired autophagy in AP manifests as activation of the initial stage but blockade of the degradation stage, resulting in harmful factors such as abnormal zymogen granules and disrupted organelles that cannot be effectively degraded[10]. Although impaired autophagy can mediate abnormal zymogen activation, inflammation, and cell death in pancreatic acinar cells[12], the specific regulatory mechanism involved is still unclear.
ZKSCAN3 is a zinc finger DNA-binding protein that simultaneously contains KRAB and SCAN domains; it is also a recognized inhibitory factor of autophagy[13,14]. Studies have shown that ZKSCAN3 can inhibit the transcription of numerous autophagy-related genes, such as LC3 and WIPI2, thereby suppressing a series of autophagy steps in various diseases[15,16]. However, the role of ZKSCAN3 in autophagy in AP has not yet been determined.
The N6-methyladenosine (m6A) modification of RNA plays an important role in the autophagy regulatory network. This process is reversible and involves mainly methyltransferases, demethylases, and methylated RNA-binding proteins[17,18]. ALKBH5 is a crucial demethylase that plays a key role in various diseases[19,20]. In ovarian cancer, the overexpression of ALKBH5 promotes the formation of the BCL-2-Beclin1 complex, and inhibits autophagy[21]. In silica-related pneumonia, ALKBH5 can mediate autophagic flux blockade through the Slam7 pathway[22]. However, in myocardial ischemia-reperfusion injury, ALKBH5 plays a role in promoting autophagic flux[23]. Although the role of m6A modification in impairing autophagy has been demonstrated in various tumors and inflammatory diseases, there is no experimental research on m6A modification in AP. Recent bioinformatics studies have shown that decreased m6A levels are related to the occurrence of severe AP[24], but whether this change is related to ALKBH5-mediated impaired autophagy in AP is unclear.
Clarifying the regulatory mechanism of autophagy in AP is crucial for early intervention. However, research on the autophagy and its regulatory mechanism in AP has not been illustrated. Therefore, in this article we aimed to explore the role and mechanism of action of ALKBH5 in ZKSCAN3 regulated autophagy. We verified the results at the cellular level through a series of molecular biology experiments, which provided a novel perspective on the research of pathogenesis and molecular mechanism of AP and highlighted new targets for therapeutic intervention.
Mouse pancreatic acinar cells (MPC-83) were cultured in RPMI-1640 supplemented with 10% FBS, 100 U/mL penicillin, and 100 mg/mL streptomycin in a 37 °C incubator with 5% CO2. The control groups were not treated, and the AP groups were pretreated with cerulein (10 nM) for 24 h.
MPC-83 cells were seeded in 6-well plates and maintained at 37 °C and 5% CO2. ALKBH5 and ZKSCAN3-siRNA (50 nM) (RiboBio, Gunagzhou, China) were transfected into MPC-83 cells. After 48 h of transfection, the cells were treated with cerulein (10 nM) for 24 h.
Total RNA was extracted from cells using the TRIzol method. All mRNAs were reverse transcribed using the PrimeScript™ RT reagent Kit (Perfect Real Time) (TaKaRa, Kyoto, Japan). Reverse transcription and quantitative real-time RT-PCR were performed with SYBR® Premix Ex Taq™ (TaKaRa, Kyoto, Japan). The results were normalized to that of β-actin and calculated via the relative quantification (2-ΔΔCt) method. The primers used were purchased from Sangon Company (Table 1).
| Genes | Sequence |
| ALKBH5 | Forward 5’- CTTTGCTTCGGCTGCAAGTT -3’ |
| Reverse 5’- CCGGCGTTCCTTAATGTCCT -3’ | |
| ZKSCAN3 | Forward 5’- CAGAGTAGGGTGGAAAGCC -3’ |
| Reverse 5’- AAGGTATGAAGGTCGGGTG -3’ | |
| Primer 1 | Forward 5’- CCAGGCGGTTCTATTGC -3’ |
| Reverse 5’- TGGCTTTCCACCCTACTCT -3’ | |
| Primer 2 | Forward: 5’- CAGAGTAGGGTGGAAAGCC-3’ |
| Reverse 5’- AGGTATGAAGGTCGGGTG-3’ | |
| Primer 3 | Forward 5’- TGGTTCGGGATGGCTAG-3’ |
| Reverse 5’- AACAGCACTGCCTTGGAG-3’ | |
| β-actin | Forward 5’- GTGGCCGAGGACTTTGATTG-3’ |
| Reverse 5’- CCTGTAACAACGCATCTCATATT-3’ |
The supernatant of MPC-83 cells was collected. The levels of interleukin 6 (IL)-6, IL-1β, and tumor necrosis factor (TNF)-α were assessed using ELISA kits (Neobioscience, Shenzhen, China).
Cell lysates were prepared using lysis buffer composed of 50 mmol/L Tris-HCl, 150 mmol/L NaCl, 0.5% sodium deoxycholate, 0.1% SDS, and 1% NP-40. The lysates were centrifuged to collect the supernatants. An equal amount of protein was denatured in SDS sample buffer and separated on 8% or 10% polyacrylamide gels based on the molecular weight of the target proteins. The separated proteins were then transferred to a PVDF membrane. The membranes were blocked with 5% nonfat milk in TBST (TBS containing 0.05% Tween 20), incubated with primary antibodies, and subsequently incubated with secondary antibodies conjugated to alkaline phosphatase. Protein expression was detected by chemiluminescence. The antibodies used were against ALKBH5 (ab195377, Abcam, Britain), ZKSCAN3 (ab223477, Abcam, Britain), LC3 (Proteintech, Wuhan, China), LAMP-2 (Proteintech, Wuhan, China) and SQSTM1 (Proteintech, Wuhan, China).
After cell fixation, the cells were treated with 0.2% Triton X-100 at room temperature. The cells were then blocked with blocking solution. Subsequently, the cells were treated with primary and secondary antibodies. DAPI dye was added to the cells, which were subsequently incubated in the dark. The cells were mounted on slides using anti-fade mounting medium, and fluorescence was observed using a fluorescence microscope. The antibodies used were against LC3 (Proteintech, Wuhan, China) and LAMP-2 (Proteintech, Wuhan, China).
The specimens were cut and fixed in a 2.5% glutaraldehyde solution with Millonig's phosphate buffer (pH = 7.3). The samples were washed three times with Millonig's phosphate buffer at 10-minute intervals. The dehydration process was performed at room temperature using a graded series of acetone (50%, 70%, and 90%) at 10-min intervals, followed by two washes with 100% acetone at 15-min intervals. The samples were then soaked and embedded in a mixture of acetone and resin (1:1) for 12 h, followed by polymerization overnight at 37 °C using 100% resin. To solidify the sample resin, the specimens were further polymerized at 37 °C overnight, followed by an additional 12 h at 60 °C. Ultrathin sections of 50-100 nm were obtained from the specimens using an ultramicrotome and a diamond knife. The sections were then stained with 3% uranyl acetate and lead nitrate, after which they were examined and photographed using a Hitachi HT-7700 electron microscope.
The M6A RNA binding protein immunoprecipitation kit was purchased from RiboBio. RNA was fragmented using RNA fragmentation buffer. Magnetic beads for m6A were prepared using magnetic beads A/G and an anti-m6A antibody. RNA immunoprecipitation was conducted by mixing the fragmented RNA with anti-m6A magnetic beads. The RNA was washed with elution buffer to remove it from the magnetic beads.
Potential m6A binding sites on ZKSCAN3 mRNA were analyzed via a website (http://www.cuilab.cn/sramp/).
All the statistical analyses were performed in GraphPad Prism 8. Independent sample t tests were used to compare the means of two samples, while one-way ANOVA was used for analyzing and comparing the means of more than two groups of samples. P values < 0.05 were considered to indicate statistical significance. The experimental results are presented as the mean ± SD.
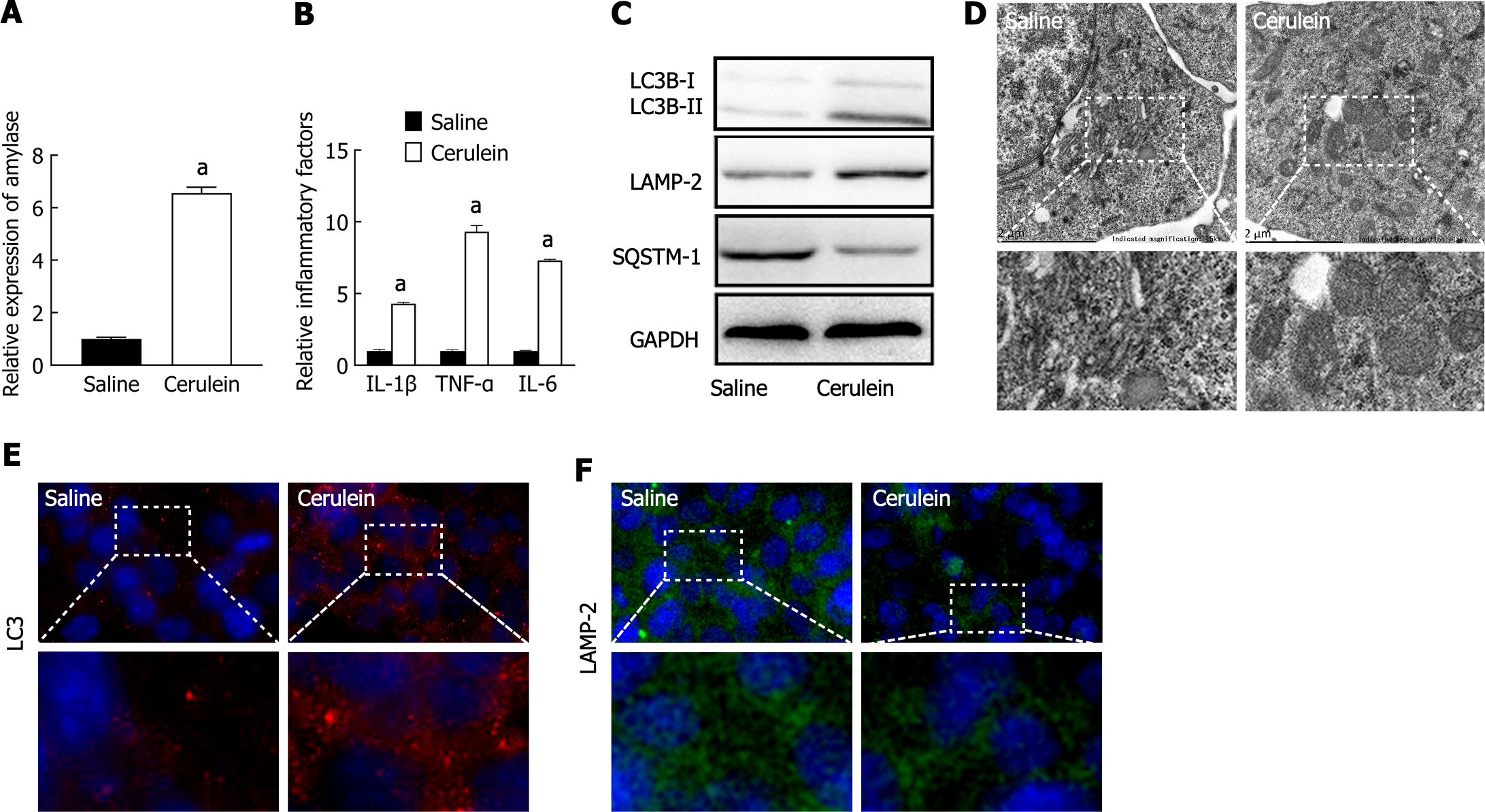
To construct a cell model of AP, MPC-83 cells were treated with 10 nM cerulein for 24 h. The levels of amylase and the inflammatory factors IL-1β, IL-6, and TNF-α in the supernatant were measured via ELISA. The results showed that the levels of amylase and inflammatory factors were significantly greater in the cerulein-treated group (Figure 1A and B), indicating that the AP cell model was successfully established.
The expression levels of autophagy-related marker proteins were detected by western blotting, which showed that the ratio of LC3B-II/I was increased in the AP group, indicating an increase in autophagosomes. The expression of LAMP-2 was decreased in the AP groups, indicating impaired lysosomal synthesis. The expression of the selective autophagy receptor SQSTM1 was increased, indicating inhibited substrate degradation (Figure 1C). Transmission electron microscopy (TEM) revealed the accumulation of circular autophagic vacuoles in the AP group, indicating impaired degradation and accumulation of autophagosomes and autolysosomes (Figure 1D). Furthermore, immunofluorescence staining revealed that LC3 was significantly increased in the AP groups (Figure 1E), while LAMP-2 expression was decreased (Figure 1F). These results demonstrated that autophagosome formation is activated, while lysosomal synthesis and function are impaired, leading to decreased substrate degradation efficiency and accumulation of autophagic vacuoles, suggesting impaired autophagy.
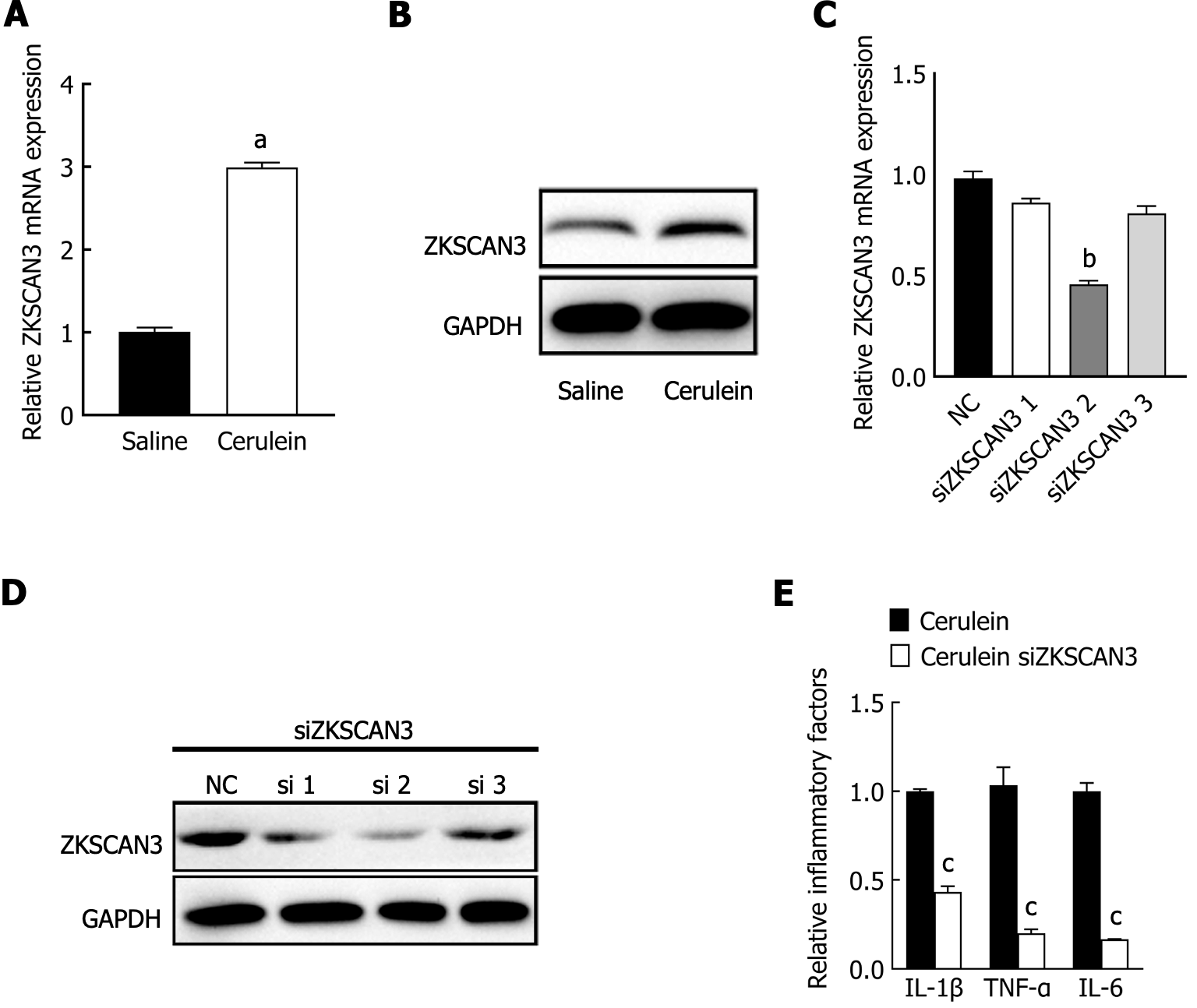
To investigate the role of ZKSCAN3 in AP, qPCR, and western blot were used to detect the expression levels of ZKSCAN3. The results showed that the mRNA and protein expression levels of ZKSCAN3 were significantly increased in the AP group (Figure 2A and B). Three different siRNAs were used to knock down the expression of ZKSCAN3, and siRNA-2 had the most significant interference effect (Figure 2C and D). Subsequent experiments were performed using siRNA-2 to knock down ZKSCAN3. After the inhibition of ZKSCAN3, the cells were treated with cerulein to construct the AP cell model. The levels of inflammatory factors IL-1β, IL-6, and TNF-α in the knocking down (KD) group were significantly lower than those in the negative control (NC) group (Figure 2E). These results suggest that ZKSCAN3 is upregulated and promotes the release of inflammatory factors in the AP mouse cell model.
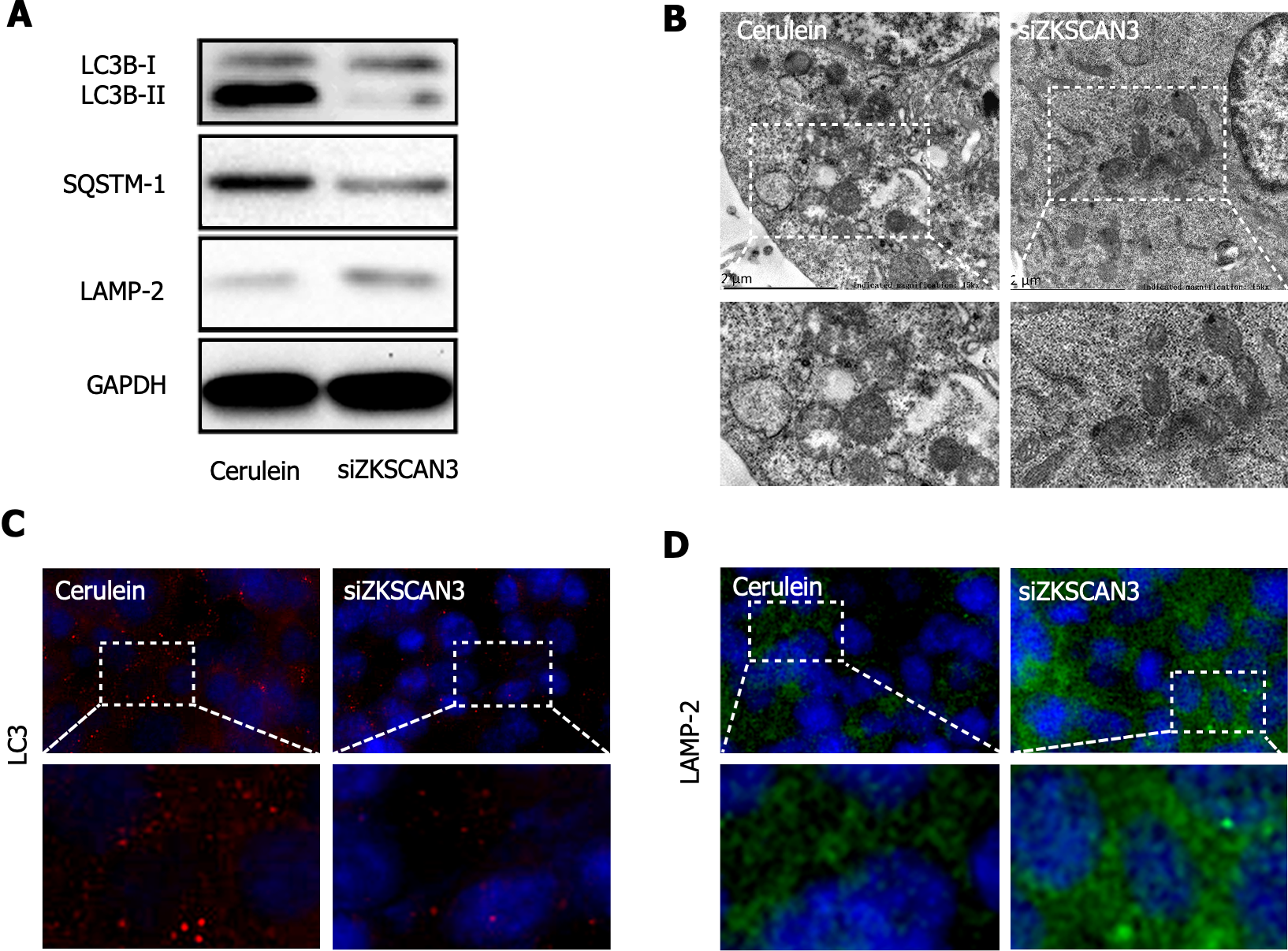
To investigate the role of ZKSCAN3 in autophagic flux in AP, western blot was used to detect the expression of autophagy marker proteins (Figure 3A). The LC3B-II/I ratio was decreased in the KD group, demonstrating the increased clearance of autophagolysosomes. The expression of LAMP-2 increased, suggesting a reduction in lysosomal biogenesis impairment, and the expression of SQSTM1 decreased, indicating an improvement in substrate degradation efficiency. TEM revealed a significant reduction in autophagosome accumulation in the KD group (Figure 3B). Immunofluorescence staining revealed decreased expression of LC3 in the KD group (Figure 3C) and increased expression of LAMP-2 (Figure 3D). These results suggest that ZKSCAN3 inhibits autophagic flux in AP and that KD ZKSCAN3 expression can impair the blockade of autophagic flux.
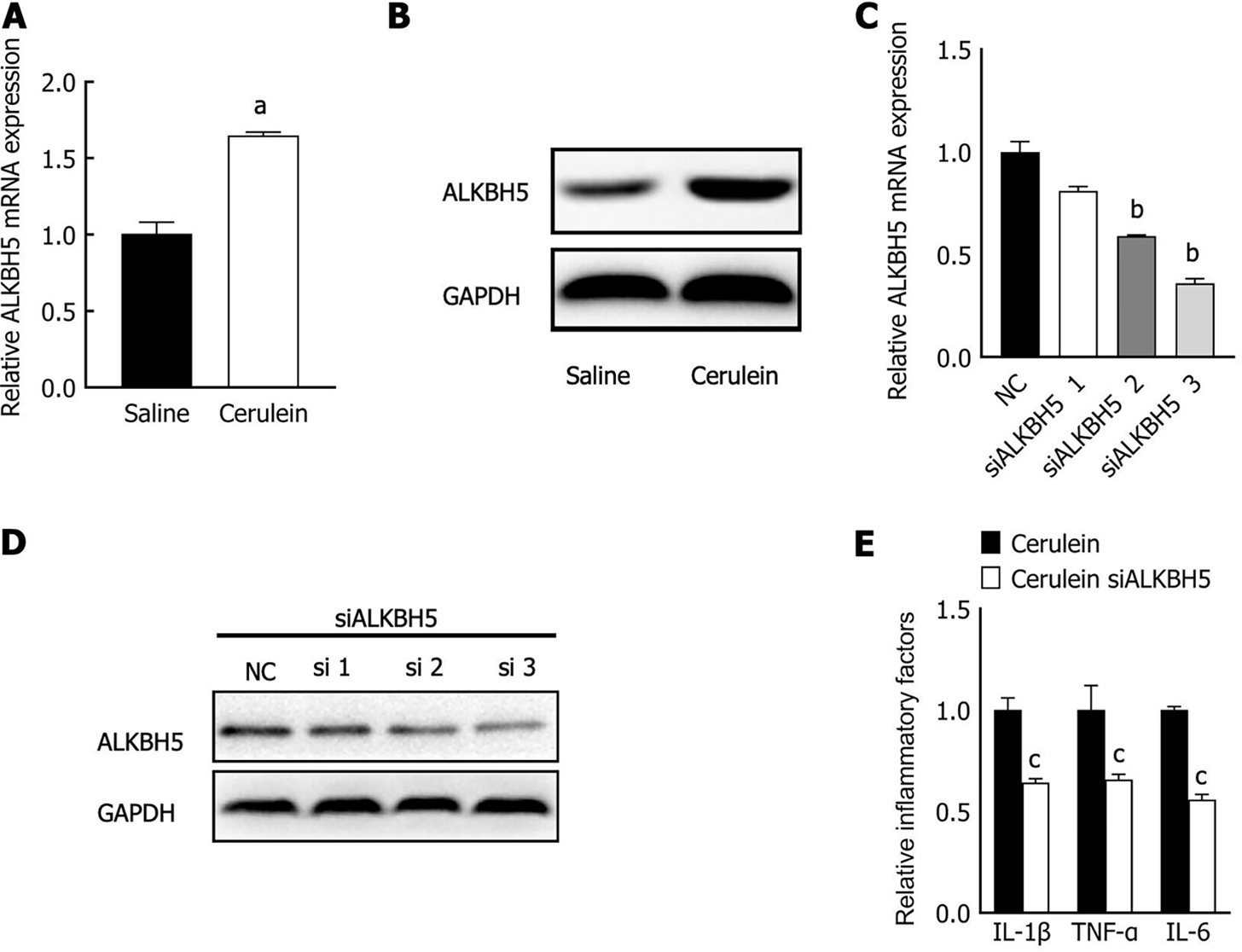
M6A methylation is widely involved in autophagy and contributes to the pathogenesis of human disease. The expression and function of ALKBH5, a primary m6A demethylase in AP, have not yet been determined. We detected ALKBH5 expression in the AP mouse cell model by qPCR and western blot analysis. The level of ALKBH5 was upregulated in the AP mouse cell model (Figure 4A and B). Furthermore, three different siRNAs were used to knock down ALKBH5 expression in MPC-83 cells, and siRNA-3 had the most effective interference effect (Figure 4C and D); therefore, siRNA-3 was used for subsequent experiments. The expression of the inflammatory factors IL-1β, IL-6, and TNF-α was significantly reduced (Figure 4E). These results suggest that ALKBH5 was upregulated in the mouse AP cell model and promoted the release of inflammatory factors.
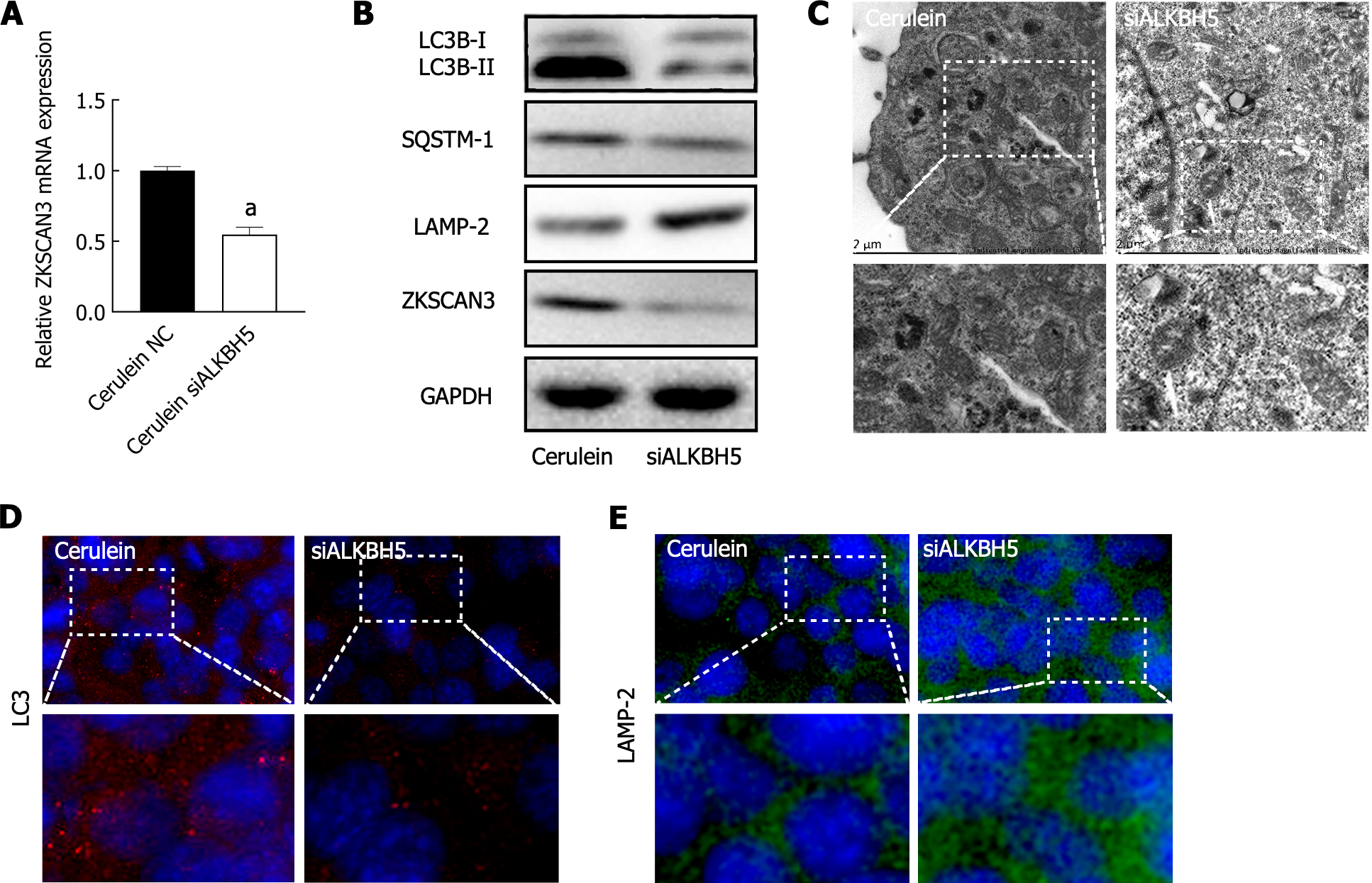
In the AP mouse cell model, knockdown of ALKBH5 downregulated the mRNA and protein expression of ZKSCAN3 (Figure 5A), indicating that ALKBH5 promotes ZKSCAN3 expression. Furthermore, the expression of LC3B-II/I and SQSTM1 decreased, while LAMP-2 expression was increased (Figure 5B), indicating that the knockdown of ALKBH5 rescued the blockade of autophagic flux in AP. TEM confirmed that autophagic vacuole accumulation was reduced after the expression of ALKBH5 was inhibited (Figure 5C). Immunofluorescence revealed that the immunoreactivity of the LC3 protein decreased (Figure 5D), while the immunoreactivity of the LAMP-2 protein increased in the AP mouse cell model transfected with the ALKBH5 target siRNA (Figure 5E). These results suggested that ALKBH5 promoted ZKSCAN3 expression, resulting in the blockade of autophagic flux in AP.
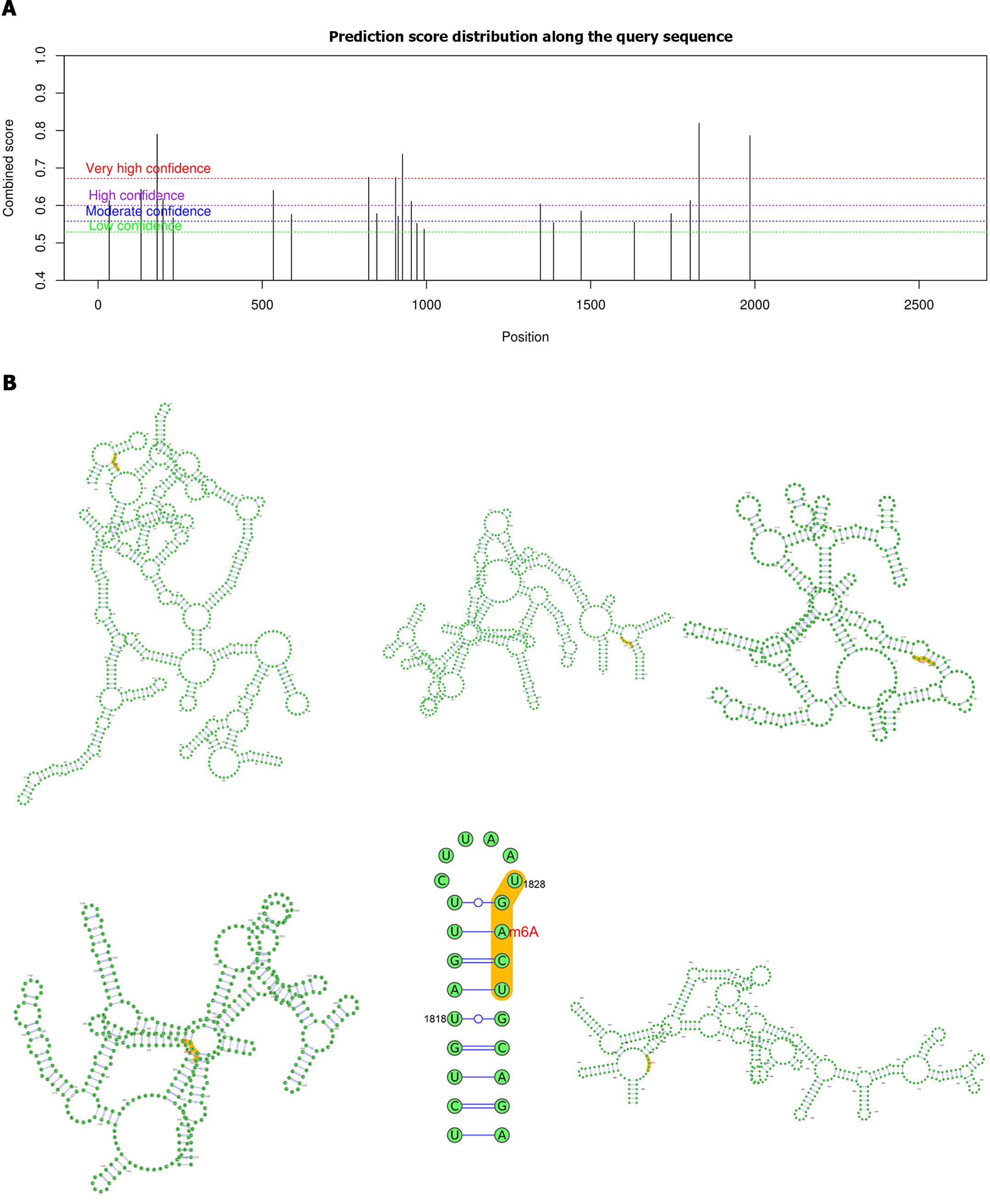
Considering that ALKBH5 is a well-known m6A demethylase, we further investigated the role of m6A modification in the regulation of ZKSCAN3 by ALKBH5. Biological software analysis revealed 23 potential m6A binding sites on ZKSCAN3 mRNA, including 6 highly credible sites, 7 highly credible sites, 6 moderately credible sites, and 4 sites with low credibility (Table 2, Figure 6A). Additionally, we constructed a secondary structure diagram of highly credible m6A binding sites (Figure 6B).
| Number | Position | Sequence context | Confidence |
| 1 | 34 | GUGCCCCGCCCCCCGGGGUCGGACUUUCGACACUUUUGUGACUGC | High |
| 2 | 131 | ACAGCUACAGUGAAACGGGAGAACUGCUUGGUUCGGGAUGGCUAG | High |
| 3 | 180 | UCAAGGGAAAGCACAACCUUGGACUCACACUCUGCAGAGGACCAG | Very high |
| 4 | 198 | UUGGACUCACACUCUGCAGAGGACCAGAUGGAGCUACUGGUCAUA | High |
| 5 | 229 | AGCUACUGGUCAUAAAGGUGGAACAAGAAGAGGCCUCCCCCUUGG | Moderate |
| 6 | 534 | GUGGCGCUGCUGGAGUACUUGGACAGGCAGCUGGAUGACACACCU | High |
| 7 | 589 | CAGAUGAUGACGAUGGGCAGGAACUCCUUUGCUCCAAGGCAGUGC | Moderate |
| 8 | 824 | CCCAGUCCUUUCCCCCAGAUGGACAGAGCAGGAUUCAUCUCAGAU | Very high |
| 9 | 849 | GAGCAGGAUUCAUCUCAGAUGAACCUCUACAAAGAUGGAAUGCAG | Moderate |
| 10 | 906 | AGCCUGGUUUCCCUGGAUCAGGACAUGCAGACUAAGGUUAGGGAC | Very high |
| 11 | 914 | UUCCCUGGAUCAGGACAUGCAGACUAAGGUUAGGGACUUGCCUCG | Moderate |
| 12 | 927 | GACAUGCAGACUAAGGUUAGGGACUUGCCUCGAGCUGAAGAAUAC | Very high |
| 13 | 954 | CCUCGAGCUGAAGAAUACAGGGACCAAAAGCCUGAGCAGACAGUG | High |
| 14 | 971 | CAGGGACCAAAAGCCUGAGCAGACAGUGUGCUUCCUGGGUGAAGA | Low |
| 15 | 993 | ACAGUGUGCUUCCUGGGUGAAGACACUGUCCCGAUUCCUACAGGU | Low |
| 16 | 1347 | GAAAAGCCCUACGAGUGUGAUGACUGUGGGAAAACCUUCACUCAG | High |
| 17 | 1387 | CUCAGAGCUGCAGCCUCCUUGAACAUCACAGAAUUCACACUGGGG | Low |
| 18 | 1471 | GGCGUAGCUCACAUCUUCUGAGACAUCAGAGGACCCAUACUGGGG | Moderate |
| 19 | 1633 | GUAGGAUUACAAGCCUUAUUGAACACCAAAAAGUACACACUGGUG | Low |
| 20 | 1745 | GAGAAGACACACGGGGAAGAAAACUUCUGUCACAGUGACCCCUGC | Moderate |
| 21 | 1803 | GUUGGUGUUCAACUGUCAUUGAACUGAAGCCACUCUGUAGUUCUU | High |
| 22 | 1830 | AGCCACUCUGUAGUUCUUAAUGACUGCAGAAGUCAUAGGCUGGGG | Very high |
| 23 | 1985 | ACAAGAGUCCUCACCCAUUGGAACUAAAUGGGCUUCCUGACUGUC | Very high |
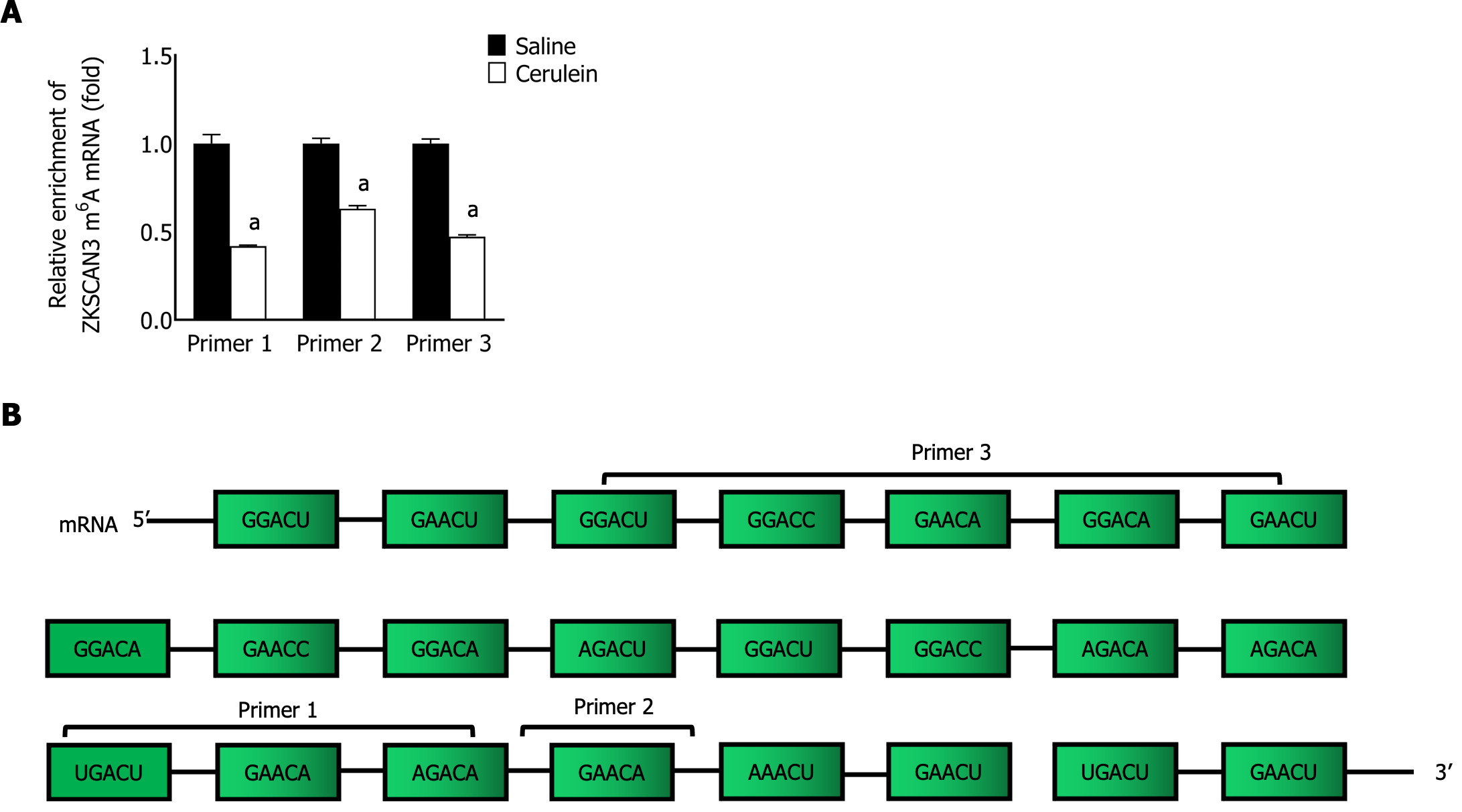
To confirm the role of m6A modification in the relationship between ALKBH5 and ZKSCAN3, MeRIP-qPCR was performed with specific primers aimed at identifying potential m6A sites, and the enrichment of m6A-modified ZKSCAN3 mRNA in the AP group was significantly lower (Figure 7). This finding suggested that ALKBH5 can decrease the m6A modification of ZKSCAN3.
This study is the first to reveal the regulatory roles of ZKSCAN3 and m6A modification in impairing autophagy in AP. We found that ALKBH5 upregulated ZKSCAN3 expression by demethylating ZKSCAN3 inhibited autophagy, and promoted the release of inflammatory factors in a mouse cell model of AP.
Impaired autophagy is one of the key pathogenic mechanisms in AP; this process affects the functions of various organelles, such as mitochondria and the endoplasmic reticulum, and disrupts the homeostasis of acinar cells[12,25]. Usually, autophagy degrades dysfunctional mitochondria during AP. Inhibition of autophagic flux by knocking out the ATG5 and ATG7 genes impaired the clearance of damaged mitochondria, further affecting generation the efficiency of ATP generation in acinar cells[26,27]. Moreover, autophagy maintains the stability of endoplasmic reticulum function. Knocking out the IκB kinase α gene leads to impaired autophagy, and the accumulated SQSTM1 further causes the accumulation of misfolded proteins in the endoplasmic reticulum, triggering endoplasmic reticulum stress and ultimately inducing AP[28]. Therefore, impaired autophagy may trigger or exacerbate other cellular pathological factors in AP. Furthermore, other pathological factors can also induce impaired autophagy. In arginine-treated mice, abnormal mitochondrial membrane leads to disrupted energy metabolism, which inhibits autophagic flux[29]. In ethanol-induced AP, endoplasmic reticulum stress causes folding and transport disorders of autophagy-related proteins[30]. Therefore, autophagy is interconnected with other pathological events during AP. Early autophagy-related intervention may help alleviate the malignant cycle caused by pathological factors.
ZKSCAN3 is currently recognized as a key autophagy inhibitor[15,31]. It affects the progression of various diseases by inhibiting autophagic flux. In hepatocellular carcinoma (HCC), ZKSCAN3 inhibits autophagy, leading to decreased degradation of local adhesion proteins and reducing the metastasis of HCC[32]. In addition, impaired autophagy mediated by ZKSCAN3 is closely related to sepsis-induced immunosuppression[33]. However, the role of ZKSCAN3 in the pathogenesis of AP is still uncertain. Our study first confirmed the high expression of ZKSCAN3 in cerulein-treated MPC-83 cells and the inhibitory effect on autophagy.
ZKSCAN3 functions mainly through nucleoplasmic translocation; when activated, it moves into the nucleus to suppress the transcription of autophagy related genes[15]. However, the upstream regulatory mechanism of ZKSCAN3 is unclear. A study revealed that SIRT1 deacetylates the lysine residues of ZKSCAN3 and promotes its shuttling between the nucleus and cytoplasm[34]. PKC and BRAF inhibitors can activate the inhibition of ZKSCAN3 via phosphorylation[35,36]. Although a few studies have revealed the upstream molecular mechanisms of ZKSCAN3, there is no related research focusing on this gene in AP.
As an important component of epigenetics, m6A modification plays an important regulatory role in autophagy. METTL3 promotes the binding of the RNA-binding protein HNRNPD to the precursor mRNA of TFEB, thus inhibiting autophagy[23]. In HCC, loss of METTL3 increases the stability of the FOXO3 mRNA 3'-UTR modification through a YTHDF1-dependent mechanism and activates autophagy[37]. Moreover, in testicular stromal cells, human chorionic gonadotropin activates autophagy flow by upregulating the expression of ALKBH5 and inhibiting the translation of the m6A-mediated protein PPM1A, thereby increasing testosterone secretion[38]. Therefore, m6A modification is widely involved in the regulation of autophagy in physiological and pathological processes. Among different types of diseases, the same type of m6A modification has different effects on autophagy, which is related to downstream molecular targets and the pathological and physiological stages of disease[39]. Currently, the regulatory role of m6A modification in autophagy has been well documented in various disorders[40], but its role in AP has rarely been studied. Bioinformatics study has shown that m6A-modified noncoding RNAs may participate in the pathological changes observed in AP, but there is still a lack of relevant experimental evidence[24]. This research is the first to demonstrate that ALKBH5 upregulates the expression of ZKSCAN3 by demethylation, thereby inhibiting autophagy in AP.
This study has several limitations. First, the experimental subjects were cell models, and further in vivo animal experiments need to be conducted. In addition, there are significant differences in homology between animal and human tissues. However, due to the lack of human pancreatic exocrine cell lines and a stable extraction method, experimental research on AP cannot be performed in depth in human tissue[41,42]. In clinical practice, identifying pancreatic, peripancreatic or infected necrotic tissues is difficult due to pancreatic juice corrosion or infection. Therefore, human pancreatic exocrine cell lines and tissues are essential for mechanistic research on AP in the future.
In summary, we first revealed the important roles of ZKSCAN3 and m6A modification in AP. In cerulein-treated MPC-83 cells, ALKBH5 upregulates ZKSCAN3 expression by demethylation, thereby inhibiting autophagic flux and aggravating the severity of AP. The results obtained in this study provide important insights into the mechanism of autophagy regulation in AP and offer reference value for future in-depth exploration and early intervention.
The incidence of acute pancreatitis (AP) is increasing annually, and its mortality rate is high. Impaired autophagy is a key factor in the occurrence and development of AP. Therefore, it is crucial to clarify the regulatory mechanism of autophagy in AP.
Evidence has shown that ALKBH5 and ZKSCAN3 can regulate autophagy in a variety of diseases, but there are no relevant studies on AP.
We aimed to explore the regulatory functions and mechanisms of autophagy mediated by ALKBH5 and ZKSCAN3 in AP.
The AP mouse cell line was constructed with cerulein, and the levels of inflammatory factors were detected via ELISA. Similarly, the expression of ALKBH5, ZKSCAN3 and autophagy-related proteins was detected via qPCR, western blot, and immunofluorescence. Microscopic manifestations of autophagy in the cell model were observed via transmission electron microscopy. Additionally, RNA binding protein immunoprecipitation was used to analyze the interaction between ALKBH5 and ZKSCAN3.
The expression of ALKBH5 and ZKSCAN3 was upregulated in the AP model, and the trend toward increased expression of autophagy-related genes suggested that autophagic flux was blocked in AP. Autophagy was improved by inhibiting the expression of ALKBH5 and ZKSCAN3. ZKSCAN3 mRNA has m6A binding sites, and ALKBH5 can upregulate its expression by demethylating ZKSCAN3, which inhibits autophagic flux, thereby aggravating inflammation in AP.
ALKBH5 suppresses autophagic flux by demethylating the m6A site on ZKSCAN3 mRNA, consequently promoting the onset and progression of AP.
We proved that ALKBH5 inhibits autophagy by upregulating ZKSCAN3, thereby promoting the occurrence and development of AP and providing new ideas for future research on autophagy regulation and early drug intervention in AP.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: SeyedAlinaghi S, Iran S-Editor: Qu XL L-Editor: A P-Editor: Chen YX
| 1. | Iannuzzi JP, King JA, Leong JH, Quan J, Windsor JW, Tanyingoh D, Coward S, Forbes N, Heitman SJ, Shaheen AA, Swain M, Buie M, Underwood FE, Kaplan GG. Global Incidence of Acute Pancreatitis Is Increasing Over Time: A Systematic Review and Meta-Analysis. Gastroenterology. 2022;162:122-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 363] [Article Influence: 121.0] [Reference Citation Analysis (1)] |
| 2. | Mederos MA, Reber HA, Girgis MD. Acute Pancreatitis: A Review. JAMA. 2021;325:382-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 499] [Article Influence: 124.8] [Reference Citation Analysis (1)] |
| 3. | Ouyang G, Pan G, Liu Q, Wu Y, Liu Z, Lu W, Li S, Zhou Z, Wen Y. The global, regional, and national burden of pancreatitis in 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. BMC Med. 2020;18:388. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 74] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 4. | Saluja A, Dudeja V, Dawra R, Sah RP. Early Intra-Acinar Events in Pathogenesis of Pancreatitis. Gastroenterology. 2019;156:1979-1993. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 179] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 5. | Lee PJ, Papachristou GI. New insights into acute pancreatitis. Nat Rev Gastroenterol Hepatol. 2019;16:479-496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 218] [Cited by in RCA: 516] [Article Influence: 86.0] [Reference Citation Analysis (0)] |
| 6. | Wen X, Yang Y, Klionsky DJ. Moments in autophagy and disease: Past and present. Mol Aspects Med. 2021;82:100966. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 7. | Mizushima N, Levine B. Autophagy in Human Diseases. N Engl J Med. 2020;383:1564-1576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 420] [Cited by in RCA: 758] [Article Influence: 151.6] [Reference Citation Analysis (0)] |
| 8. | Klionsky DJ, Petroni G, Amaravadi RK, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cadwell K, Cecconi F, Choi AMK, Choi ME, Chu CT, Codogno P, Colombo MI, Cuervo AM, Deretic V, Dikic I, Elazar Z, Eskelinen EL, Fimia GM, Gewirtz DA, Green DR, Hansen M, Jäättelä M, Johansen T, Juhász G, Karantza V, Kraft C, Kroemer G, Ktistakis NT, Kumar S, Lopez-Otin C, Macleod KF, Madeo F, Martinez J, Meléndez A, Mizushima N, Münz C, Penninger JM, Perera RM, Piacentini M, Reggiori F, Rubinsztein DC, Ryan KM, Sadoshima J, Santambrogio L, Scorrano L, Simon HU, Simon AK, Simonsen A, Stolz A, Tavernarakis N, Tooze SA, Yoshimori T, Yuan J, Yue Z, Zhong Q, Galluzzi L, Pietrocola F. Autophagy in major human diseases. EMBO J. 2021;40:e108863. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 555] [Cited by in RCA: 985] [Article Influence: 246.3] [Reference Citation Analysis (0)] |
| 9. | Chen T, Tu S, Ding L, Jin M, Chen H, Zhou H. The role of autophagy in viral infections. J Biomed Sci. 2023;30:5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 106] [Reference Citation Analysis (0)] |
| 10. | Gukovskaya AS, Gukovsky I, Algül H, Habtezion A. Autophagy, Inflammation, and Immune Dysfunction in the Pathogenesis of Pancreatitis. Gastroenterology. 2017;153:1212-1226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 260] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 11. | Wang S, Ni HM, Chao X, Wang H, Bridges B, Kumer S, Schmitt T, Mareninova O, Gukovskaya A, De Lisle RC, Ballabio A, Pacher P, Ding WX. Impaired TFEB-mediated lysosomal biogenesis promotes the development of pancreatitis in mice and is associated with human pancreatitis. Autophagy. 2019;15:1954-1969. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 85] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 12. | Yuan X, Wu J, Guo X, Li W, Luo C, Li S, Wang B, Tang L, Sun H. Autophagy in Acute Pancreatitis: Organelle Interaction and microRNA Regulation. Oxid Med Cell Longev. 2021;2021:8811935. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 13. | Saftig P, Haas A. Turn up the lysosome. Nat Cell Biol. 2016;18:1025-1027. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 74] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 14. | Füllgrabe J, Ghislat G, Cho DH, Rubinsztein DC. Transcriptional regulation of mammalian autophagy at a glance. J Cell Sci. 2016;129:3059-3066. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 157] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 15. | Chauhan S, Goodwin JG, Chauhan S, Manyam G, Wang J, Kamat AM, Boyd DD. ZKSCAN3 is a master transcriptional repressor of autophagy. Mol Cell. 2013;50:16-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 218] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 16. | Pan H, Yan Y, Liu C, Finkel T. The role of ZKSCAN3 in the transcriptional regulation of autophagy. Autophagy. 2017;13:1235-1238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 17. | Liang J, Sun J, Zhang W, Wang X, Xu Y, Peng Y, Zhang L, Xiong W, Liu Y, Liu H. Novel Insights into The Roles of N(6)-methyladenosine (m(6)A) Modification and Autophagy in Human Diseases. Int J Biol Sci. 2023;19:705-720. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 18. | Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. 2019;20:608-624. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1100] [Cited by in RCA: 1625] [Article Influence: 270.8] [Reference Citation Analysis (0)] |
| 19. | Qu J, Yan H, Hou Y, Cao W, Liu Y, Zhang E, He J, Cai Z. RNA demethylase ALKBH5 in cancer: from mechanisms to therapeutic potential. J Hematol Oncol. 2022;15:8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 142] [Article Influence: 47.3] [Reference Citation Analysis (0)] |
| 20. | Jiang X, Liu B, Nie Z, Duan L, Xiong Q, Jin Z, Yang C, Chen Y. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther. 2021;6:74. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 265] [Cited by in RCA: 1314] [Article Influence: 328.5] [Reference Citation Analysis (0)] |
| 21. | Zhu H, Gan X, Jiang X, Diao S, Wu H, Hu J. ALKBH5 inhibited autophagy of epithelial ovarian cancer through miR-7 and BCL-2. J Exp Clin Cancer Res. 2019;38:163. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 181] [Article Influence: 30.2] [Reference Citation Analysis (0)] |
| 22. | Yin H, Gu P, Xie Y, You X, Zhang Y, Yao Y, Yang S, Wang D, Chen W, Ma J. ALKBH5 mediates silica particles-induced pulmonary inflammation through increased m(6)A modification of Slamf7 and autophagy dysfunction. J Hazard Mater. 2024;462:132736. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 23. | Song H, Feng X, Zhang H, Luo Y, Huang J, Lin M, Jin J, Ding X, Wu S, Huang H, Yu T, Zhang M, Hong H, Yao S, Zhao Y, Zhang Z. METTL3 and ALKBH5 oppositely regulate m(6)A modification of TFEB mRNA, which dictates the fate of hypoxia/reoxygenation-treated cardiomyocytes. Autophagy. 2019;15:1419-1437. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 334] [Cited by in RCA: 379] [Article Influence: 63.2] [Reference Citation Analysis (0)] |
| 24. | Wu J, Yuan XH, Jiang W, Lu YC, Huang QL, Yang Y, Qie HJ, Liu JT, Sun HY, Tang LJ. Genome-wide map of N(6)-methyladenosine circular RNAs identified in mice model of severe acute pancreatitis. World J Gastroenterol. 2021;27:7530-7545. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 3] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (1)] |
| 25. | Voronina S, Chvanov M, De Faveri F, Mayer U, Wileman T, Criddle D, Tepikin A. Autophagy, Acute Pancreatitis and the Metamorphoses of a Trypsinogen-Activating Organelle. Cells. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 18] [Reference Citation Analysis (0)] |
| 26. | Shirihai OS, Song M, Dorn GW 2nd. How mitochondrial dynamism orchestrates mitophagy. Circ Res. 2015;116:1835-1849. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 244] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 27. | Diakopoulos KN, Lesina M, Wörmann S, Song L, Aichler M, Schild L, Artati A, Römisch-Margl W, Wartmann T, Fischer R, Kabiri Y, Zischka H, Halangk W, Demir IE, Pilsak C, Walch A, Mantzoros CS, Steiner JM, Erkan M, Schmid RM, Witt H, Adamski J, Algül H. Impaired autophagy induces chronic atrophic pancreatitis in mice via sex- and nutrition-dependent processes. Gastroenterology. 2015;148:626-638.e17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 132] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 28. | Li N, Wu X, Holzer RG, Lee JH, Todoric J, Park EJ, Ogata H, Gukovskaya AS, Gukovsky I, Pizzo DP, VandenBerg S, Tarin D, Atay C, Arkan MC, Deerinck TJ, Moscat J, Diaz-Meco M, Dawson D, Erkan M, Kleeff J, Karin M. Loss of acinar cell IKKα triggers spontaneous pancreatitis in mice. J Clin Invest. 2013;123:2231-2243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 102] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 29. | Biczo G, Vegh ET, Shalbueva N, Mareninova OA, Elperin J, Lotshaw E, Gretler S, Lugea A, Malla SR, Dawson D, Ruchala P, Whitelegge J, French SW, Wen L, Husain SZ, Gorelick FS, Hegyi P, Rakonczay Z Jr, Gukovsky I, Gukovskaya AS. Mitochondrial Dysfunction, Through Impaired Autophagy, Leads to Endoplasmic Reticulum Stress, Deregulated Lipid Metabolism, and Pancreatitis in Animal Models. Gastroenterology. 2018;154:689-703. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 294] [Cited by in RCA: 280] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 30. | Lugea A, Waldron RT, French SW, Pandol SJ. Drinking and driving pancreatitis: links between endoplasmic reticulum stress and autophagy. Autophagy. 2011;7:783-785. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 20] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 31. | Hu H, Ji Q, Song M, Ren J, Liu Z, Wang Z, Liu X, Yan K, Hu J, Jing Y, Wang S, Zhang W, Liu GH, Qu J. ZKSCAN3 counteracts cellular senescence by stabilizing heterochromatin. Nucleic Acids Res. 2020;48:6001-6018. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 69] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 32. | Zhang X, Bai Y, Huang L, Liu S, Mo Y, Cheng W, Wang G, Cao Z, Chen X, Cui H, Qi L, Ma L, Liu M, Guan XY, Ma NF. CHD1L augments autophagy-mediated migration of hepatocellular carcinoma through targeting ZKSCAN3. Cell Death Dis. 2021;12:950. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 33. | Ouyang X, Becker E Jr, Bone NB, Johnson MS, Craver J, Zong WX, Darley-Usmar VM, Zmijewski JW, Zhang J. ZKSCAN3 in severe bacterial lung infection and sepsis-induced immunosuppression. Lab Invest. 2021;101:1467-1474. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 34. | Wu X, Ren Y, Wen Y, Lu S, Li H, Yu H, Li W, Zou F. Deacetylation of ZKSCAN3 by SIRT1 induces autophagy and protects SN4741 cells against MPP(+)-induced oxidative stress. Free Radic Biol Med. 2022;181:82-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 35. | Li Y, Xu M, Ding X, Yan C, Song Z, Chen L, Huang X, Wang X, Jian Y, Tang G, Tang C, Di Y, Mu S, Liu X, Liu K, Li T, Wang Y, Miao L, Guo W, Hao X, Yang C. Protein kinase C controls lysosome biogenesis independently of mTORC1. Nat Cell Biol. 2016;18:1065-1077. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 272] [Article Influence: 30.2] [Reference Citation Analysis (0)] |
| 36. | Li S, Song Y, Quach C, Guo H, Jang GB, Maazi H, Zhao S, Sands NA, Liu Q, In GK, Peng D, Yuan W, Machida K, Yu M, Akbari O, Hagiya A, Yang Y, Punj V, Tang L, Liang C. Transcriptional regulation of autophagy-lysosomal function in BRAF-driven melanoma progression and chemoresistance. Nat Commun. 2019;10:1693. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 121] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 37. | Lin Z, Niu Y, Wan A, Chen D, Liang H, Chen X, Sun L, Zhan S, Chen L, Cheng C, Zhang X, Bu X, He W, Wan G. RNA m(6) A methylation regulates sorafenib resistance in liver cancer through FOXO3-mediated autophagy. EMBO J. 2020;39:e103181. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 117] [Cited by in RCA: 338] [Article Influence: 67.6] [Reference Citation Analysis (0)] |
| 38. | Chen Y, Wang J, Xu D, Xiang Z, Ding J, Yang X, Li D, Han X. m(6)A mRNA methylation regulates testosterone synthesis through modulating autophagy in Leydig cells. Autophagy. 2021;17:457-475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 118] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 39. | Ma Q, Long S, Gan Z, Tettamanti G, Li K, Tian L. Transcriptional and Post-Transcriptional Regulation of Autophagy. Cells. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 27] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 40. | Füllgrabe J, Klionsky DJ, Joseph B. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat Rev Mol Cell Biol. 2014;15:65-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 318] [Cited by in RCA: 376] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 41. | Case RM. Is the rat pancreas an appropriate model of the human pancreas? Pancreatology. 2006;6:180-190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 27] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 42. | Saloman JL, Albers KM, Cruz-Monserrate Z, Davis BM, Edderkaoui M, Eibl G, Epouhe AY, Gedeon JY, Gorelick FS, Grippo PJ, Groblewski GE, Husain SZ, Lai KKY, Pandol SJ, Uc A, Wen L, Whitcomb DC. Animal Models: Challenges and Opportunities to Determine Optimal Experimental Models of Pancreatitis and Pancreatic Cancer. Pancreas. 2019;48:759-779. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |