Published online Mar 21, 2024. doi: 10.3748/wjg.v30.i11.1524
Peer-review started: November 9, 2023
First decision: December 15, 2023
Revised: January 5, 2024
Accepted: February 20, 2024
Article in press: March 21, 2024
Published online: March 21, 2024
Processing time: 133 Days and 6 Hours
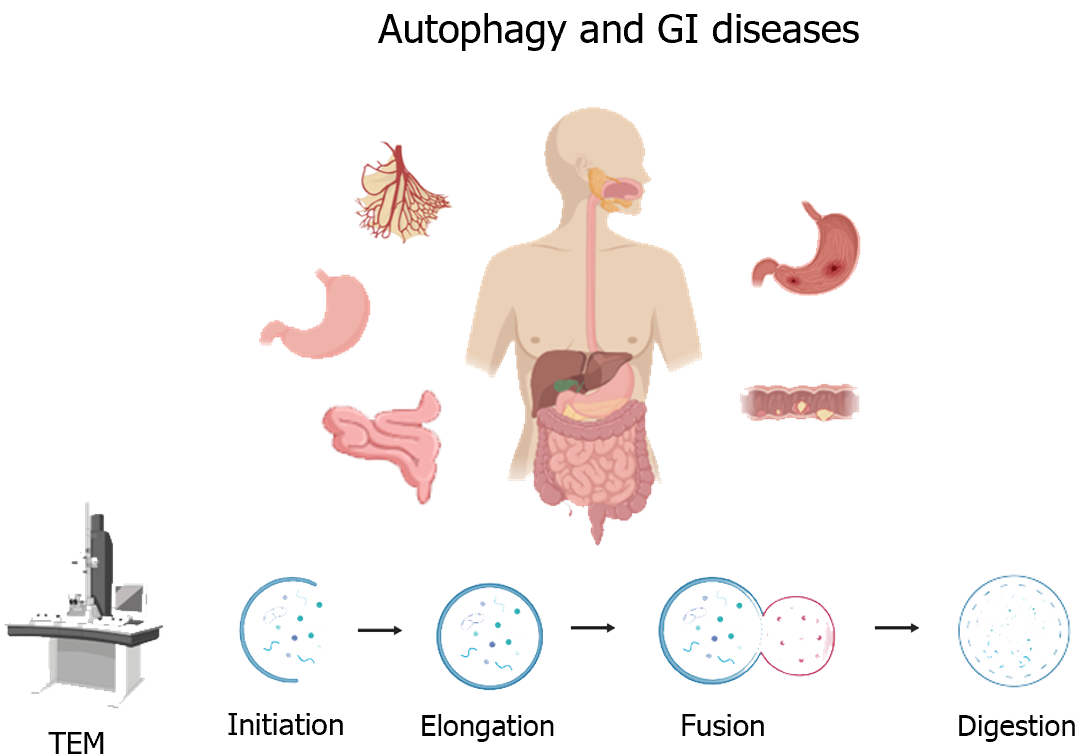
Autophagy is a cellular catabolic process characterized by the formation of double-membrane autophagosomes. Transmission electron microscopy is the most rigorous method to clearly visualize autophagic engulfment and degradation. A large number of studies have shown that autophagy is closely related to the digestion, secretion, and regeneration of gastrointestinal (GI) cells. However, the role of autophagy in GI diseases remains controversial. This article focuses on the morphological and biochemical characteristics of autophagy in GI diseases, in order to provide new ideas for their diagnosis and treatment.
Core Tip: Autophagy, from a morphological standpoint, shares similarities with other biological processes such as phagocytosis and apoptosis. As an intracellular catabolic mechanism, autophagy, along with the ubiquitin-proteasome system, contributes to maintaining cellular homeostasis. Moreover, autophagy also assumes a role in programmed cell death when apoptosis is absent. Numerous studies have established the close association between autophagy and the physiological functions of different gastrointestinal (GI) cells. Morphological investigations have furnished substantial evidence highlighting autophagy's pro-survival role in benign conditions like intestinal ischemia-reperfusion injury, inflammatory bowel disease, and GI motility disorders. Further research into the involvement of autophagy in GI tumors is necessary to unravel these unresolved mysteries in the future.
- Citation: Chang YF, Li JJ, Liu T, Wei CQ, Ma LW, Nikolenko VN, Chang WL. Morphological and biochemical characteristics associated with autophagy in gastrointestinal diseases. World J Gastroenterol 2024; 30(11): 1524-1532
- URL: https://www.wjgnet.com/1007-9327/full/v30/i11/1524.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i11.1524
Autophagy, as a cellular catabolic process, is closely related to the digestion, secretion, and regeneration of gastrointestinal (GI) cells. Morphological studies have shown that autophagy is similar to other biological phenomena such as phagocytosis and apoptosis, and it is involved in maintaining cellular homeostasis and programmed cell death, as well as cell growth, development, and differentiation. Autophagy has been found to play a pro-survival role in benign GI diseases like intestinal ischemia-reperfusion (I/R) injury, inflammatory bowel disease (IBD), and GI motility disorders. However, under pathological conditions, the role of autophagy in GI diseases varies, possibly due to the different degrees of autophagy or the presence of other factors. Therefore, more studies on the role of autophagy in GI tumors are required to address these unresolved questions in the future.
Autophagy occurs in all eukaryotic cells, including plant and animal cells, and is an evolutionarily conserved cellular catabolic process. The occurrence of autophagy cannot be separated from the existence of lysosomes. However, autophagy is rare in cells in a state of normal proliferation. Taking gastric tissue as an example, autolysosomes are difficult to observe by transmission electron microscopy (TEM) under normal circumstances[1]. Autophagy is elevated only when cells lack energy sources (starvation), face external stimuli (invasion by pathogens), and be in disease states (degenerative lesions, cancer, etc.). Thus, autophagy is also thought to often play a pro-survival role. However, in some cases, inhibiting autophagy can actually help to cure diseases. For example, studies have found that autophagy enhances the drug resistance of tumor cells to chemotherapy in kidney cancer, prostate cancer, and other cancers. The combination of autophagy inhibitor drugs and chemotherapy drugs can achieve good therapeutic effects. In addition, autophagy is also considered as a programmed death process. Excessive autophagy is thought to cause cell death. Thus, the effects of autophagy on cells in different states are complex (Figure 1).
Before the advent of electron microscopy, a variety of particle-containing vesicles could only be observed by ordinary light microscopy. Since 1933, the advent of TEM has accelerated the study of morphology to the subcellular level[2]. Compared with ordinary microscopes, electron microscopes can magnify tens of thousands of times, so submicroscopic structures within cells can be observed. Thus, electron microscopy is the "gold standard" for studying autophagic morphology. TEM images can provide information such as autophagosome integrity, changes in the number and volume of autophagic vesicles, and autophagosome-lysosomal interactions. In addition, this technique allows visualization of organelles inside autophagic chamber to distinguish whether autophagy is selective autophagy. Observing by TEM, we can clearly capture the moment of fusion of autophagosomes and lysosomes and the morphological changes of organelles during degradation.
The process of autophagy can be divided into five stages: Initiation, elongation, closure, fusion, and decomposition. Morphological studies of autophagy have found that a bilayer membrane structure derived from the endoplasmic reticulum without ribosomes is first formed in the cell, and the degenerate organelles form distinct aggregates, which are gradually surrounded by this bilayer membrane structure. The membrane of the autophagosome is continuously elongated and gradually envelops the aggregates. Eventually, autophagosomes fuse with lysosomes to release acid-lysozymes to break down the contents. Generally, typical features of different stages of autophagy can be observed simultaneously by TEM. According to the type of autophagic body contents, autophagy can be divided into selective autophagy and non-selective autophagy. Non-selective autophagy occurs when various organelles such as the endoplasmic reticulum and mitochondria accumulate in autophagosomes. When selective autophagy occurs, aggregation of only one type of content can be observed in autophagosomes. Common inclusions include mitochondria, lipids, and foreign pathogens (such as bacteria and viruses). Autophagy is also divided into macro-autophagy, micro-autophagy, and chaperone-mediated autophagy. The autophagy mentioned in this article mainly refers to macroscopic autophagy.
The GI tract is the largest contact area with the external environment of the cavity organs. Many biochemical reactions occur in the GI tract every day. GI epithelial cells together with a variety of microorganisms constitute the first barrier of the human digestive system. GI cells are made up of three types of cells: Digestive cells (master cells and absorptive cells), secretory cells (goblet cells and Paneth cells), and regenerative cells (stem cells). Goblet cells are mucus-secreting cells that form a physical barrier between intestinal epithelial cells (IECs) and the external environment. One study found that autophagy produced a thicker, less penetrating mucus layer in mice, which enhanced intestinal anti-inflammatory function[3]. Mucus production protects gastric mucosal epithelial cells from chemicals (e.g., alcohol and nonsteroidal anti-inflammatory drugs) and microorganisms. In that study, Naama et al[3] also found that autophagy relieves endoplasmic reticulum stress through autophagy-related protein Beclin1, thereby promoting goblet cell mucus secretion. Similarly, Paneth cells secrete antimicrobial proteins that are highly dependent on endoplasmic reticulum stress and autophagy levels[4]. Gorbunov et al[5] found that autophagy plays a role not only in secretory cells, but also in intestinal stem cells. Yang et al[6] demonstrated that autophagy is required for ileal stem cell maintenance and mammalian survival. In addition, recent studies have shown that autophagy is required to maintain increased enterocyte proliferation in honeybees[7].
According to reports, amino acid deficiency can regulate autophagy activity in IECs[8]. The researchers found that autophagic vacuoles increased by TEM and confocal microscopy[9]. In addition, exposure of IECs to hypoxia and lipopolysaccharide for 24 h not only increased the number of autophagic vesicles, but also significantly increased their diameter[10]. Interestingly, in the midgut epithelial cells of shrimp, approximately 40% of cells show signs of autophagy. The endoplasmic reticulum pool, electron transparent content, vacuoles, poly-vesicles, lamellar bodies, vesicles of autophagosome in lipids, and electron dense particles were observed. In addition, the researchers observed that degenerated mitochondria were mainly concentrated in autophagosomes (mitochondrial autophagy). A study has found that the reduction of intestinal cell volume in shrimp involves a programmed process that requires autophagy. In addition, UBA1 knockout significantly reduced the size of midgut cells, and double membrane autophagosomes containing mitochondria or ribosomes were observed in the cytoplasm[11].
GI epithelial cells constitute the first barrier to protect the alimentary tract from injury. The intestinal epithelial tight junction (TJ), which is the second line of defense in the intestinal mucosa, protects against permeation of luminal antigens, endotoxins, and bacteria into the blood stream. Recent research found that autophagy promotes membrane localization of occluding protein, a principal TJ component involved in TJ barrier enhancement, which could protect against inflammation-induced barrier loss[12]. Furthermore, Kim et al[13] discovered that protease-activated receptor 2 regulates autophagy and intestinal epithelial TJs, thus reducing intestinal epithelial permeability. Additionally, another study discovered that rapamycin (autophagy inducer) dramatically improved intestinal damage in benzo[a]pyrene induced intestinal epithelial TJ disruption[14]. In conclusion, the activation of autophagy plays an important role in maintaining intestinal barrier function against toxic chemicals, intestinal inflammation, and intestinal permeability.
The intestinal epithelium is frequently exposed to the invasion of many foreign pathogens, leading to increased permeability and intestinal barrier loss. When bacteria infect host cells, selective autophagy initially engulfs the pathogens to limit the access to nutrients. Although autophagy initially triggers an innate immune response that induces intestinal immune cells to produce interferon and clear harmful pathogens, some bacteria (such as Escherichia coli, Salmonella, and Listeria) have evolved strategies to inhibit or escape it. For example, Escherichia coli hinders the autophagosome-lysosome fusion to inhibit autophagic flux, thus preventing the clearance of acidic hydrolase[15]. Besides that, Yang et al[16] suggested that Salmonella escapes host immune responses by inhibiting autophagy degradation. Previously, a large number of bacteria have been shown to evade NOD pathway-mediated intestinal immune surveillance by inhibiting autophagy[17,18]. Molecule evidence has been found that autophagy is involved in the secretion of membrane vesicles from Listeria monocytogenes in vitro[19]. In addition, one similar study discovered that Fusobacterium modulates autophagy to survive, thus aggravating experimental colitis via the miR-574-5p/CARD3 axis[20]. The latest findings show that bacterial extracellular vesicles induced mitophagy through mTOR pathways relieve oxidative stress in colonic epithelial cells[21]. Libertellenone T, a compound isolated from Endolichenic fungus, also induces autophagy to strengthen the epithelial barrier function of the colon[22].
In contrast, some viruses exploit autophagy for replication to survive inside intestinal cells. Recently, the effect of autophagy on SARS-CoV-2 infection has drawn much attention. Some studies showed that SARS-CoV-2 exploits host autophagy machinery for intestinal dissemination[23,24]. Furthermore, Cloherty et al[25] proofed that Berbamine, a selection of autophagy-blocking drugs, can suppress intestinal SARS-CoV-2 infection as well as prevent SARS-CoV-2-mediated disruption of the intestinal barrier via an autophagy-mediated BNIP3 mechanism. However, not all viruses have evolved such an escape mechanism. One study discovered that autophagy induced by urolithin A, an intestinal metabolite of ellagic acid, inhibits enterovirus 71 replication in infected cells[26]. In addition, another study discovered that the autophagy gene (ATG) Epg5 plays an important role in intestinal antiviral signaling by modulating interferon-γ responses[27].
Autophagy dysfunction can lead to disruption of intestinal barrier function, triggering an immune response and leading to chronic intestinal inflammation. Genome-wide association studies have found that mutations in ATGs are associated with IBD. At present, many autophagy-related genes (such as ATG16L1, ULK1, NOD2, LRRK2, and IRGM) have been shown to be susceptibility genes for IBD[28,29]. One study found that ATG5 expression in intestinal myeloid cells modulates IL-12, thereby preventing uncontrolled IFN-γ-driven intestinal inflammation[30]. Furthermore, mice with specific deletion of ATG16L1 in IECs have aggravated intestinal injury[31]. ATG16L1T300A is a single nucleotide polymorphism of the susceptibility gene for Crohn's disease (CD)[32]. Further studies have shown that autophagy disorder caused by the ATG16L1T300A polymorphism contributes to the increased risk of CD through NF-κB-mediated inflammation[33]. In addition, researchers have found that ATG16L1 interferes with Paneth cell secretion of antimicrobial agents and dendritic cell antigen presentation, which leads to intestinal mucosal barrier dysfunction and the development of CD.
In recent years, more and more animal experiments have revealed the presence of a large number of autophagic vesicles accompanied by mitochondrial vacuolization in DSS-induced colitis. In Wistar rats, vitamin D has been shown to alleviate stress colitis through mTOR-STAT3 signaling and regulation of autophagy[34]. Similarly, we found that activation of estrogen receptor β, which is highly expressed in intestinal tissues, can inhibit colitis by promoting NLRP6-mediated autophagy[35]. In addition, Ma and collaborators demonstrated that Parkin loss may lead to high drug resistance in DSS-induced colitis[36].
Intestinal I/R injury is a common GI barrier dysfunction. The ultrastructural changes of the intestinal epithelium under the transmission electron microscope can provide information about the early changes of intestinal I/R, including the ischemia phase and reperfusion phase. One study showed that a large number of autophagosomes were found in the cytoplasm of colonic epithelial cells after 1 h of ischemia, with organelle damage, cytolysis, and lysosome formation[37]. However, in another study, a significant reduction in autophagic vacuoles was observed in intestinal tissues 4 h after reperfusion by TEM[38]. Another study found that the number of autophagosomes and autolysosomes increased at 4 h and decreased at 20 h after I/R upon electron microscopy analysis of intestinal epithelial tissues taken at 0, 4, and 20 h after I/R[39]. Thus, based on morphological evidence, autophagy has a conflicting role in the pathology of I/R-induced intestinal injury. In addition to TEM results, several studies have found that the autophagy-related marker LC3BII/I ratio and the mitophagy-related PINK1/Parkin pathway are significantly up-regulated during intestinal I/R injury[40-42]. Consistent with this, Liu et al[43] demonstrated in rat experiments that inhibition of autophagy alleviated intestinal I/R injury through the miR-146a/TXNIP axis. Similarly, upregulation of miR-182 in I/R mice leading to a significant reduction in autophagosomes has also found morphological evidence observed by TEM[44]. Studies have found that selenium nanoparticles can effectively alleviate intestinal epithelial barrier damage by inhibiting autophagy mediated by the TBC1D15/Rab7 signaling pathway[45]. In contrast to the above studies, activation of the AMPK/ SIRT1-autophagy pathway alleviated intestinal I/R injury[46,47]. These studies seem to suggest that autophagic changes during the ischemic phase play a more decisive role in the course of the disease. Therefore, studying the role of autophagy in intestinal I/R injury may require a more unified modeling approach and further analysis of the morphological changes of autophagy in different periods. Another common intestinal barrier dysfunction is necrotizing enterocolitis. The ultrastructure of rapamycin-treated IEC-6 and Caco2 cells was observed by TEM, and the formation of autophagic vacuoles was significantly accumulated, which could be reduced by human β-defensin-3 (hBD3) treatment[48].
Functional dyspepsia (FD) is a common GI motility disorder, affecting 11.5%-29.2% of people worldwide. Interstitial cells of Cajal (ICC), especially muscle ICC (ICC-MY), are the key cells to GI motility. Early studies found that the impaired autophagy of ICC was closely related to gastric hypomotility in rats with gastroparesis[49], especially with the reduction and structural abnormalities of ICC-MY cells. Zhang et al[50] observed a large number of autophagosomes in the ultrastructure of ICC-MY in the FD model group by electron microscopy, and even degeneration or reduction of organelles. This suggests that increased autophagy and decreased differentiation of ICC-MY play an important role in FD. In addition, Drp-1 mediated mitophagy in ICC significantly promoted gastric motility in FD rats. Lee et al[51] also found that the traditional Chinese medicine compound Chaihu Shugan powder inhibits ICC autophagy through the PI3K/PDK1 pathway, thus playing a role in promoting GI motility. In addition, many studies have found that electroacupuncture can improve GI motility disorders by activating autophagy[52-54]. In addition, Fu et al[55] demonstrated that exosomes derived from patients with irritable bowel syndrome have an inhibitory effect on autophagy in human colonic epithelial cells by promoting ATG14. Although there are still many mysteries about how autophagy is impaired in GI motility disorders, with the further accumulation and analysis of morphological evidence, it is believed that more new regulatory mechanisms will be discovered in the future.
GI cancers have attracted much attention due to their high recurrence and metastasis rates, difficult diagnosis, and poor prognosis. More and more evidence has shown that although chemotherapy drugs are clinically effective, it has become a common phenomenon that many patients develop chemotherapy resistance in GI cancers during treatment.
Gastric cancer: Gastric cancer has attracted much attention due to its high recurrence and metastasis rates, difficult diagnosis, and poor prognosis. Common treatments include surgical resection, radiotherapy[56], and chemotherapy. Helicobacter pylori infection is a common cause in patients with gastric cancer. A study of H. pylori-positive human biopsy specimens revealed onion-like (autophagosome-like) structures containing intact bacteria as well as autolysosomes enclosing degraded material[57]. A number of studies have confirmed that autophagy is related to the chemoresistance in gastric cancer, including resistance to oxaliplatin, cisplatin, and paclitaxel[58-62]. It was found that in paclitaxel-pretreated BGC gastric cancer cells, typical double-membrane autophagic vacuoles and residual organelles around the nucleus could be clearly captured by TEM[62]. Further morphological studies revealed that overexpression of SIRT5, Sec62, and TOB1 genes can induce autophagy in gastric cancer cells[63-65]. Of course, autophagy activation is not present in all drug-resistant gastric cancer cell lines. He et al[66] observed multiple autophagosomes (double-membrane structure) and autolysosomes (single-membrane structure) in the cytoplasm of BGC gastric cancer cells treated with 5-FU. Moreover, the ratio of autophagosome area to that of the cytoplasm was significantly different from that of the control group. However, in AGS cells treated with 5-FU, few autophagosomes and autolysosomes were observed by TEM. In addition, gastric cancer cell-derived exosomes (GC-Ex) have been found to have the ability to induce neutrophil autophagy[67]. The number of autophagosomes was increased in treated neutrophils. TEM and immunofluorescence staining showed that neutrophils treated with GC-Ex had more autophagosomes than those in the control group. Further study showed that FTO silencing reduced the number of autophagosomes in SGC-7901/DDP cells[68].
Colorectal cancer: Colorectal cancer (CRC) is the third most common malignancy and the second leading cause of cancer death in the world. Multiple clinicopathological studies have confirmed that several autophagy-related genes, such as ATG9B, ATG4B, and ULK1, are CRC prognostic markers[69-72]. Accumulating evidence suggests that cytoprotective autophagy not only increases cancer cell survival, but also enhances tumor drug resistance in CRC[73-75]. One study showed that inhibition of autophagy enhanced doxorubicin hydrochloride-induced apoptosis in human colon cancer cells[76]. Further studies found that MTOR signaling dependent mitochondrial dysfunction promotes colorectal cancer cell death[77]. Regulation of the Beclin1/Beclin2 signaling pathway may be the key to inducing autophagic death of colorectal cancer cells[78-80]. In addition, a study on the mechanism of lipopolysaccharide-induced injury in the colon adenoma cell lines Caco-2 and HT-29 showed that autophagic flow was blocked at the autolysosome stage in vitro and in vivo[81]. Moreover, Bacillus Calmette-Guerin has been shown to induce autophagic cell death through TLR2 and TLR4 signaling pathways in a radiosensitive colorectal cell line[82]. In addition, Liu et al[83] found that induction of autophagy-related ferroptosis through the MEK1/2/ERK/c-FOS axis enhanced the sensitivity of colon cancer cells to chemotherapy. TEM showed mitochondrial destruction and increased number of autophagosomes in the diabetic group compared with the non-diabetic group[40].
Autophagy and GI drugs: Autophagy is closely related to the occurrence and development of GI cancer and drug resistance. A large number of studies have found that a variety of natural compounds can induce autophagy to exert anti-cancer effects. For example, salidroside was found to induce autophagy in AGS cells[84]. Moreover, autolysosome accumulation in gastric cancer cells treated with narcicycline and galangin was observed under the electron microscope. TEM showed that the number of autophagosomes increased in lutein-treated IEC-6 cells[8]. In addition, several Chinese herbs such as ononin, celastrol, licorice, and Jianpi-Qingchang decoction have been shown to protect IECs and treat experimental colitis by activating mitophagy[85-88]. Subsequently, Truzzi and colleagues demonstrated that stimulation of autophagy by a combination of spermidine and eugenol supplements reduced intestinal inflammatory parameters[89].
From the perspective of morphology, autophagy is similar to the biological phenomena such as phagocytosis and apoptosis. As an intracellular catabolic mechanism, autophagy and the ubiquitin-proteasome system jointly assume the role of maintaining cellular homeostasis. Not only that, autophagy also plays a role in programmed cell death in cells lacking apoptosis. Autophagy is inextricably linked to cell growth, development, and differentiation. A large number of studies have confirmed that autophagy is closely related to the physiological functions of the GI tract in different types of GI cells. Morphological studies have provided us with a large amount of evidence that autophagy plays a pro-survival role in benign diseases such as intestinal I/R injury, IBD, and GI motility disorders. However, under pathological conditions, the role of autophagy is not the same, which may be due to the different degrees of autophagy or the existence of other factors. Therefore, more studies on the role of autophagy in GI tumors are needed to solve these unsolved mysteries in the future.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Machado NC, Brazil S-Editor: Yan JP L-Editor: Wang TQ P-Editor: Cai YX
| 1. | Jia Q, Li L, Wang X, Wang Y, Jiang K, Yang K, Cong J, Cai G, Ling J. Hesperidin promotes gastric motility in rats with functional dyspepsia by regulating Drp1-mediated ICC mitophagy. Front Pharmacol. 2022;13:945624. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 23] [Reference Citation Analysis (0)] |
| 2. | Ruska E. Nobel lecture. The development of the electron microscope and of electron microscopy. Biosci Rep. 1987;7:607-629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 34] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 3. | Naama M, Telpaz S, Awad A, Ben-Simon S, Harshuk-Shabso S, Modilevsky S, Rubin E, Sawaed J, Zelik L, Zigdon M, Asulin N, Turjeman S, Werbner M, Wongkuna S, Feeney R, Schroeder BO, Nyska A, Nuriel-Ohayon M, Bel S. Autophagy controls mucus secretion from intestinal goblet cells by alleviating ER stress. Cell Host Microbe. 2023;31:433-446.e4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 88] [Article Influence: 44.0] [Reference Citation Analysis (0)] |
| 4. | Lu R, Zhang YG, Xia Y, Zhang J, Kaser A, Blumberg R, Sun J. Paneth Cell Alertness to Pathogens Maintained by Vitamin D Receptors. Gastroenterology. 2021;160:1269-1283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 73] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 5. | Gorbunov NV, Kiang JG. Up-regulation of autophagy in small intestine Paneth cells in response to total-body gamma-irradiation. J Pathol. 2009;219:242-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 37] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 6. | Yang Y, White E. Autophagy in PDGFRA(+) mesenchymal cells is required for intestinal homeostasis and mammalian survival. Autophagy. 2023;19:726-728. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 7. | Guo Y, Hu R, Li N, Wu J, Yu H, Tan J, Li Z, Xu S. Autophagy Is Required to Sustain Increased Intestinal Cell Proliferation during Phenotypic Plasticity Changes in Honey Bee (Apis mellifera). Int J Mol Sci. 2023;24. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 8. | Chang CJ, Lin JF, Hsiao CY, Chang HH, Li HJ, Lee GA, Hung CF. Lutein Induces Autophagy via Beclin-1 Upregulation in IEC-6 Rat Intestinal Epithelial Cells. Am J Chin Med. 2017;45:1273-1291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 9. | Shi H, Zhao X, Ding Z, Han C, Jiang Y, Qian W, Lin R, Hou X. Na+/H+ Exchanger Regulates Amino Acid-Mediated Autophagy in Intestinal Epithelial Cells. Cell Physiol Biochem. 2017;42:2418-2429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 10. | Yamoto M, Lee C, Chusilp S, Yazaki Y, Alganabi M, Li B, Pierro A. The role of autophagy in intestinal epithelial injury. Pediatr Surg Int. 2019;35:1389-1394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 11. | Chang TK, Shravage BV, Hayes SD, Powers CM, Simin RT, Wade Harper J, Baehrecke EH. Uba1 functions in Atg7- and Atg3-independent autophagy. Nat Cell Biol. 2013;15:1067-1078. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 131] [Cited by in RCA: 154] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 12. | Saha K, Subramenium Ganapathy A, Wang A, Michael Morris N, Suchanec E, Ding W, Yochum G, Koltun W, Nighot M, Ma T, Nighot P. Autophagy Reduces the Degradation and Promotes Membrane Localization of Occludin to Enhance the Intestinal Epithelial Tight Junction Barrier against Paracellular Macromolecule Flux. J Crohns Colitis. 2023;17:433-449. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 13.5] [Reference Citation Analysis (1)] |
| 13. | Kim Y, Lee Y, Heo G, Jeong S, Park S, Yoo JW, Jung Y, Im E. Modulation of Intestinal Epithelial Permeability via Protease-Activated Receptor-2-Induced Autophagy. Cells. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 14. | Li J, Bai J, Si X, Jia H, Wu Z. Benzo[a]pyrene induces epithelial tight junction disruption and apoptosis via inhibiting the initiation of autophagy in intestinal porcine epithelial cells. Chem Biol Interact. 2023;374:110386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 15. | David L, Taieb F, Pénary M, Bordignon PJ, Planès R, Bagayoko S, Duplan-Eche V, Meunier E, Oswald E. Outer membrane vesicles produced by pathogenic strains of Escherichia coli block autophagic flux and exacerbate inflammasome activation. Autophagy. 2022;18:2913-2925. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 38] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 16. | Yang L, Wang JF, Liu N, Wang X, Wang J, Yang GH, Yang GY, Zhu YH. Lactobacillusjohnsonii L531 Protects against Salmonella Infantis-Induced Intestinal Damage by Regulating the NOD Activation, Endoplasmic Reticulum Stress, and Autophagy. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 17. | Liu W, Zhou Y, Peng T, Zhou P, Ding X, Li Z, Zhong H, Xu Y, Chen S, Hang HC, Shao F. N(ε)-fatty acylation of multiple membrane-associated proteins by Shigella IcsB effector to modulate host function. Nat Microbiol. 2018;3:996-1009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 68] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 18. | Ge P, Lei Z, Yu Y, Lu Z, Qiang L, Chai Q, Zhang Y, Zhao D, Li B, Pang Y, Liu CH, Wang J. M. tuberculosis PknG manipulates host autophagy flux to promote pathogen intracellular survival. Autophagy. 2022;18:576-594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 57] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 19. | Karthikeyan R, Gayathri P, Ramasamy S, Suvekbala V, Jagannadham MV, Rajendhran J. Transcriptome responses of intestinal epithelial cells induced by membrane vesicles of Listeria monocytogenes. Curr Res Microb Sci. 2023;4:100185. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 20. | Wei S, Zhang J, Wu X, Chen M, Huang H, Zeng S, Xiang Z, Li X, Dong W. Fusobacterium nucleatum Extracellular Vesicles Promote Experimental Colitis by Modulating Autophagy via the miR-574-5p/CARD3 Axis. Inflamm Bowel Dis. 2023;29:9-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 25] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 21. | Marzoog TR, Jabir MS, Ibraheem S, Jawad SF, Hamzah SS, Sulaiman GM, Mohammed HA, Khan RA. Bacterial extracellular vesicles induced oxidative stress and mitophagy through mTOR pathways in colon cancer cells, HT-29: Implications for bioactivity. Biochim Biophys Acta Mol Cell Res. 2023;1870:119486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 22. | Gamage CDB, Kim JH, Yang Y, Taş İ, Park SY, Zhou R, Pulat S, Varlı M, Hur JS, Nam SJ, Kim H. Libertellenone T, a Novel Compound Isolated from Endolichenic Fungus, Induces G2/M Phase Arrest, Apoptosis, and Autophagy by Activating the ROS/JNK Pathway in Colorectal Cancer Cells. Cancers (Basel). 2023;15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 23. | Han L, Zheng Y, Deng J, Nan ML, Xiao Y, Zhuang MW, Zhang J, Wang W, Gao C, Wang PH. SARS-CoV-2 ORF10 antagonizes STING-dependent interferon activation and autophagy. J Med Virol. 2022;94:5174-5188. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 70] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 24. | Gassen NC, Papies J, Bajaj T, Emanuel J, Dethloff F, Chua RL, Trimpert J, Heinemann N, Niemeyer C, Weege F, Hönzke K, Aschman T, Heinz DE, Weckmann K, Ebert T, Zellner A, Lennarz M, Wyler E, Schroeder S, Richter A, Niemeyer D, Hoffmann K, Meyer TF, Heppner FL, Corman VM, Landthaler M, Hocke AC, Morkel M, Osterrieder N, Conrad C, Eils R, Radbruch H, Giavalisco P, Drosten C, Müller MA. SARS-CoV-2-mediated dysregulation of metabolism and autophagy uncovers host-targeting antivirals. Nat Commun. 2021;12:3818. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 170] [Cited by in RCA: 204] [Article Influence: 51.0] [Reference Citation Analysis (0)] |
| 25. | Cloherty APM, Rader AG, Patel KS, Pérez-Vargas J, Thompson CAH, Ennis S, Niikura M, Wildenberg ME, Muncan V, Schreurs RRCE, Jean F, Ribeiro CMS. Berbamine suppresses intestinal SARS-CoV-2 infection via a BNIP3-dependent autophagy blockade. Emerg Microbes Infect. 2023;12:2195020. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 26. | Wang S, Qiao J, Chen Y, Tian L, Sun X. Urolithin A inhibits enterovirus 71 replication and promotes autophagy and apoptosis of infected cells in vitro. Arch Virol. 2022;167:1989-1997. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 27. | Lee S, Kalugotla G, Ingle H, Rodgers R, Wu C, Wang Y, Li Y, Yang X, Zhang J, Borella NR, Deng H, Droit L, Hill R, Peterson ST, Desai C, Lawrence D, Lu Q, Baldridge MT. Intestinal antiviral signaling is controlled by autophagy gene Epg5 independent of the microbiota. Autophagy. 2022;18:1062-1077. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 28. | Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, Ripke S, Lee JC, Jostins L, Shah T, Abedian S, Cheon JH, Cho J, Dayani NE, Franke L, Fuyuno Y, Hart A, Juyal RC, Juyal G, Kim WH, Morris AP, Poustchi H, Newman WG, Midha V, Orchard TR, Vahedi H, Sood A, Sung JY, Malekzadeh R, Westra HJ, Yamazaki K, Yang SK; International Multiple Sclerosis Genetics Consortium; International IBD Genetics Consortium, Barrett JC, Alizadeh BZ, Parkes M, Bk T, Daly MJ, Kubo M, Anderson CA, Weersma RK. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979-986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1898] [Cited by in RCA: 1861] [Article Influence: 186.1] [Reference Citation Analysis (0)] |
| 29. | Chauhan S, Mandell MA, Deretic V. IRGM governs the core autophagy machinery to conduct antimicrobial defense. Mol Cell. 2015;58:507-521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 189] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 30. | Merkley SD, Goodfellow SM, Guo Y, Wilton ZER, Byrum JR, Schwalm KC, Dinwiddie DL, Gullapalli RR, Deretic V, Jimenez Hernandez A, Bradfute SB, In JG, Castillo EF. Non-autophagy Role of Atg5 and NBR1 in Unconventional Secretion of IL-12 Prevents Gut Dysbiosis and Inflammation. J Crohns Colitis. 2022;16:259-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 31. | Foerster EG, Tsang DKL, Goyal S, Robertson SJ, Robert LM, Maughan H, Streutker CJ, Girardin SE, Philpott DJ. ATG16L1 protects from interferon-γ-induced cell death in the small intestinal crypt. Mucosal Immunol. 2023;16:135-152. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 32. | Baradaran Ghavami S, Kabiri F, Nourian M, Balaii H, Shahrokh S, Chaleshi V, Sherkat G, Shalileh F, Asadzadeh Aghdaei H. Association between variants of the autophagy related gene ATG16L1 in inflammatory bowel diseases and clinical statues. Gastroenterol Hepatol Bed Bench. 2019;12:S94-S100. [PubMed] |
| 33. | Gao P, Liu H, Huang H, Sun Y, Jia B, Hou B, Zhou X, Strober W, Zhang F. The Crohn Disease-associated ATG16L1(T300A) polymorphism regulates inflammatory responses by modulating TLR- and NLR-mediated signaling. Autophagy. 2022;18:2561-2575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 24] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 34. | Abdelmalak MFL, Abdelrahim DS, George Michael TMA, Abdel-Maksoud OM, Labib JMW. Vitamin D and lactoferrin attenuate stress-induced colitis in Wistar rats via enhancing AMPK expression with inhibiting mTOR-STAT3 signaling and modulating autophagy. Cell Biochem Funct. 2023;41:211-222. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 35. | Zilleruelo I, Espinoza E, Ruiz I. Influence of the assessment of the severity on the frequency of adverse drug reactions (ADRs). Int J Clin Pharmacol Ther Toxicol. 1987;25:328-333. [PubMed] |
| 36. | Ma Z, Wu J, Wu Y, Sun X, Rao Z, Sun N, Fu Y, Zhang Z, Li J, Xiao M, Zeng Q, Han C, Ding D, Zhang H, Yuan H, Zhang J, Yang S, Chen Y. Parkin increases the risk of colitis by downregulation of VDR via autophagy-lysosome degradation. Int J Biol Sci. 2023;19:1633-1644. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 37. | Grosche A, Morton AJ, Graham AS, Sanchez LC, Blikslager AT, Polyak MM, Freeman DE. Ultrastructural changes in the equine colonic mucosa after ischaemia and reperfusion. Equine Vet J Suppl. 2011;8-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 38. | Li Z, Wang G, Feng D, Zu G, Li Y, Shi X, Zhao Y, Jing H, Ning S, Le W, Yao J, Tian X. Targeting the miR-665-3p-ATG4B-autophagy axis relieves inflammation and apoptosis in intestinal ischemia/reperfusion. Cell Death Dis. 2018;9:483. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 76] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 39. | Jiang M, Wan S, Dai X, Ye Y, Hua W, Ma G, Pang X, Wang H, Shi B. Protective effect of ghrelin on intestinal I/R injury in rats. Open Med (Wars). 2022;17:1308-1317. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 40. | Zeng Z, Liu HM, Zhang YY, Chen R, Sun T, Li W, Sun Q, Xia ZY, Meng QT. Aggravated intestinal ischemiareperfusion injury is associated with activated mitochondrial autophagy in a mouse model of diabetes. Mol Med Rep. 2020;22:1892-1900. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 41. | Li S, Zhou Y, Gu X, Zhang X, Jia Z. NLRX1/FUNDC1/NIPSNAP1-2 axis regulates mitophagy and alleviates intestinal ischaemia/reperfusion injury. Cell Prolif. 2021;54:e12986. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 65] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 42. | Zhang Q, Liu XM, Hu Q, Liu ZR, Liu ZY, Zhang HG, Huang YL, Chen QH, Wang WX, Zhang XK. Dexmedetomidine inhibits mitochondria damage and apoptosis of enteric glial cells in experimental intestinal ischemia/reperfusion injury via SIRT3-dependent PINK1/HDAC3/p53 pathway. J Transl Med. 2021;19:463. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 38] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 43. | Zhenzhen L, Wenting L, Jianmin Z, Guangru Z, Disheng L, Zhiyu Z, Feng C, Yajing S, Yingxiang H, Jipeng L, Zhanhai W, Yan Z, Xin L, Yongqiang L, Yufang L. miR-146a-5p/TXNIP axis attenuates intestinal ischemia-reperfusion injury by inhibiting autophagy via the PRKAA/mTOR signaling pathway. Biochem Pharmacol. 2022;197:114839. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 44. | Li Y, Luo Y, Li B, Niu L, Liu J, Duan X. miRNA-182/Deptor/mTOR axis regulates autophagy to reduce intestinal ischaemia/reperfusion injury. J Cell Mol Med. 2020;24:7873-7883. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 45. | Dou X, Qiao L, Song X, Chang J, Pi S, Zhang X, Zeng X, Zhu L, Xu C. Biogenic selenium nanoparticles alleviate intestinal epithelial barrier injury by regulating mitochondria-lysosome crosstalk. Food Funct. 2023;14:4891-4904. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 46. | Li B, Li W, Zheng M, Wang Y, Diao Y, Mou X, Liu J. Corilagin alleviates intestinal ischemia/reperfusion injury by relieving oxidative stress and apoptosis via AMPK/Sirt1-autophagy pathway. Exp Biol Med (Maywood). 2023;248:317-326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 47. | Liu X, Yang B, Tan YF, Feng JG, Jia J, Yang CJ, Chen Y, Wang MH, Zhou J. The role of AMPK-Sirt1-autophagy pathway in the intestinal protection process by propofol against regional ischemia/reperfusion injury in rats. Int Immunopharmacol. 2022;111:109114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 48. | Chen L, Lv Z, Gao Z, Ge G, Wang X, Zhou J, Sheng Q. Human β-defensin-3 reduces excessive autophagy in intestinal epithelial cells and in experimental necrotizing enterocolitis. Sci Rep. 2019;9:19890. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 49. | Wei X, Lin Y, Zhao D, Xiao X, Chen Q, Chen S, Peng Y. Electroacupuncture Relieves Suppression of Autophagy in Interstitial Cells of Cajal of Diabetic Gastroparesis Rats. Can J Gastroenterol Hepatol. 2020;2020:7920715. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 50. | Zhang LM, Zeng LJ, Deng J, Zhang YQ, Wang YJ, Xie TY, Ling JH. Investigation of autophagy and differentiation of myenteric interstitial cells of Cajal in the pathogenesis of gastric motility disorders in rats with functional dyspepsia. Biotechnol Appl Biochem. 2018;65:533-539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 51. | Li L, Jia Q, Wang X, Wang Y, Wu C, Cong J, Ling J. Chaihu Shugan San promotes gastric motility in rats with functional dyspepsia by regulating Drp-1-mediated ICC mitophagy. Pharm Biol. 2023;61:249-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 14] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 52. | Wang L, Chen Y, Xu MM, Cao W, Zheng QH, Zhou SY, Yao JP, Xi MH, Qin HY, Li Y, Zhang W. Electroacupuncture Alleviates Functional Constipation in Mice by Activating Enteric Glial Cell Autophagy via PI3K/AKT/mTOR Signaling. Chin J Integr Med. 2023;29:459-469. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 53. | Song LZ, Xu N, Yu Z, Yang H, Xu CC, Qiu Z, Dai JW, Xu B, Hu XM. The effect of electroacupuncture at ST25 on Parkinson's disease constipation through regulation of autophagy in the enteric nervous system. Anat Rec (Hoboken). 2023;306:3214-3228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 54. | Yang J, Wang L, Mei M, Guo J, Yang X, Liu S. Electroacupuncture repairs intestinal barrier by upregulating CB1 through gut microbiota in DSS-induced acute colitis. Chin Med. 2023;18:24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 55. | Fu R, Liu S, Zhu M, Zhu J, Chen M. Apigenin reduces the suppressive effect of exosomes derived from irritable bowel syndrome patients on the autophagy of human colon epithelial cells by promoting ATG14. World J Surg Oncol. 2023;21:95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 56. | Shi X, Zou J, Wang Y, Zhao J, Ye B, Qi Q, Liu F, Hu J, Li S, Tian Y. MST4 as a novel therapeutic target for autophagy and radiosensitivity in gastric cancer. IUBMB Life. 2023;75:117-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 57. | Hu W, Zhang L, Li MX, Shen J, Liu XD, Xiao ZG, Wu DL, Ho IHT, Wu JCY, Cheung CKY, Zhang YC, Lau AHY, Ashktorab H, Smoot DT, Fang EF, Chan MTV, Gin T, Gong W, Wu WKK, Cho CH. Vitamin D3 activates the autolysosomal degradation function against Helicobacter pylori through the PDIA3 receptor in gastric epithelial cells. Autophagy. 2019;15:707-725. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 114] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 58. | Ren J, Hu Z, Niu G, Xia J, Wang X, Hong R, Gu J, Wang D, Ke C. Annexin A1 induces oxaliplatin resistance of gastric cancer through autophagy by targeting PI3K/AKT/mTOR. FASEB J. 2023;37:e22790. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 27] [Reference Citation Analysis (0)] |
| 59. | Wang J, Sun Y, Zhang X, Cai H, Zhang C, Qu H, Liu L, Zhang M, Fu J, Zhang J, Wang J, Zhang G. Oxidative stress activates NORAD expression by H3K27ac and promotes oxaliplatin resistance in gastric cancer by enhancing autophagy flux via targeting the miR-433-3p. Cell Death Dis. 2021;12:90. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 55] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 60. | Dai X, Chen Y, Chen N, Dou J, Zhuang H, Wang J, Zhao X, Zhang X, Zhao H. KLF5-mediated aquaporin 3 activated autophagy to facilitate cisplatin resistance of gastric cancer. Immunopharmacol Immunotoxicol. 2023;45:140-152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 61. | Nong ZL, Zhao K, Wang Y, Yu Z, Wang CJ, Chen JQ. CLIC1-mediated autophagy confers resistance to DDP in gastric cancer. Anticancer Drugs. 2024;35:1-11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 3] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 62. | Yu YF, Hu PC, Wang Y, Xu XL, Rushworth GM, Zhang Z, Wei L, Zhang JW. Paclitaxel induces autophagy in gastric cancer BGC823 cells. Ultrastruct Pathol. 2017;41:284-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 63. | Gu W, Qian Q, Xu Y, Xu X, Zhang L, He S, Li D. SIRT5 regulates autophagy and apoptosis in gastric cancer cells. J Int Med Res. 2021;49:300060520986355. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 64. | Su S, Shi YT, Chu Y, Jiang MZ, Wu N, Xu B, Zhou H, Lin JC, Jin YR, Li XF, Liang J. Sec62 promotes gastric cancer metastasis through mediating UPR-induced autophagy activation. Cell Mol Life Sci. 2022;79:133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 20] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 65. | Zang WJ, Hu YL, Qian CY, Feng Y, Liu JZ, Yang JL, Huang H, Zhu YZ, Xue WJ. HDAC4 promotes the growth and metastasis of gastric cancer via autophagic degradation of MEKK3. Br J Cancer. 2022;127:237-248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 10] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 66. | He XX, Huang CK, Xie BS. Autophagy inhibition enhanced 5FUinduced cell death in human gastric carcinoma BGC823 cells. Mol Med Rep. 2018;17:6768-6776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 67. | Zhang X, Shi H, Yuan X, Jiang P, Qian H, Xu W. Tumor-derived exosomes induce N2 polarization of neutrophils to promote gastric cancer cell migration. Mol Cancer. 2018;17:146. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 115] [Cited by in RCA: 251] [Article Influence: 35.9] [Reference Citation Analysis (0)] |
| 68. | Zhang Y, Gao LX, Wang W, Zhang T, Dong FY, Ding WP. M(6) A demethylase fat mass and obesity-associated protein regulates cisplatin resistance of gastric cancer by modulating autophagy activation through ULK1. Cancer Sci. 2022;113:3085-3096. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 37] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 69. | Kim IS, Cho SY, Yang M, Han S, Lee KH, Kim JY, Kim JM, Kang S, Jo EK, Ryu H. ATG9B Is a Poor Prognostic Marker Associated With Immune Evasion in Colon Adenocarcinoma. Anticancer Res. 2023;43:1943-1957. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 70. | Hu WH, Liu TT, Liu PF, Morgan P, Lin IL, Tsai WL, Cheng YY, Hsieh AT, Hu TH, Shu CW. ATG4B and pS383/392-ATG4B serve as potential biomarkers and therapeutic targets of colorectal cancer. Cancer Cell Int. 2023;23:63. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 71. | Lv P, Wu Z, Lai L, Zhang Y, Pei B. The clinicopathological significance and potential function of ULK1 in colon cancer. Biotechnol Genet Eng Rev. 2023;1-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 72. | Bednarczyk M, Muc-Wierzgoń M, Dzięgielewska-Gęsiak S, Fatyga E, Waniczek D. Transcription of Autophagy Associated Gene Expression as Possible Predictors of a Colorectal Cancer Prognosis. Biomedicines. 2023;11. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 73. | Yue Y, Zhang Q, Wang X, Sun Z. STAT3 regulates 5-Fu resistance in human colorectal cancer cells by promoting Mcl-1-dependent cytoprotective autophagy. Cancer Sci. 2023;114:2293-2305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 74. | Manzoor S, Saber-Ayad M, Maghazachi AA, Hamid Q, Muhammad JS. MLH1 mediates cytoprotective nucleophagy to resist 5-Fluorouracil-induced cell death in colorectal carcinoma. Neoplasia. 2022;24:76-85. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 75. | Saini H, Dave R, Chatterjee S, Mandloi A, Sharma H, Daiya A, Mukherjee S, Chowdhury R, Chowdhury S. Transcriptomic analysis reveals differential adaptation of colorectal cancer cells to low and acute doses of cisplatin. Gene. 2023;864:147304. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 76. | Tang H, Wu D, Yang H, Yang J, Zhang Y, Li M, Liu H, Li Q. [Inhibition of autophagy enhances apoptosis induced by doxorubicin hydrochloride in human colon cancer cells]. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2022;38:237-243. [PubMed] |
| 77. | Zheng Y, Yang W, Jia Y, Ji J, Wu L, Feng J, Li Y, Cheng Z, Zhang J, Li J, Dai W, Xu X, Wu J, Zhou Y, Guo C. Promotion of colorectal cancer cell death by ezetimibe via mTOR signaling-dependent mitochondrial dysfunction. Front Pharmacol. 2023;14:1081980. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 78. | Kong W, Zhu H, Zheng S, Yin G, Yu P, Shan Y, Liu X, Ying R, Ma S. Larotrectinib induces autophagic cell death through AMPK/mTOR signalling in colon cancer. J Cell Mol Med. 2022;26:5539-5550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 79. | Xu W, Nie C, Chen X. DUSP4 inhibits autophagic cell death and apoptosis in colorectal cancer by regulating BCL2-Beclin1/Bax signaling. Mol Biol Rep. 2023;50:3229-3239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 80. | Qian J, Cao Y, Zhang J, Li L, Wu J, Yu J, Huo J. Tanshinone IIA Alleviates the Biological Characteristics of Colorectal Cancer via Activating the ROS/JNK Signaling Pathway. Anticancer Agents Med Chem. 2023;23:227-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 81. | Zhou M, Xu W, Wang J, Yan J, Shi Y, Zhang C, Ge W, Wu J, Du P, Chen Y. Boosting mTOR-dependent autophagy via upstream TLR4-MyD88-MAPK signalling and downstream NF-κB pathway quenches intestinal inflammation and oxidative stress injury. EBioMedicine. 2018;35:345-360. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 260] [Cited by in RCA: 254] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 82. | Yuk JM, Shin DM, Song KS, Lim K, Kim KH, Lee SH, Kim JM, Lee JS, Paik TH, Kim JS, Jo EK. Bacillus calmette-guerin cell wall cytoskeleton enhances colon cancer radiosensitivity through autophagy. Autophagy. 2010;6:46-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 62] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 83. | Liu Z, Xu Y, Liu X, Wang B. PCDH7 knockdown potentiates colon cancer cells to chemotherapy via inducing ferroptosis and changes in autophagy through restraining MEK1/2/ERK/c-Fos axis. Biochem Cell Biol. 2022;100:445-457. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 84. | Rong L, Li Z, Leng X, Li H, Ma Y, Chen Y, Song F. Salidroside induces apoptosis and protective autophagy in human gastric cancer AGS cells through the PI3K/Akt/mTOR pathway. Biomed Pharmacother. 2020;122:109726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 151] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 85. | Yu T, Lu X, Liang Y, Yang L, Yin Y, Chen H. Ononin alleviates DSS-induced colitis through inhibiting NLRP3 inflammasome via triggering mitophagy. Immun Inflamm Dis. 2023;11:e776. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 86. | Chen D, Ye L, Liu Y, Yu J, Ni S, Chen Y, Zhong J. Celastrol activates mitochondrial autophagy through Nur77-TRAF2-p62/SQSTM1 pathway in the treatment of experimental colitis. Minerva Pediatr (Torino). 2023;75:760-762. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 87. | Kong J, Xiang Q, Shi G, Xu Z, Ma X, Wang Y, Xuan Z, Xu F. Licorice protects against ulcerative colitis via the Nrf2/PINK1-mediated mitochondrial autophagy. Immun Inflamm Dis. 2023;11:e757. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 7] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 88. | Qiao D, Liu X, Zhang Y, Zhang Z, Tang Y, Chen Q, Shi Y, Chen Y, Tang Z, Dai Y. Jianpi-Qingchang decoction alleviates ulcerative colitis by modulating endoplasmic reticulum stress-related autophagy in intestinal epithelial cells. Biomed Pharmacother. 2023;158:114133. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 89. | Truzzi F, Whittaker A, D'Amen E, Valerii MC, Abduazizova V, Spisni E, Dinelli G. Spermidine-Eugenol Supplement Preserved Inflammation-Challenged Intestinal Cells by Stimulating Autophagy. Int J Mol Sci. 2023;24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |