Published online Feb 21, 2023. doi: 10.3748/wjg.v29.i7.1202
Peer-review started: October 27, 2022
First decision: November 14, 2022
Revised: November 19, 2022
Accepted: December 21, 2022
Article in press: December 21, 2022
Published online: February 21, 2023
Processing time: 116 Days and 18.3 Hours
Helicobacter pylori and the stomach microbiome play a crucial role in gastric car
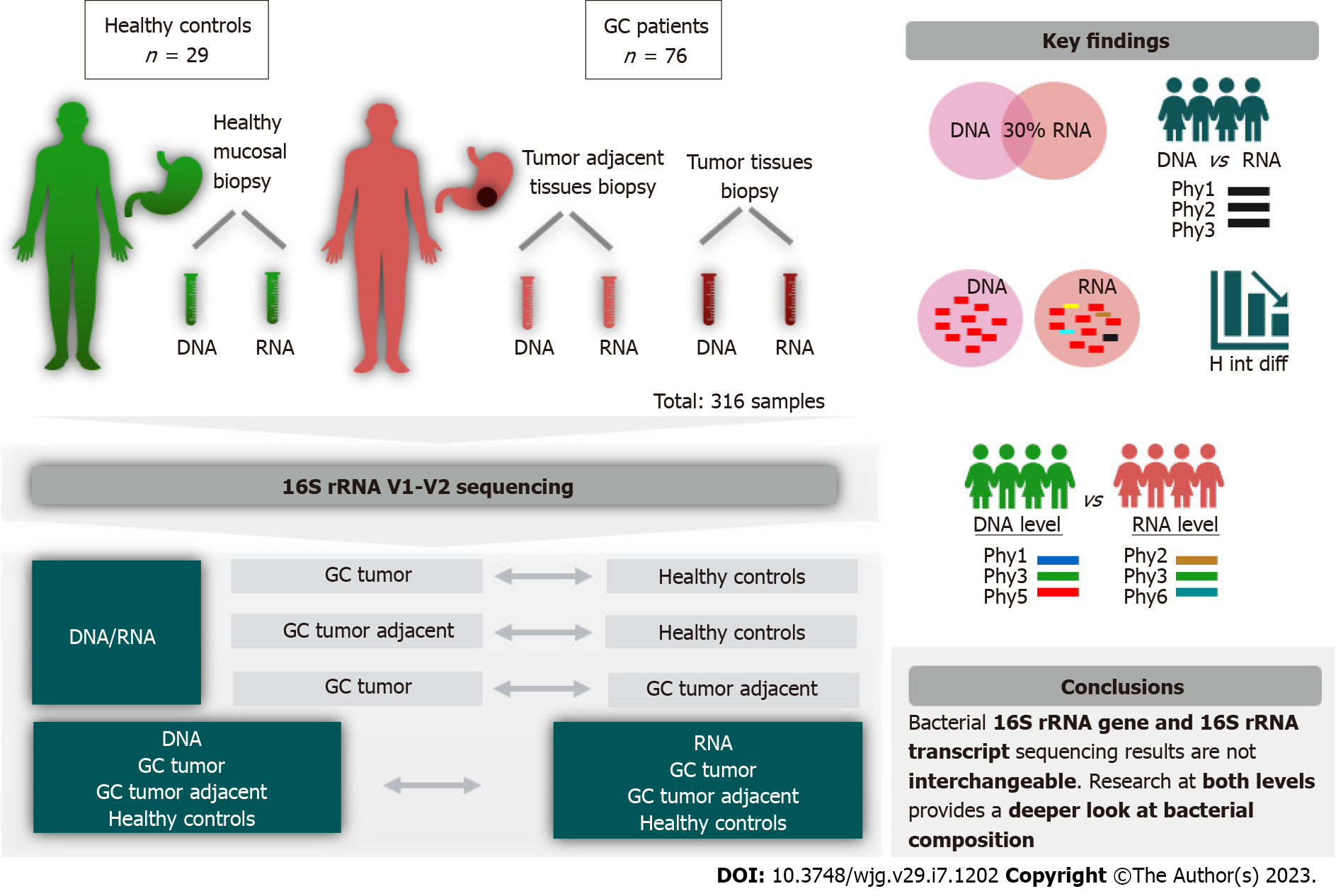
To characterize the microbiota of GC using 16S rRNA gene and its transcript and determine difference in the bacterial composition.
In this study, 316 DNA and RNA samples extracted from 105 individual stomach biopsies were included. The study cohort consisted of 29 healthy control in
Microbial analysis revealed that only a portion of phylotypes (18%-30%) overlapped between microbial profiles obtained from DNA and RNA samples. Detailed analysis revealed differences between GC and controls depending on the chosen modality, identifying 17 genera at the DNA level and 27 genera at the RNA level. Ten of those bacteria were found to be different from the control group at both levels. The key taxa showed congruent results in various tests used; however, differences in 7 bacteria taxa were found uniquely only at the DNA level, and 17 uniquely only at the RNA level. Furthermore, RNA sequencing was more sensitive for detecting differences in bacterial richness, as well as differences in the relative abundance of Reyranella and Sediminibacterium according to the type of GC. In each study group (control, tumor, and tumor adjacent) were found differences between DNA and RNA bacterial profiles.
Comprehensive microbial study provides evidence for the effect of choice of sequencing modality on the microbiota profile, as well as on the identified differences between case and control.
Core Tip: In this study, we aimed to characterize the microbiota of gastric cancer (GC) on two levels: 16S rRNA gene and its transcript. Our study showed that only a small portion of bacterial sequences overlapped using those two approaches. Moreover, our study revealed that obtained results comparing the case group with the controls depend on the chosen modality. We also showed that Reyranella and Sediminibacterium was associated with the Lauren classification and RNA level was more sensitive to detect low abundant bacteria. This study provides novel insights into microbiome study as well as new founding related to complex GC pathogenesis.
- Citation: Nikitina D, Lehr K, Vilchez-Vargas R, Jonaitis LV, Urba M, Kupcinskas J, Skieceviciene J, Link A. Comparison of genomic and transcriptional microbiome analysis in gastric cancer patients and healthy individuals. World J Gastroenterol 2023; 29(7): 1202-1218
- URL: https://www.wjgnet.com/1007-9327/full/v29/i7/1202.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i7.1202
Microbiota analyses are becoming increasingly relevant in scientific and clinical studies. Most modern microbiome studies use 16S rRNA gene sequence analysis at the DNA level, thereby enabling the identification of bacteria at all stages of their existence (active, dead, and inactive bacteria in the form of endospores) simultaneously. However, some of the more recent studies use RNA samples, which are subsequently reverse transcribed into cDNA for sequencing, giving us knowledge about the metabolic state of the microbial community[1]. RNA has a shorter half-life than DNA and turns over in cells more rapidly, providing a deeper look at bacterial activity[2].
The stomach has long been considered an almost sterile organ due to its acidic environment and enzymatic effects[3]. Since its identification, it is known that Helicobacter pylori (H. pylori) is perfectly adapted not only to survive in the acidic environment of the stomach, but also to colonize this part of the gastrointestinal tract[4]. H. pylori is the major cause of peptic ulcer disease and the most significant risk factor for gastric cancer (GC). GC remains one of the most common cancers in the world and the fourth leading cause of cancer-related death[5]. However, only a minority of people infected with H. pylori develop GC, which may be linked to non-H. pylori microbiota-associated alterations in the stomach[6]. Studies in insulin-gastrin (INS-GAS) mice and in humans indicated the importance of other members of the stomach bacterial community in the development of gastric carcinogenesis[7-10].
There is only one study that has compared DNA and RNA profiles of the stomach microbiota[11]. However, the profiles of the active and standing microbiota in GC have not been studied. In this study, we systematically characterized the microbiota of GC on both levels using the 16S rRNA gene (DNA level) and its transcript (RNA level). GC tumor and tumor adjacent tissue samples, as well as healthy mucosa samples from the young control group, were used for the comparison. We obtained detailed data on bacterial composition within groups depending on study modality (DNA or RNA) and performed association analysis with clinical characteristics to question the potential impact of approach on the outcome.
In total, 316 DNA and RNA samples from a group of 105 individuals were included in the study (Figure 1). The study cohort consisted of 29 healthy control individuals and 76 patients with GC. Participants did not report any antibiotic intake at least a month before endoscopy. Gastric tissue biopsy samples from damaged mucosa and healthy mucosa at least 5 cm from the tumor tissue were collected from GC patients using single-bite biopsy forceps. From the controls, healthy stomach mucosa biopsies were collected. Tissue samples were placed in sterile cryotubes (Thermo Fisher Scientific, United States), snap-frozen in liquid nitrogen, and stored at -86 °C until further study. Clinical data obtained from histological examination, such as tumor size, number of lymph nodes damaged by tumor cells, presence of metastases (TNM classification), cell differentiation (grading), type of GC (Lauren classification) and stage of GC, were included in the analysis. An overview of the demographic and clinical characteristics of the study cohort is given in Supplementary Table 1.
Study individuals were recruited at the Department of Gastroenterology at the Hospital of Lithuanian University of Health Sciences Kaunas Clinics during the years 2012-2018. This study was approved by the local ethics committee (BE-2-10), and all participants gave their written informed consent.
Total DNA and RNA were extracted from gastric biopsy samples using an AllPrep DNA/RNA Mini kit (Qiagen, Germany) according to the manufacturer’s recommendations. RNA was reverse transcribed into cDNA using the Superscript IV First-Strand Synthesis System Purification Kit (Invitrogen, Carlsbad, CA) and random hexamer primers, following the manufacturer’s instructions. Amplicon libraries were generated as described previously[12,13]. The bacterial 16S rRNA gene V1-V2 region was amplified using the 27F and 338R polymerase chain reaction primers and sequenced on a MiSeq (2 × 250 bp; Illumina, Hayward, CA).
Bioinformatic processing was performed as described previously[14]. FastQ files were analyzed using the dada 2 package[15], version 1.10.1, in R. In total, 7735281 paired-end reads were received, with an average of 22953 per sample. Samples that did not reach 5000 reads were discarded from the analysis (21 samples out of initial 337). All samples were rarefied to an equal sequencing depth of 5047 reads using the phyloseq package[16], with returning 10496 phylotypes (Supplementary Table 2). Phylotypes were annotated to a taxonomic affiliation based on the naive Bayesian classification[17] with a pseudobootstrap threshold of 80%. The relative abundances (expressed as percentages) of different microbial communities’ phylogenetic ranks (from phylum to class, order, family, genus and phylotype) were used for downstream analyses.
The phylogenetic tree was built using the online tool iTOL[18], after hierarchical clustering using the Bray-Curtis algorithm[19] at the phylotype level in Past 3[20]. Bacterial richness and Shannon diversity indices were calculated using the vegan[21] package from R. The data matrices comprising the percentage of abundances of each of the abovementioned taxa were used to construct sample-similarity matrices by the Bray-Curtis algorithm, where samples were ordinated by principal coordinate analysis (PCoA) at the phylotype level using the patchwork[22] package from R.
Differences in relative abundance of detected bacteria (at all taxonomic ranks) between study groups were evaluated by PERMANOVA and ANOSIM statistical tests, using 9999 permutations. Groups were considered significantly different if the P value was < 0.05, considering an estimate effect-size F values for PERMANOVA and R values for ANOSIM tests. Calculation was made by Past 3 program. The distributions of taxa abundance values were compared by Mann-Whitney test followed by Benjamini-Hochberg correction for multiple comparisons, named as false discovery rate value. Differences were considered significant when the corrected p value (q value) was < 0.05.
The bacterial networks were visualized using Cytoscape 3.8.0[23], after the Spearman correlation test performed with the psych[24] package from R, with threshold of 0.2 in absolute value and P value < 0.05. Phylotypes that accounted for at least 1% of the total number of phylotypes and at least 10% of the samples in each group were used for correlation analysis.
The bacterial contents of 180 biopsy samples taken from 105 individuals were characterized as described above (Supplementary Table 1). After sequencing and rarefying library size to the minimum sequencing depth, 10496 different phylotypes belonging to 23 phyla, 40 classes, 82 orders, 169 families, and 463 genera were retrieved and taxonomically annotated.
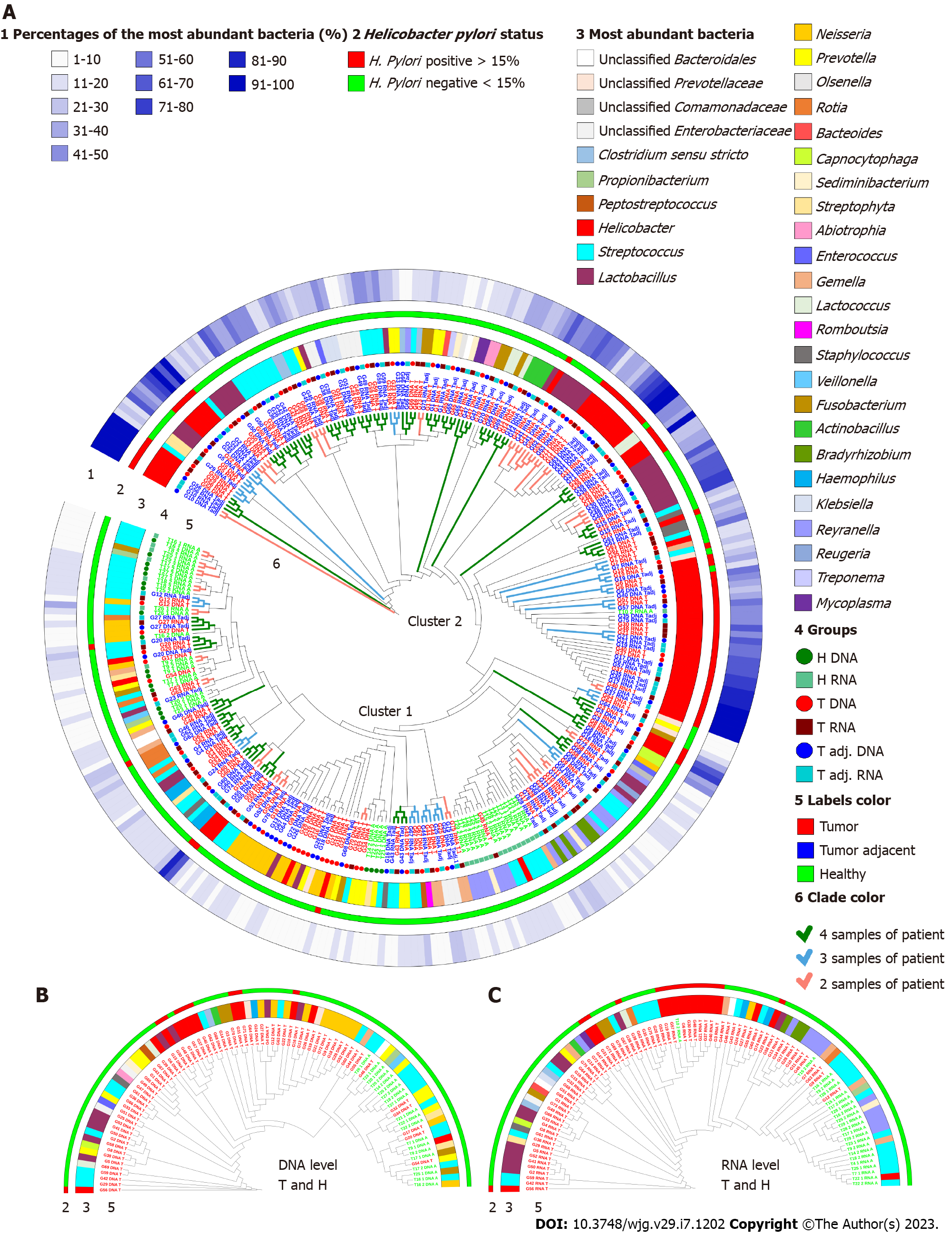
The global bacterial profiles were grouped into two clusters based on their Bray-Curtis similarities as percentages (Figure 2A). Analyzing all samples together, the main factor for clustering was bacterial heterogeneity. The first cluster consisted of samples with a more heterogeneous microbiome profile, where the most abundant bacteria accounted for less than 30%. All control samples (except T10_2) were located in this cluster. The second cluster - where the most abundant bacteria accounted for more than 30% of GC patient samples - was shaped by the most abundant bacteria Helicobacter, the abundance of which reached 98%-100% in some samples (Figure 2A, Supplementary Table 2).
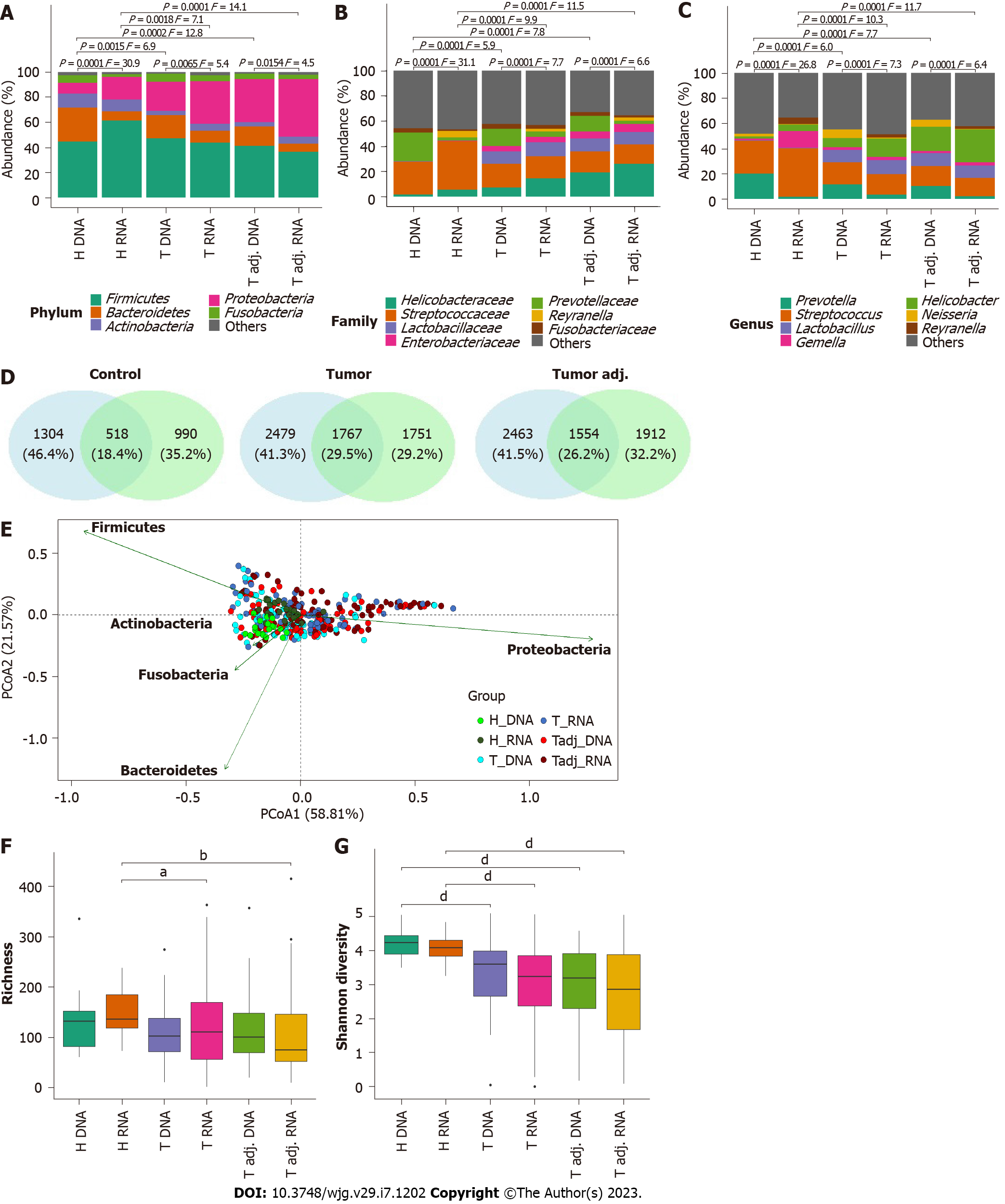
Further PERMANOVA and ANOSIM analyses showed that DNA and RNA groups of the same study individuals were significantly different in all taxonomic ranks (Figures 3A-C; Supplementary Table 3). Differences between DNA and RNA samples were noticeable even at the phylum level. Although Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, and Fusobacteria were the main bacterial phyla in all groups, Bacteroides and Fusobacteria were significantly more abundant at the DNA level. However, in the control group, Firmicutes and Proteobacteria were more abundant at the RNA level. Bacterial profile analysis indicated that only a portion of phylotypes (18%-30%) were common between bacterial profiles obtained from DNA and RNA samples (Figure 3D). PCoA supported the distinction of bacterial communities at the DNA and RNA levels, especially in the control group (Figure 3E). Firmicutes, Bacteroidetes, Proteobacteria, and Fusobacteria were major phyla determinants for sample differentiation.
More detailed analysis revealed that DNA and RNA samples differed from each other by 12, 10, and 30 phylotypes and by 18, 17, and 35 genera in the tumor, tumor adjacent, and control groups, respectively (Table 1, Supplementary Table 4). In all study groups, bacteria such as Neisseria, Peptostreptococcus, Prevotella, Veillonella, and Oribacterium were significantly more abundant at the DNA level, while Staphylococcus, Methyloversatilis, Pseudomonas, Reyranella, Corynebacterium, and Sediminibacterium were significantly enriched at the RNA level. Interestingly, most of these bacteria founded in the RNA samples were not observed in the DNA samples at all, or their relative abundance was low. Some changes in the relative abundance of bacteria between DNA and RNA samples were specific for the study group. For instance, in the control group, Helicobacter, Gemella, and Streptococcus were enriched at the RNA level, while Actinomyces and Alloprevotella were enriched at the DNA level. In the GC groups (tumor and tumor adjacent), Fusobacterium, Granulicatella, Solobacterium, and Porphyromonas were enriched at the DNA level. No bacteria were enriched at the RNA level in this group.
| Increased or decreased at DNA level | |||
| H | T | Tadj | |
| H & T & Tadj | |||
| Haemophilus | Increased | Decreased | Increased |
| Methyloversatilis | Decreased | Decreased | Decreased |
| Neisseria | Increased | Increased | Increased |
| Peptostreptococcus | Increased | Increased | Increased |
| Prevotella | Increased | Increased | Increased |
| Pseudomonas | Decreased | Decreased | Decreased |
| Reyranella | Decreased | Decreased | Decreased |
| Sediminibacterium | Decreased | Decreased | Decreased |
| Staphylococcus | Decreased | Decreased | Decreased |
| Veillonella | Increased | Increased | Increased |
| H | |||
| Actinomyces | Increased | ||
| Gemella | Decreased | ||
| Helicobacter | Decreased | ||
| Streptococcus | Decreased | ||
| T & Tadj | |||
| Fusobacterium | Increased | Increased | |
| Granulicatella | Increased | Increased | |
| Porphyromonas | Increased | Increased | |
| Solobacterium | Increased | Increased | |
Nevertheless, despite the found differences between DNA and RNA, samples of the same origin tended to cluster together in each of the study groups (Figure 2). Paired samples, 46 pairs out of 64 (72%) in the tumor group and 38 pairs out of 58 (66%) in the tumor adjacent tissue group, clustered next to each other, indicating their global similarity (Supplementary Figure 1). Paired samples from the control group were not added to this analysis due to the small number of paired samples.
The GC samples had lower bacterial richness and diversity compared to control samples (Figures 3F and 3G). While differences in diversity were found both at the DNA and RNA levels, differences in bacterial richness were found only at the RNA level. Group-average agglomerative hierarchical clustering analysis showed that it was possible to distinguish patients with GC from controls by their bacterial profile, as samples tended to cluster based on clinical status (both at the DNA and RNA levels) (Figures 2B and 2C). These results were supported by the phylogenetic analysis of global stomach bacteria, which revealed significant differences between the GC group and control groups at all taxonomic ranks (Figures 3A-C, Supplementary Table 3).
Bacterial abundance differential analysis revealed 15 phylotypes and 17 genera that differed between the GC and control groups at the DNA level (Table 2, Supplementary Figure 2, Supplementary Table 4). Meanwhile, at the RNA level, there were twice as many differences: 40 at the phylotype level and 27 at the genus level. Half of the differences detected at the DNA level were also found at the RNA level (58% of genera and 46% of phylotypes). These bacteria include previously described bacteria, such as Lactobacillus, Propionibacterium, Streptococcus, and Veillonella, among others[25-29].
| T vs H | Increased or decreased in T | Tadj vs H | Increased or decreased in Tadj | ||
| DNA | RNA | DNA | RNA | ||
| Lactobacillus | Increased | Increased | Lactobacillus | Increased | Increased |
| Actinomyces | Decreased | Decreased | Actinomyces | Decreased | Decreased |
| Atopobium | Decreased | Decreased | Atopobium | Decreased | Decreased |
| Granulicatella | Decreased | Decreased | Granulicatella | Decreased | Decreased |
| Propionibacterium | Decreased | Decreased | Propionibacterium | Decreased | Decreased |
| Streptococcus | Decreased | Decreased | Streptococcus | Decreased | Decreased |
| Veillonella | Decreased | Decreased | Veillonella | Decreased | Decreased |
| Rothia | Decreased | Increased | Rothia | Decreased | Decreased |
| Clostridium sensu stricto | Increased | Leptotrichia | Decreased | ||
| Prevotella | Decreased | Prevotella | Decreased | ||
| Pseudomonas | Increased | Pseudomonas | Increased | ||
| Staphylococcus | Increased | Staphylococcus | Decreased | ||
| Gemella | Decreased | Gemella | Decreased | ||
| Methyloversatilis | Decreased | Methyloversatilis | Decreased | ||
| Parvimonas | Decreased | Parvimonas | Decreased | ||
| Reyranella | Decreased | Reyranella | Decreased | ||
| Sediminibacterium | Decreased | Sediminibacterium | Decreased | ||
Although fewer unique bacteria were identified only at the DNA level (8 phylotypes and 7 genera), they were more studied and more frequently discussed in the literature as being associated with various human health conditions (Supplementary Table 4). These bacteria include Campylobacter, Clostridium sensu stricto, Prevotella, and Saccharibacteria, among others. Of the listed bacteria, Clostridium sensu stricto was enriched, and others were decreased in GC patients. Uniquely, only at the RNA level were 33 and 17 differences at the phylotype and genus levels, respectively, found between the GC and control groups (Supplementary Table 4). Essentially, this group included such bacteria that were not established or their abundance at the DNA level was negligible, for example, Limnohabitans, Methylobacterium, Methyloversatilis, Pseudomonas, Reyranella, Rhodoluna, Sediminibacterium, and Staphylococcus. Of the listed bacteria, only Pseudomonas and Staphylococcus were more abundant in GC samples, while all the others were more abundant in healthy individuals.
Bacterial diversity and profile comparison analysis between tumor and tumor adjacent tissues did not reveal significant differences at either the DNA or RNA level (Figures 3A-C, 3F and 3G). Moreover, assemblages of approach from each individual typically clustered together irrespective of tissue type (tumor or tumor adjacent tissue) (Figure 2A).
Analysis of the bacterial network similarity revealed that the main network holding bacteria with the highest betweenness centrality score was different between DNA and RNA levels in all study groups (Supplementary Figure 3). In the bacterial network of the control group at the DNA level, phylotypes depending on the Streptococcus, Prevotella, and Actinomyces genera accounted for 68% (58 out of 85) of the total number of bacteria and formed the core network keeping bacteria, while at the RNA level, core bacteria were Streptococcus and Gemella, making up to 62% (47 out of 75) (Supplementary Figures3A and 3B). The GC groups showed different DNA/RNA networks as well: The main network forming bacteria in the tumor adjacent tissue at the DNA level was Prevotella, Gemella, and Granulicatella, while the RNA network was shaped by Streptococcus, Reyranella, and Fusobacterium (Supplementary Figures 3C and 3D). The most critical network-forming bacteria in tumor tissue were: Granulicatella, Veillonella, and Neisseria at the DNA level and Reyranella, Acinetobacter, and Prevotellaceae at the RNA level (Supplementary Figures 3E and 3F).
Two common bacterial clusters (one at the DNA level and another at the RNA level) with strong positive correlations for tumor and tumor adjacent tissues were discovered (Supplementary Figures3C-F), which confirms the absence of significant differences between tumor and tumor adjacent tissue microbiome profiles. At the DNA level, the common cluster consisted of Phy6 (Neisseria), Phy15 (unclassified Prevotellaceae), Phy23 (Neisseria perflava), Phy29 (Prevotella melaninogenica), Phy87 (Solobacterium), and Phy98 (Prevotella). The common cluster at the RNA level included phylotypes such as Phy7 (Reyranella), Phy33 (Sediminibacterium), Phy46 (Propionibacterium acnes), Phy94 (Methyloversatilis), Phy107 (Pseudomonas aeruginosa), and Phy108 (Sphingomonas echinoides). Detected clusters were not found in the control group.
Generally, under the same analysis conditions, GC patients displayed a simpler bacterial network at both the DNA and RNA levels. At the DNA level, control, the tumor, and tumor adjacent groups had 85, 25, and 23 bacteria, respectively; at the RNA level, they had 75, 21, and 18, respectively. Moreover, analysis of bacterial interactions in controls had not only positive but also negative correlations, while GC analysis showed mostly positive correlations.
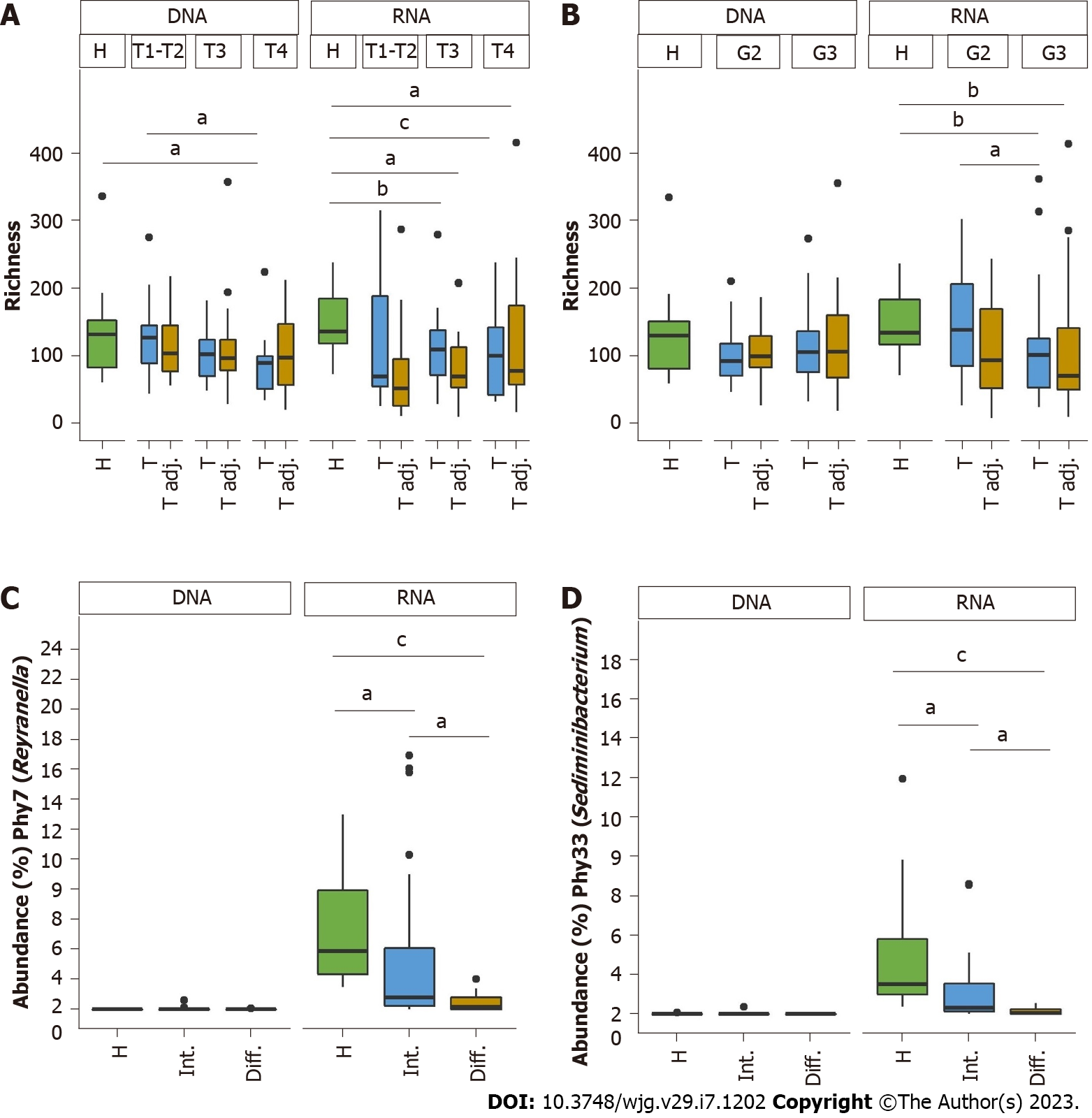
At the DNA level, according to clinical parameters, statistically significant differences were found only in the decrease in bacterial richness between smaller tumors (T1-T2) and extended tumors (T4). The RNA level turned out to be more sensitive and allowed us to detect richness differences between grade II and grade III (Figures 4A and 4B). Moreover, at the RNA level, the relative abundance of the Phy7 (Reyranella) and Phy33 (Sediminibacterium) phylotypes was lower in the diffuse type of GC than in the intestinal type (Figures 4C and 4D). No differences were found between subgroups at the DNA level.
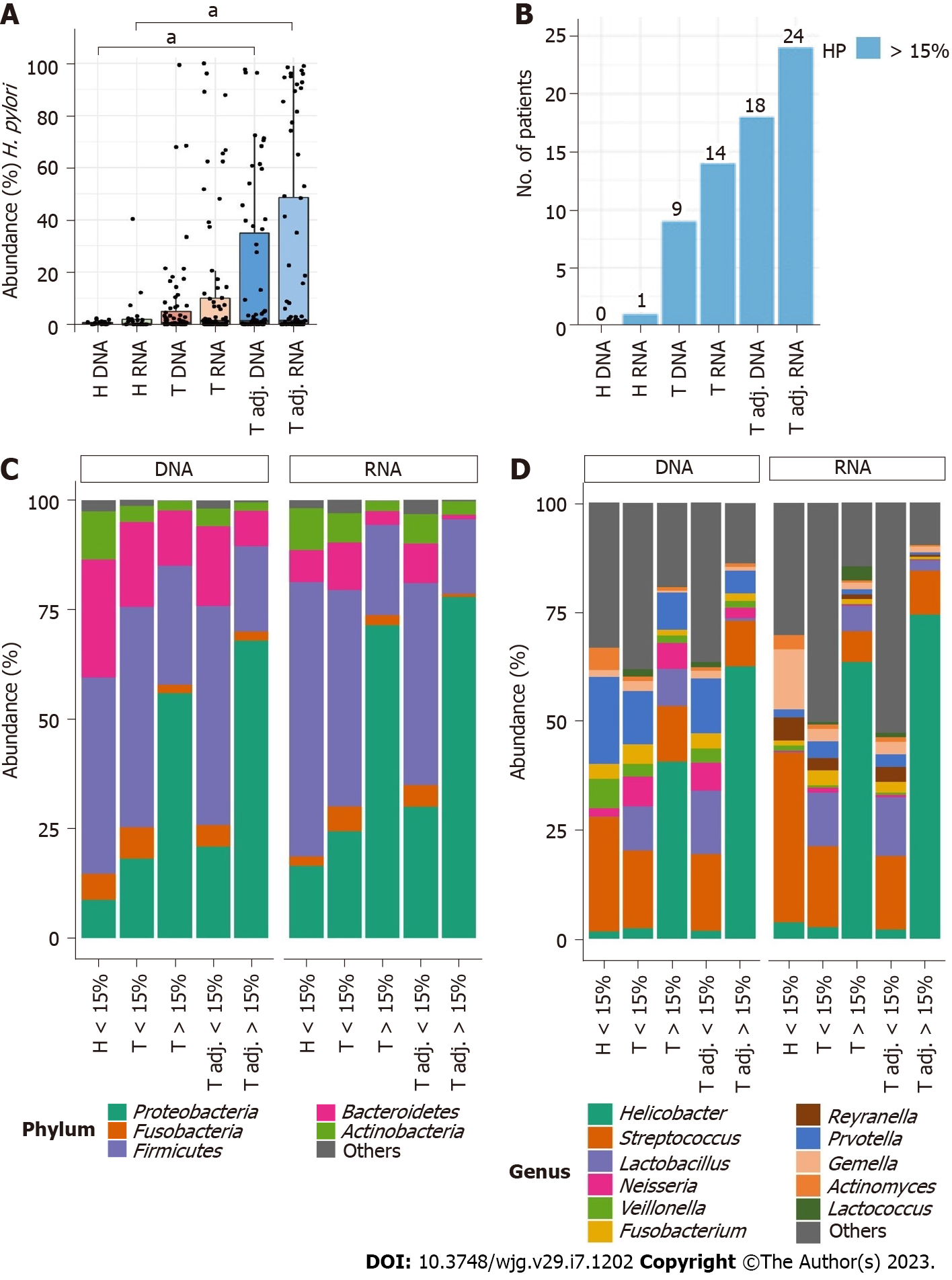
PCoA showed Helicobacter to be the major determinant for differentiating samples based on their bacterial composition in the stomach (Supplementary Figure 4). Overall, H. pylori was detected in 115 and 117 DNA and RNA samples, respectively. In the control group, H. pylori was lower than that in the GC groups at both the DNA and RNA levels (Figure 5A). The tumor adjacent sample group showed the highest number of samples with high H. pylori abundance (Figure 5B). Both in tumor and tumor adjacent groups the mean abundance of H. pylori was increased at the RNA level, although no significant differences between DNA and RNA samples were found.
High H. pylori relative abundance (> 15%) led to an increase in the relative abundance of Proteobacteria and a decrease in other major bacterial phyla, such as Firmicutes, Bacteroidetes, and Fusobacteria (Figure 5C). In tumor tissues analyzed at the DNA and RNA level, Helicobacter was only one genus which changed significantly between samples with high and low H. pylori abundance (Figure 5D, Supplementary Table 5). On the other hand, in tumor adjacent tissues, more bacteria were found, the number of which changed together with Helicobacter. At the DNA level, as the relative abundance of H. pylori increased, the abundance of Porphyromonas and Prevotella significantly decreased. In line with our previous results, more significant differences were found at the RNA level: Staphylococcus significantly increased and seven bacteria (Campylobacter, Fusobacterium, Prevotella, Pseudomonas, Reyranella, Sediminibacterium, Streptococcus) decreased (Figure 5C, Supplementary Table 5). Porphyromonas tended to decrease in tumor adjacent RNA samples as well, although it did not reach a statistically significant level (Supplementary Table 5).
Despite growing interest in the study of microbiota, there is still limited agreement on the most appropriate standard for such studies, especially using 16S rRNA sequencing. Here, we performed systematic analysis of bacterial communities at both the 16S rRNA gene and 16S rRNA transcript levels. To estimate the impact of the different approaches, we used the GC model and considered not only healthy gastric tissues but also GC tumor and adjacent tissues.
The analysis of the study results showed that there were significant differences in the relative abundance of the gastric tissue microbiome between 16S rRNA gene transcript and 16S rRNA gene levels in all study groups (control, tumor, and tumor adjacent). This is the first GC study indicating that active and standing gastric microbiomes are distinct even at the largest taxonomic levels.
Differences in bacterial communities at the DNA and RNA levels could be explained by several possibilities. Using DNA as a research material summed up all bacteria, both biologically active passive in the form of endospores, and DNA sequences of already destroyed and dead bacteria[2,30]. The presence and number of ribosomes in bacteria reflects their metabolic activity; thus, the analysis at the RNA level shows the metabolic activity of live and active bacteria in the community[31-33]. For instance, previously GC-associated bacteria such as Prevotella, Veillonella, and Neisseria in our study were present in high abundance in all analyzed groups at the DNA level but were greatly reduced at the RNA level. In contrast, Pseudomonas, Reyranella, and Staphylococcus were present in higher abundance at the RNA level in all groups. However, it is erroneous to assume that only active bacteria can influence host responses. Many studies have shown that inactivated bacteria or parts of their cells can also influence inflammatory processes or other responses in host tissues. For example, Rabie et al[34] showed that thermally inactivated Salmonella, Staphylococcus, Escherichia, and Pseudomonas strains with unchangeable surface proteins cause colon and breast cancer cell proliferation. In Suprewicz et al[35]’s study, heat-inactivated Enterococcus faecalis, Actinomyces odontolyticus, and Propionibacterium acnes caused cell proliferation changes in lung, breast, and ovarian carcinoma. Postbiotics work based on the same principle. To avoid possible bacterial infection during therapy, instead of active bacteria, their metabolites, which are involved in anti-inflammatory and anticancer mechanisms, are used[36].
It is also cannot be excluded that the shift in bacterial abundance between DNA and RNA levels might stem from varying numbers of copies of the 16S rRNA gene[37] or target sequence quantity inequality[38]. Bacterial rRNAs (16S rRNA, 23S rRNA and 5S rRNA) are typically organized into one operon, and their transcription occurs together, with the number of such operons varying from 1 to 15[38]. In the case of active bacteria, an increase in 16S rRNA gene copies proportionally increases the pool of 16S rRNA transcripts. However, in the case of inactive bacteria, a larger number of 16S gene copies enables the detection of some bacteria, which could not be detected at the RNA level.
The amount of target sequences using the 16S rRNA gene and its transcript are not the same. Of all types of RNA molecules present in the cell, the most common (80%-90%) are included in the ribosome structure rRNAs[39]. 16S rRNAs make up one-third of the total rRNAs. On the other hand, when analyzing the microbiota using the 16S rRNA gene, only one gene is amplified out of the total number of genes, which in different bacteria varies from 1500 to 7000[37]. Thus, the initial larger amount of the bacterial target sequence at the RNA level makes it possible to increase the depth of sequencing and detect more rare bacteria that would be lost during DNA-level analysis. In addition, a shift toward DNA or RNA levels can also be caused by ingestion of bacterial parts from the higher parts of the digestive tract.
Our analysis revealed that the profile of differences found between GC and control tissue depended on the chosen modality: At the DNA level, 17 bacterial genera were detected, and at the RNA level, 27 bacterial genera were detected. Ten of those bacteria (Actinomyces, Alloprevotella, Atopobium, Granulicatella, Lactobacillus, Megasphaera, Propionibacterium, Rothia, Streptococcus, Veillonella) were found to be different from the control group at both levels of sequencing; seven bacterial taxa (Campylobacter, Clostridium sensu stricto, Leptotrichia, Oribacterium, Prevotella, Saccharibacteria genera incertae sedis, Stomatobaculum) were found uniquely only at the DNA level; and 17 (Anaerococcus, Corynebacterium, Eubacterium, Flavobacterium, Gemella, Legionella, Limnohabitans, Massilia, Methylobacterium, Methyloversatilis, Parvimonas, Pseudomonas, Reyranella, Rhodoluna, Sediminibacterium, Solobacterium, Staphylococcus) were found uniquely only at the RNA level. These results confirm the importance of unifying the procedures for studying the microbiota.
Although our study focused on differences in methodology, it did reveal several important findings for the GC study as well. Fourteen bacteria genera were identified to be decreased in patients with GC. Eleven of these bacteria (Actinomyces, Atopobium, Propionibacterium, Streptococcus, Granulicatella, Veillonella, Rothia, Parvimonas, Gemella, Prevotella, Leptotrichia) were previously established in the stomach of healthy people in the absence of gastrointestinal diseases[40]. Most of them are common members of the upper gastrointestinal tract and have strong enzymatic activities. Our study also found four bacteria genera, which were significantly increased in GC patients’ stomach biopsy: Lactobacillus, Clostridium sensu stricto, Staphylococcus, and Pseudomonas.
Lactobacillus is commonly used as a probiotic; however, it has been verified in multiple studies to be enriched in GC[41]. Lactobacillus strains, as well as Clostridium and Staphylococcus, can reduce nitrate to nitrite[42,43]. During the nitrate-reducing process, many N-nitroso compounds are formed that inhibit cell apoptosis and promote mutagenesis and protooncogene expression[44-47]. Clostridium is part of the normal gastrointestinal tract; however, in several previous studies, as in ours, an increase in the number of Clostridium sensu stricto was found[48-50]. Interestingly, Lertpiriyapong et al[9] showed earlier onset and faster progression of GC in INS-GAS mice with restricted microbiota (including Clostridium, Lactobacillus, and Bacteroides), highlighting a possible role of these bacteria in GC. Additionally, several studies have detected increased levels of Staphylococcus in patients with upper gastrointestinal diseases[51-53]. One of the reasons for this may be that stains of Staphylococcus have the enzyme urease, and are able to catalyze the hydrolysis of urea to carbon dioxide and ammonia, which can neutralize gastric hydrochloric acid, thus promoting bacterial existence. Although we found an increased number of Pseudomonas, this bacterial infection affects people with weakened immune systems (including patients with cancer), and thus, it is more likely that this finding is the result of already developed pathological processes.
We did not detect significant bacterial abundance, richness, or diversity alterations at either the DNA or RNA level between tumor and tumor adjacent tissues. This result is consistent with two previous studies[54,55] but contradicts recent GC studies where significant differences between tumor-affected and nearby healthy tissues were found[27,56]. Moreover, we found the same clusters of bacterial networks in tumor and tumor adjacent tissues at both the DNA and RNA levels. These results may suggest that with the onset and development of carcinogenic processes, local changes in stomach tissues lead not only to a change in the bacterial composition but are also precise uniformity between cancer-affected and still healthy tissues (at least within a radius of 5 cm from the tumor area).
Studying GC samples at the RNA level, we managed to identify microbiome associations with clinical data. Analysis revealed two phylotypes (Phy7 and Phy33) related to Reyranella and Sediminibacterium, respectively. The relative number of those phylotypes gradually decreased from healthy to GC patients through intestinal growth type (considered as less aggressive cell growing type) to GC patients with diffuse growth type of cancer cells with worse outcome prognosis. To our knowledge, this is the first mention of these bacteria associated with the GC cell growth type. Reyranella is part of Proteobacteria and has previously been associated with the main chemokine expression, which is involved in T-cell attraction during cancerogenesis[57,58]. In another study, it was shown that there are significantly lower amounts of circulating natural killer and Treg cells in patients with diffuse/mixed-type GC compared to intestinal-type GC[59]. Taken together, these results suggest that Reyranella may be involved in the decrease in T-cell number and thus stimulation of cell growth of diffuse-type GC. Sediminibacterium was reported to be associated with GC, but there is no knowledge about the possible role of this bacteria in the pathophysiological processes[60,61]. Therefore, more detailed research on the effects of Reyranella and Sediminibacterium on GC cells is needed to be able to use these bacterial phylotypes as potential biomarkers.
H. pylori is the most common bacterial infection worldwide, as well as the main risk factor for GC[40]. It has been shown that during the transition from H. pylori-induced inflammation to the growth and development of carcinogenic cells, H. pylori is no longer detected in the affected areas in such large abundance[62]. Our results, showing that more H. pylori were found in tumor adjacent tissue than in tumor tissue, both at the DNA and RNA levels, confirm this. According to some previous reports, infection with H. pylori promotes the proliferation of non-Helicobacter bacteria from Proteobacteria, Spirochetes, and Acidobacteria and limits the spread of bacteria such as Actinobacteria, Bacteroidetes and Firmicutes[63,64]. Although most of the bacteria we found with altered numbers in GC were not associated with H. pylori, changes in the number of bacteria, such as Granulicatella, Lactobacillus, Rothia, Pseudomonas, Gemella, Prevotella, Leptotrichia, Clostridium sensu stricto, and Fusobacterium, were associated with high H. pylori abundance.
The question regarding the causality in the gastric microbiome is still partially unanswered. On the one hand, alterations in gastric microbiota have a causal role in the progression of carcinogenesis (e.g., H. pylori). On the other hand, the role of other bacteria is less understood. However, there are new studies that strongly suggest the impact of the gastric microbiome on inflammation and carcinogenesis. For instance, a recent study by Kwon et al[65] showed that intestinal metaplasia or GC patient gastric microbiome transplantation contributes to changes in the phenotype of premalignant lesions. In this regard, a detailed understanding of the output of different sequencing technologies and comparability between RNA/DNA-based analyses is critical.
Since systematic analysis to assess the differences with respect to GC has not been performed before, we would like to point out some limitations of this work. While the primary focus of the work was related to technical differences, thus food preferences, sex, and aging can be potential contributing factors that have not been thoroughly considered in this study. Overall, the focus was on providing a truly confirmed healthy cohort for the most precise comparison to strengthen the differences. Nevertheless, PERMANOVA and Mann-Whitney analyses performed in each of the study groups (tumor DNA, tumor RNA, tumor adjacent DNA, tumor adjacent RNA, control DNA, control RNA) did not reveal significant differences between the sexes and age (divided by median) (Supple
In conclusion, our study provides evidence that the tumor microbiome of GC patients has a distinct pattern compared to healthy controls, while the difference analyzed from adjacent tissue was rather low. Despite some overlap between the data obtained from the 16S rRNA transcript and 16S rRNA gene, our results showed the critical importance of the chosen study material on the resulting bacterial profile. Thus, researchers comparing their results with previous studies might take into consideration which initial material was used, either the 16S rRNA gene or 16S rRNA transcript. Our results showed that the RNA level was more sensitive for detecting low abundance bacteria and allowed us to detect differences according to GC clinical data.
There is currently no gold standard for analyzing the microbiome in 16S rRNA studies. Two common modalities are: Sequencing of DNA (16S rRNA gene) and sequencing of RNA (16S rRNA transcript). Gastric cancer (GC) remains one of the most common cancers in the world and microbiome takes important place in its carcinogenesis.
Microbiota studies are becoming more relevant and widespread. Comparison of different approaches for microbiome studying is necessary for correct interpretation of other studies results, as well as for a deeper understanding of bacterial composition.
To investigate how the choice of sequencing modality affects the bacterial profile of differences between case and controls as well as to characterize the microbiota of GC tissues using 16S rRNA gene and its transcript.
The study included healthy tissues from the control group, as well as tumor and tumor adjacent tissues from GC patients. From all biopsies RNA and DNA were extracted. 16S rRNA V1-V2 region was sequenced for all samples. For significant differences between groups permutational multivariate analysis of variance and Mann-Whitney test followed by false-discovery rate test were used.
Only a small portion of bacterial sequences overlapped on DNA and RNA levels in all groups. Differences between GC and control groups also only partially overlayed on DNA and RNA levels. RNA sequencing was more sensitive for detecting differences in bacterial richness, low abundance bacteria, and changes in the relative abundance of Reyranella and Sediminibacterium according to the type of GC. In each study group differences between DNA and RNA bacterial profiles were identified.
Chosen study material (16S rRNA transcript or 16S rRNA gene) greatly affects detectable microbiome profile as well as the differences between cases and controls.
This study provides microbiome analysis applying two different methodologies using GC gastric tissues as example and could serve as a reference for future research.
We would like to thank Justina Arstikyte, Ilka Kramer, and Susan Engerlberg for the technical assistance in this study. The authors would also like to thank Prof. Martin Zenker and Dr. Denny Schanze for their support in sequencing the samples. Additionally, we are grateful to the Team of Data Integration Center of University Medicine Magdeburg for local data-analysis solutions, supported by MIRACUM and funded by the German Federal Ministry of Education and Research (BMBF) within the “Medical Informatics Funding Scheme” (FKZ 01ZZ1801H).
Provenance and peer review: Unsolicited article; Externally peer reviewed
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Lithuania
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Gao W, China; Li J, China S-Editor: Wang JJ L-Editor: A P-Editor: Wang JJ
| 1. | Pichon M, Burucoa C. Impact of the Gastro-Intestinal Bacterial Microbiome on Helicobacter-Associated Diseases. Healthcare (Basel). 2019;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 2. | Blazewicz SJ, Barnard RL, Daly RA, Firestone MK. Evaluating rRNA as an indicator of microbial activity in environmental communities: limitations and uses. ISME J. 2013;7:2061-2068. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 591] [Cited by in RCA: 472] [Article Influence: 39.3] [Reference Citation Analysis (0)] |
| 3. | Sheh A, Fox JG. The role of the gastrointestinal microbiome in Helicobacter pylori pathogenesis. Gut Microbes. 2013;4:505-531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 156] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 4. | Stewart OA, Wu F, Chen Y. The role of gastric microbiota in gastric cancer. Gut Microbes. 2020;11:1220-1230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 125] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 5. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64542] [Article Influence: 16135.5] [Reference Citation Analysis (176)] |
| 6. | Björkholm B, Falk P, Engstrand L, Nyrén O. Helicobacter pylori: resurrection of the cancer link. J Intern Med. 2003;253:102-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 53] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 7. | Lofgren JL, Whary MT, Ge Z, Muthupalani S, Taylor NS, Mobley M, Potter A, Varro A, Eibach D, Suerbaum S, Wang TC, Fox JG. Lack of commensal flora in Helicobacter pylori-infected INS-GAS mice reduces gastritis and delays intraepithelial neoplasia. Gastroenterology. 2011;140:210-220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 320] [Cited by in RCA: 303] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 8. | Lee CW, Rickman B, Rogers AB, Ge Z, Wang TC, Fox JG. Helicobacter pylori eradication prevents progression of gastric cancer in hypergastrinemic INS-GAS mice. Cancer Res. 2008;68:3540-3548. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 107] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 9. | Lertpiriyapong K, Whary MT, Muthupalani S, Lofgren JL, Gamazon ER, Feng Y, Ge Z, Wang TC, Fox JG. Gastric colonisation with a restricted commensal microbiota replicates the promotion of neoplastic lesions by diverse intestinal microbiota in the Helicobacter pylori INS-GAS mouse model of gastric carcinogenesis. Gut. 2014;63:54-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 230] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 10. | Whary MT, Muthupalani S, Ge Z, Feng Y, Lofgren J, Shi HN, Taylor NS, Correa P, Versalovic J, Wang TC, Fox JG. Helminth co-infection in Helicobacter pylori infected INS-GAS mice attenuates gastric premalignant lesions of epithelial dysplasia and glandular atrophy and preserves colonization resistance of the stomach to lower bowel microbiota. Microbes Infect. 2014;16:345-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 11. | Wurm P, Dörner E, Kremer C, Spranger J, Maddox C, Halwachs B, Harrison U, Blanchard T, Haas R, Högenauer C, Gorkiewicz G, Fricke WF. Qualitative and Quantitative DNA- and RNA-Based Analysis of the Bacterial Stomach Microbiota in Humans, Mice, and Gerbils. mSystems. 2018;3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 12. | Schulz C, Schütte K, Koch N, Vilchez-Vargas R, Wos-Oxley ML, Oxley APA, Vital M, Malfertheiner P, Pieper DH. The active bacterial assemblages of the upper GI tract in individuals with and without Helicobacter infection. Gut. 2018;67:216-225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 146] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 13. | Vasapolli R, Schütte K, Schulz C, Vital M, Schomburg D, Pieper DH, Vilchez-Vargas R, Malfertheiner P. Analysis of Transcriptionally Active Bacteria Throughout the Gastrointestinal Tract of Healthy Individuals. Gastroenterology. 2019;157:1081-1092.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 115] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 14. | Camarinha-Silva A, Jáuregui R, Chaves-Moreno D, Oxley AP, Schaumburg F, Becker K, Wos-Oxley ML, Pieper DH. Comparing the anterior nare bacterial community of two discrete human populations using Illumina amplicon sequencing. Environ Microbiol. 2014;16:2939-2952. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 149] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 15. | Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581-583. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18515] [Cited by in RCA: 17716] [Article Influence: 1968.4] [Reference Citation Analysis (0)] |
| 16. | McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8462] [Cited by in RCA: 11651] [Article Influence: 970.9] [Reference Citation Analysis (0)] |
| 17. | Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261-5267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12842] [Cited by in RCA: 13180] [Article Influence: 732.2] [Reference Citation Analysis (0)] |
| 18. | Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1906] [Cited by in RCA: 2204] [Article Influence: 116.0] [Reference Citation Analysis (0)] |
| 19. | Bray JR, Curtis JT. An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol Monogr. 1957;326-349. [RCA] [DOI] [Full Text] [Cited by in Crossref: 6387] [Cited by in RCA: 6451] [Article Influence: 94.9] [Reference Citation Analysis (0)] |
| 20. | Hammer Ø, Harper DAT, Ryan PD. Past: Paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;. |
| 21. | Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB, Simpson GL, Sólymos P, Stevens MHH, Wagner H. vegan: Community Ecology Package. Software. 2012;. |
| 23. | Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498-2504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24663] [Cited by in RCA: 33512] [Article Influence: 1595.8] [Reference Citation Analysis (0)] |
| 24. | Revelle W. psych: Procedures for Personality and Psychological Research. Available from: https://www.scholars.northwestern.edu/en/publications/psych-procedures-for-personality-and-psychological-research. |
| 25. | Castaño-Rodríguez N, Goh KL, Fock KM, Mitchell HM, Kaakoush NO. Dysbiosis of the microbiome in gastric carcinogenesis. Sci Rep. 2017;7:15957. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 182] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 26. | Wang L, Zhou J, Xin Y, Geng C, Tian Z, Yu X, Dong Q. Bacterial overgrowth and diversification of microbiota in gastric cancer. Eur J Gastroenterol Hepatol. 2016;28:261-266. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 171] [Cited by in RCA: 158] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 27. | Liu X, Shao L, Liu X, Ji F, Mei Y, Cheng Y, Liu F, Yan C, Li L, Ling Z. Alterations of gastric mucosal microbiota across different stomach microhabitats in a cohort of 276 patients with gastric cancer. EBioMedicine. 2019;40:336-348. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 222] [Cited by in RCA: 208] [Article Influence: 34.7] [Reference Citation Analysis (0)] |
| 28. | Eun CS, Kim BK, Han DS, Kim SY, Kim KM, Choi BY, Song KS, Kim YS, Kim JF. Differences in gastric mucosal microbiota profiling in patients with chronic gastritis, intestinal metaplasia, and gastric cancer using pyrosequencing methods. Helicobacter. 2014;19:407-416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 220] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 29. | Hu YL, Pang W, Huang Y, Zhang Y, Zhang CJ. The Gastric Microbiome Is Perturbed in Advanced Gastric Adenocarcinoma Identified Through Shotgun Metagenomics. Front Cell Infect Microbiol. 2018;8:433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 89] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 30. | Gralla JD. Escherichia coli ribosomal RNA transcription: regulatory roles for ppGpp, NTPs, architectural proteins and a polymerase-binding protein. Mol Microbiol. 2005;55:973-977. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 55] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 31. | Brettar I, Christen R, Höfle MG. Analysis of bacterial core communities in the central Baltic by comparative RNA-DNA-based fingerprinting provides links to structure-function relationships. ISME J. 2012;6:195-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 34] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 32. | Egert M, Schmidt I, Höhne HM, Lachnit T, Schmitz RA, Breves R. rRNA-based profiling of bacteria in the axilla of healthy males suggests right-left asymmetry in bacterial activity. FEMS Microbiol Ecol. 2011;77:146-153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 34] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 33. | Gaidos E, Rusch A, Ilardo M. Ribosomal tag pyrosequencing of DNA and RNA from benthic coral reef microbiota: community spatial structure, rare members and nitrogen-cycling guilds. Environ Microbiol. 2011;13:1138-1152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 78] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 34. | Rabiei P, Mohabatkar H, Behbahani M. Studying the effects of several heat-inactivated bacteria on colon and breast cancer cells. Mol Biol Res Commun. 2019;8:91-98. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 35. | Suprewicz Ł, Tokajuk G, Cieśluk M, Deptuła P. Bacteria Residing at Root Canals Can Induce Cell Proliferation and Alter the Mechanical Properties of Gingival and Cancer Cells. Int J Mol Sci. 2020;21:7914. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 36. | Rad AH, Aghebati-Maleki L, Kafil HS, Abbasi A. Molecular mechanisms of postbiotics in colorectal cancer prevention and treatment. Crit Rev Food Sci Nutr. 2021;61:1787-1803. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 81] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 37. | Gregory TR. Synergy between sequence and size in large-scale genomics. Nat Rev Genet. 2005;6:699-708. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 222] [Cited by in RCA: 202] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 38. | Větrovský T, Baldrian P. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS One. 2013;8:e57923. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 665] [Cited by in RCA: 695] [Article Influence: 57.9] [Reference Citation Analysis (0)] |
| 39. | Karpinets TV, Greenwood DJ, Sams CE, Ammons JT. RNA:protein ratio of the unicellular organism as a characteristic of phosphorous and nitrogen stoichiometry and of the cellular requirement of ribosomes for protein synthesis. BMC Biol. 2006;4:30. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 79] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 40. | Rajilic-Stojanovic M, Figueiredo C, Smet A, Hansen R, Kupcinskas J, Rokkas T, Andersen L, Machado JC, Ianiro G, Gasbarrini A, Leja M, Gisbert JP, Hold GL. Systematic review: gastric microbiota in health and disease. Aliment Pharmacol Ther. 2020;51:582-602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 121] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 41. | Vinasco K, Mitchell HM, Kaakoush NO, Castaño-Rodríguez N. Microbial carcinogenesis: Lactic acid bacteria in gastric cancer. Biochim Biophys Acta Rev Cancer. 2019;1872:188309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 124] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 42. | Doherty JR, Cleveland JL. Targeting lactate metabolism for cancer therapeutics. J Clin Invest. 2013;123:3685-3692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 640] [Cited by in RCA: 895] [Article Influence: 74.6] [Reference Citation Analysis (0)] |
| 43. | Poehlein A, Schilling T, Bhaskar Sathya Narayanan U, Daniel R. First Insights into the Draft Genome of Clostridium colicanis DSM 13634, Isolated from Canine Feces. Genome Announc. 2016;4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 44. | Ambs S, Ogunfusika MO, Merriam WG, Bennett WP, Billiar TR, Harris CC. Up-regulation of inducible nitric oxide synthase expression in cancer-prone p53 knockout mice. Proc Natl Acad Sci U S A. 1998;95:8823-8828. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 121] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 45. | Li J, Billiar TR, Talanian RV, Kim YM. Nitric oxide reversibly inhibits seven members of the caspase family via S-nitrosylation. Biochem Biophys Res Commun. 1997;240:419-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 395] [Cited by in RCA: 387] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 46. | Forsythe SJ, Cole JA. Nitrite accumulation during anaerobic nitrate reduction by binary suspensions of bacteria isolated from the achlorhydric stomach. J Gen Microbiol. 1987;133:1845-1849. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 47. | Feng CW, Wang LD, Jiao LH, Liu B, Zheng S, Xie XJ. Expression of p53, inducible nitric oxide synthase and vascular endothelial growth factor in gastric precancerous and cancerous lesions: correlation with clinical features. BMC Cancer. 2002;2:8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 98] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 48. | Lopetuso LR, Scaldaferri F, Petito V, Gasbarrini A. Commensal Clostridia: leading players in the maintenance of gut homeostasis. Gut Pathog. 2013;5:23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 445] [Cited by in RCA: 552] [Article Influence: 46.0] [Reference Citation Analysis (0)] |
| 49. | Larsen B, Galask RP. Vaginal microbial flora: practical and theoretic relevance. Obstet Gynecol. 1980;55:100S-113S. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 64] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 50. | Hsieh YY, Tung SY, Pan HY, Yen CW, Xu HW, Lin YJ, Deng YF, Hsu WT, Wu CS, Li C. Increased Abundance of Clostridium and Fusobacterium in Gastric Microbiota of Patients with Gastric Cancer in Taiwan. Sci Rep. 2018;8:158. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 197] [Article Influence: 28.1] [Reference Citation Analysis (0)] |
| 51. | Parlet CP, Brown MM, Horswill AR. Commensal Staphylococci Influence Staphylococcus aureus Skin Colonization and Disease. Trends Microbiol. 2019;27:497-507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 168] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 52. | Aviles-Jimenez F, Vazquez-Jimenez F, Medrano-Guzman R, Mantilla A, Torres J. Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type of gastric cancer. Sci Rep. 2014;4:4202. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 179] [Cited by in RCA: 264] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 53. | Delgado S, Cabrera-Rubio R, Mira A, Suárez A, Mayo B. Microbiological survey of the human gastric ecosystem using culturing and pyrosequencing methods. Microb Ecol. 2013;65:763-772. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 162] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 54. | Tseng CH, Lin JT, Ho HJ, Lai ZL, Wang CB, Tang SL, Wu CY. Gastric microbiota and predicted gene functions are altered after subtotal gastrectomy in patients with gastric cancer. Sci Rep. 2016;6:20701. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 67] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 55. | Li XX, Wong GL, To KF, Wong VW, Lai LH, Chow DK, Lau JY, Sung JJ, Ding C. Bacterial microbiota profiling in gastritis without Helicobacter pylori infection or non-steroidal anti-inflammatory drug use. PLoS One. 2009;4:e7985. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 160] [Cited by in RCA: 189] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 56. | Chen XH, Wang A, Chu AN, Gong YH, Yuan Y. Mucosa-Associated Microbiota in Gastric Cancer Tissues Compared With Non-cancer Tissues. Front Microbiol. 2019;10:1261. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 103] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 57. | Cremonesi E, Governa V, Garzon JFG, Mele V, Amicarella F, Muraro MG, Trella E, Galati-Fournier V, Oertli D, Däster SR, Droeser RA, Weixler B, Bolli M, Rosso R, Nitsche U, Khanna N, Egli A, Keck S, Slotta-Huspenina J, Terracciano LM, Zajac P, Spagnoli GC, Eppenberger-Castori S, Janssen KP, Borsig L, Iezzi G. Gut microbiota modulate T cell trafficking into human colorectal cancer. Gut. 2018;67:1984-1994. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 195] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 58. | Iftekhar A, Sperlich A, Janssen KP, Sigal M. Microbiome and Diseases: Colorectal Cancer. In: Haller D. The Gut Microbiome in Health and Disease. Cham: Springer, 2018: 231-249. |
| 59. | Pernot S, Terme M, Radosevic-Robin N, Castan F, Badoual C, Marcheteau E, Penault-Llorca F, Bouche O, Bennouna J, Francois E, Ghiringhelli F, De La Fouchardiere C, Samalin E, Baptiste Bachet J, Borg C, Boige V, Voron T, Stanbury T, Tartour E, Gourgou S, Malka D, Taieb J. Infiltrating and peripheral immune cell analysis in advanced gastric cancer according to the Lauren classification and its prognostic significance. Gastric Cancer. 2020;23:73-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 84] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 60. | Ferreira RM, Pereira-Marques J, Pinto-Ribeiro I, Costa JL, Carneiro F, Machado JC, Figueiredo C. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut. 2018;67:226-236. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 341] [Cited by in RCA: 476] [Article Influence: 68.0] [Reference Citation Analysis (1)] |
| 61. | Wu ZF, Zou K, Wu GN, Jin ZJ, Xiang CJ, Xu S, Wang YH, Wu XY, Chen C, Xu Z, Li WS, Yao XQ, Zhang JF, Liu FK. A Comparison of Tumor-Associated and Non-Tumor-Associated Gastric Microbiota in Gastric Cancer Patients. Dig Dis Sci. 2021;66:1673-1682. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 31] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 62. | Ota H, Katsuyama T, Nakajima S, El-Zimaity H, Kim JG, Graham DY, Genta RM. Intestinal metaplasia with adherent Helicobacter pylori: a hybrid epithelium with both gastric and intestinal features. Hum Pathol. 1998;29:846-850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 23] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 63. | Wroblewski LE, Peek RM Jr, Wilson KT. Helicobacter pylori and gastric cancer: factors that modulate disease risk. Clin Microbiol Rev. 2010;23:713-739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 817] [Cited by in RCA: 1012] [Article Influence: 67.5] [Reference Citation Analysis (1)] |
| 64. | Dias-Jácome E, Libânio D, Borges-Canha M, Galaghar A, Pimentel-Nunes P. Gastric microbiota and carcinogenesis: the role of non-Helicobacter pylori bacteria - A systematic review. Rev Esp Enferm Dig. 2016;108:530-540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 51] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 65. | Kwon SK, Park JC, Kim KH, Yoon J, Cho Y, Lee B, Lee JJ, Jeong H, Oh Y, Kim SH, Lee SD, Hwang BR, Chung Y, Kim JF, Nam KT, Lee YC. Human gastric microbiota transplantation recapitulates premalignant lesions in germ-free mice. Gut. 2022;71:1266-1276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 54] [Article Influence: 18.0] [Reference Citation Analysis (0)] |