Published online Feb 7, 2023. doi: 10.3748/wjg.v29.i5.879
Peer-review started: September 25, 2022
First decision: October 18, 2022
Revised: November 26, 2022
Accepted: January 11, 2023
Article in press: January 11, 2023
Published online: February 7, 2023
Processing time: 134 Days and 3.6 Hours
Small intestinal vascular malformations (angiodysplasias) are common causes of small intestinal bleeding. While capsule endoscopy has become the primary diagnostic method for angiodysplasia, manual reading of the entire gastrointes
To evaluate whether artificial intelligence can assist the diagnosis and increase the detection rate of angiodysplasias in the small intestine, achieve automatic disease detection, and shorten the capsule endoscopy (CE) reading time.
A convolutional neural network semantic segmentation model with a feature fusion method, which automatically recognizes the category of vascular dysplasia under CE and draws the lesion contour, thus improving the efficiency and accuracy of identifying small intestinal vascular malformation lesions, was proposed. Resnet-50 was used as the skeleton network to design the fusion mechanism, fuse the shallow and depth features, and classify the images at the pixel level to achieve the segmentation and recognition of vascular dysplasia. The training set and test set were constructed and compared with PSPNet, Deeplab3+, and UperNet.
The test set constructed in the study achieved satisfactory results, where pixel accuracy was 99%, mean intersection over union was 0.69, negative predictive value was 98.74%, and positive predictive value was 94.27%. The model parameter was 46.38 M, the float calculation was 467.2 G, and the time length to segment and recognize a picture was 0.6 s.
Constructing a segmentation network based on deep learning to segment and recognize angiodysplasias lesions is an effective and feasible method for diagnosing angiodysplasias lesions.
Core Tip: Small intestinal vascular malformation (vascular dysplasia) is a common cause of small intestinal bleeding. Herein, we proposed a semantic recognition segmentation network to recognize small intestinal vascular malformation lesions. This method can assist doctors in identifying lesions, improving the detection rate of intestinal vascular dysplasia, realizing automatic disease detection, and shortening the capsule endoscopy reading time.
- Citation: Chu Y, Huang F, Gao M, Zou DW, Zhong J, Wu W, Wang Q, Shen XN, Gong TT, Li YY, Wang LF. Convolutional neural network-based segmentation network applied to image recognition of angiodysplasias lesion under capsule endoscopy. World J Gastroenterol 2023; 29(5): 879-889
- URL: https://www.wjgnet.com/1007-9327/full/v29/i5/879.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i5.879
Small intestinal vascular malformations (angiodysplasias) are common causes of small intestinal bleeding[1,2]. Angiodysplasias are degenerative lesions that manifest as abnormalities of arteries, veins, or capillaries of the original normal blood vessels. Occasionally, the term angiodysplasias include various synonymous disease concepts, such as angioectasia (AE), Dieulafoy’s lesion (DL), and arteriovenous malformation. According to the Yano-Yamamoto classification, small bowel vascular lesions are classified into four types under endoscopy[3]. AE includes small erythemas and can be defined as type 1a: punctuate (< 1 mm), or type 1b: patchy (a few mm). They are characterized by thin, dilated, and tortuous veins lacking smooth muscle layers, which explain their weakness and tendency to bleed. Typically, DLs consist of small mucosal defects and can be classified as type 2a: punctuate lesions with pulsatile bleeding or type 2b: pulsatile red protrusions without surrounding venous dilatation[4]. Some arteriovenous malformations and pulsatile red protrusions with dilated peripheral veins are defined as type 3. Congenital intestinal arteriovenous malformations manifest as polypoid or cluster type[5,6] and are classified as type 4. Nevertheless, the Yano-Yamamoto classification cannot fully reflect the histopathological findings.
Capsule endoscopy (CE) is a painless and well-tolerated approach that can achieve complete visualization of the small intestine[7]. It captures images for > 8 h[8]. Previous studies have demonstrated the probability of CE diagnosis of angiodysplasias was 30%-70%, and > 50% of obscure gastrointestinal bleeding patients have angiodysplasias[9-11]. The detection rate of CE was reported to be higher than other diagnostic methods, such as small bowel computed tomography, mesenteric angiography, and enteroscopy. Therefore, using CE as a first-line inspection tool for the diagnosis of angiodysplasias is recommended[12]. Nonetheless, CE has some limitations, and only 69% of angiodysplasias can be diagnosed by gastroenterologists[13]. Less relevant lesions, such as erosions or tiny red spots, are regarded as negative results; however, distinguishing highly relevant lesions from less relevant lesions could be challenging. In addition, the diagnostic efficiency of CE decreases when the presence of bile pigments, food residues, or bubbles affects the observation of the intestinal mucosa. The doctor’s manual reading of the entire gastrointestinal tract is time-consuming, and the heavy workload affects the accuracy of the diagnosis. Therefore, making diagnosis of angiodysplasias solely based on CE is challenging.
The detection rate of angiodysplasias in the small intestine can be increased by using artificial intelligence (AI) to assess the effect of automatic diagnosis, which has been successfully applied for the recognition and diagnosis of gastrointestinal endoscopic images[14]. AI assists in the recognition and diagnosis of CE images, eliminates errors in manual reading, reduces the workload of doctors, and improves diagnosis efficiency. The clinical application of AI-based deep learning technology in wireless CE has been a research focus, which has gained increasing interest in the past two years[15-32]. Several studies[15,23-26] have used deep learning to identify ulcers from CE data. Pogorelov et al[27] used the color texture features to detect small intestinal bleeding in CE data. Blanes-Vidal et al[28] constructed a classification network to identify intestinal polyp lesions in CE data. Kundu et al[29] and Hajabdollahi et al[30] identified small bowel bleeding in CE data using a classification neural network. Obscure gastrointestinal bleeding is the main indication for small intestinal CE, and the potential risk of bleeding from vascular malformations is high[14]. Therefore, we focused on AI-assisted recognition technology for angiodysplasias in the present study. Hitherto, there are few semantic segmentation networks based on deep learning to segment and recognize angiodysplasias lesions in CE, which prompted us to introduce a segmentation model in the study. Compared with the classification model and target detection model in deep learning, the segmentation model based on deep learning can more accurately locate the focus of small intestinal vascular malformation, better assist doctors in diagnosing small intestinal vascular malformation, and improve the accuracy and efficiency of doctors' diagnosis.
Currently, significant progress has been made in semantic segmentation in the field of deep learning. To the best of our knowledge, this is the first paper that proposed using a semantic segmentation network to solve the pixel-level small intestinal vascular malformation focus recognition and location. Resnet-50 was used as the skeleton network, and the fusion mechanism based on shallow features and deep features was introduced so that the segmentation model could accurately locate the location and category of lesions. Shallow features can perceive the texture details of lesions, while deep features can perceive the semantic information between lesions. By combining these two features to segment the image, the phenomenon where the lesion area is divided into uncorrelated small areas is reduced, the pixel accuracy (PA) is improved, and the missed detection rate of the lesion is reduced. This paper introduced the proposed network structure in detail and compared three common segmentation models, i.e., PSPNet[31], Deeplabv3+[32], and UperNet[33]. The obtained results confirmed that the model proposed in this paper had high-performance indicators.
ResNet was introduced in 2015 and won first place in the classification task of the ImageNet competition on account of being "simple and practical". Afterward, many methods, which were based on ResNet50 or ResNet101, have been widely used in detection, segmentation, recognition, and other fields. This method makes a reference (X) for the input of each layer, learning to form residual functions rather than learning some functions without reference (X). This residual function is easier to optimize and can greatly deepen the number of network layers. Moreover, the extracted image features have strong robustness. ResNet50 is faster than ResNet100. Therefore, ResNet50 is selected as the skeleton network of the semantic segmentation network in this paper. Based on the fusion of shallow and deep features, Resnet-50 was used as the skeleton network to construct an improved convolutional neural network (CNN) segmentation network model that automatically recognizes the type of angiodysplasias under CE and draws the outline of the lesion in the study. The present study aimed to assist doctors in diagnosing angiodysplasias lesions with CE.
The model proposed in this study was composed of three sub-units, i.e., down-sampling, up-sampling, and classifier. CE small intestine data were used as input in the module, and the final output was image lesion category information and lesion boundary information.
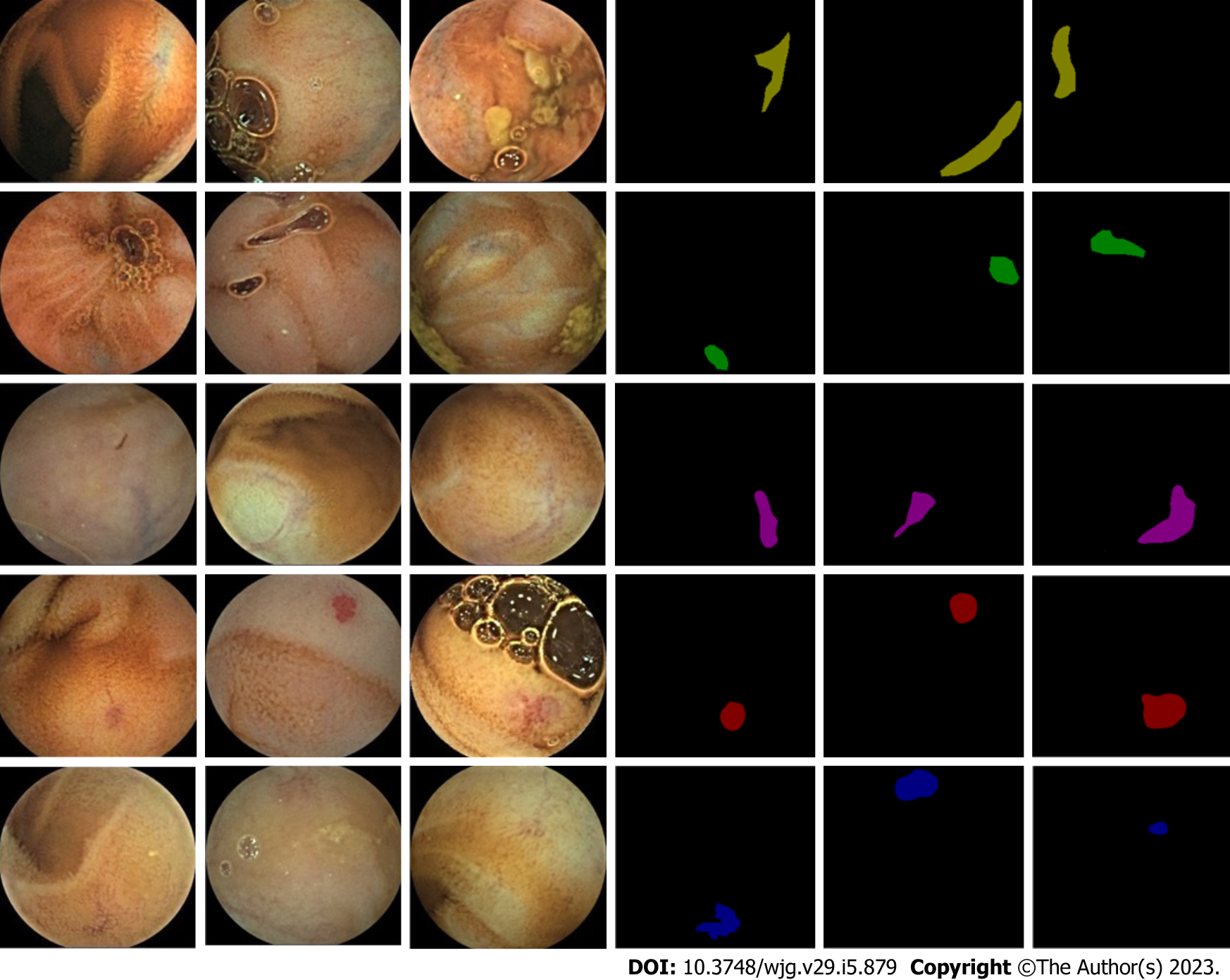
In order to train and evaluate the segmentation model, 378 patients with angiodysplasias who underwent OMOM CE (China Chongqing Kingsoft Technology Co., Ltd) at the Ruijin Hospital between January 2014 and December 2020 were recruited in this study. The sampling frequency of OMOM capsules of 2fps, the working time of > 12 h, and the apex field of view of 150° were used to diagnose the patients. A total of 12403 pictures were identified with an image resolution of 256 × 240. The patient data were anonymized, any personal identification information was omitted, and examination information (such as examination date and patient name) was deleted from the original image. All patients provided written informed consent, and the ethics committee approved the study [the certification number was (2017) provisional ethics review No. 138]. The annotated data were marked by an experienced endoscopy group that included three experts from Ruijin Hospital Affiliated to Shanghai Jiao Tong University. The average age of the experts was 35 years, and their average CE reading experience was 5 years, with an average of 150 CE cases each year. The five types of lesions of vascular malformation were annotated, and 12403 image data and 12403 annotated mask image data were generated. The data sample map is shown in Figure 1.
This project used the image data of 178 cases as the training set and the remaining 200 cases as the test set. The training set was divided into training and verification data at a ratio of 7:3 during the training process. The test set contained 1500 images without lesions and 1500 images with lesions. The training set and test set image data are summarized in Table 1.
| Lesion type | Lesion morphology | Number of pictures/pieces | |
| Training set | Test set | ||
| Telangiectasia | Red cluster | 838 | 38 |
| Red spider nevus | 162 | 4 | |
| Venous dilatation | Red branched | 752 | 38 |
| Blue branched | 2583 | 1088 | |
| Vein tumor | Blue cluster | 3058 | 332 |
The training data were preprocessed to meet the requirements of the deep learning model. The preprocessing steps of the model constructed in the study were as follows: (1) Resizing the image to 256 × 240 × 3; (2) using enhancement methods (rotation, flip, and tilt) on the resized image; and (3) normalizing all images. In order to train a deep learning model, the dataset was split. The dataset image was randomly divided into two parts: 70% for training and 30% for verification.
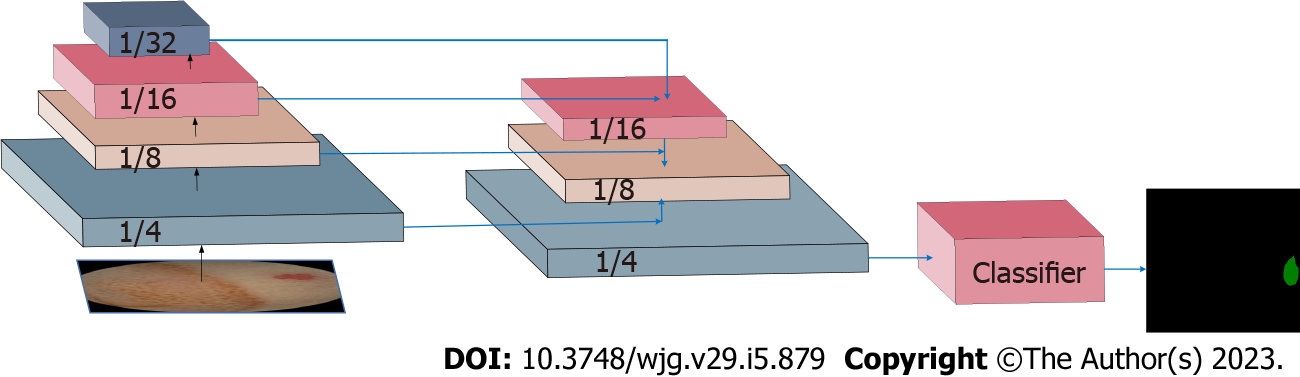
The network structure proposed in this study is shown in Figure 2. The construction of the network model was inspired by the UperNet model. ResNet-50 was used as the skeleton network. The fusion mechanism of shallow features and deep features were introduced. Subsequently, the feature with the same size as the original image was obtained through the down-sampling operation. Finally, the classifier was connected to realize the pixel-level segmentation task of the image.
Based on the new semantic segmentation recognition network framework, a single end-to-end network could be trained to capture and analyze the semantic information of the CE small intestine data. In order to fuse the shallow features and deep feature information, the last feature mapping set output by each stage in ResNet was expressed as C1, C2, C3, and C4, and the two-by-two fusion of features were utilized as down-sampling operation input, where the down-sampling rates were 4, 8, 16, and 32, respectively. The texture features of the lesion were captured at the highest layer, and the pixel-level segmentation of the lesion was completed based on the lowest layer features.
The last down-sampling operation generated a feature map with the same resolution as the original image, with a size of 256 × 240. After the feature was operated by Flatten, a classifier composed of a fully connected layer was connected to complete the segmentation and recognition tasks of the capsule data.
In order to assess the fusion of features of different scales, bilinear interpolation was used to adjust them according to the size, after which a non-evolutionary layer was applied to fuse the features of different levels and reduce the channel size. All non-classifier convolutional layers underwent batch normalization Relu operations after output. The learning rate of the current iteration was equivalent to the initial learning rate multiplied by (1-iter/max-iter_size)power, and the initial learning rate and power were set to 0.02 and 0.9, respectively.
The performance of the segmentation model of angiodysplasias lesions in CE was evaluated based on the following indicators: Positive predictive value (PPV), negative predictive value (NPV), mean intersection over union (mIOU), and PA. PPV and NPV were calculated using formulae 1 and 2, respectively.
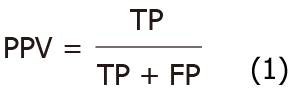
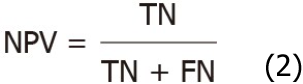
Where true positive (TP) and true negative (TN) are the true number of positive samples and the true number of negative samples, respectively; FP and FN are false positives and false negatives, respectively. IOU and mIOU calculation formulae are shown as formulae 3 and 4, respectively.

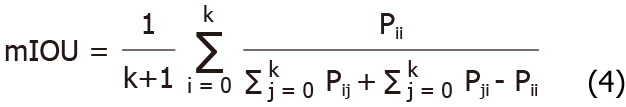
Supposedly, there were K+1 categories (including an empty category or background) in semantic segmentation, which indicated that class i is predicted as i, and class j is predicted as j. The PA is calculated by formula 5.
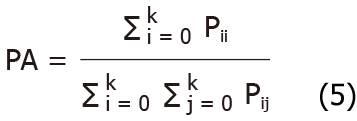
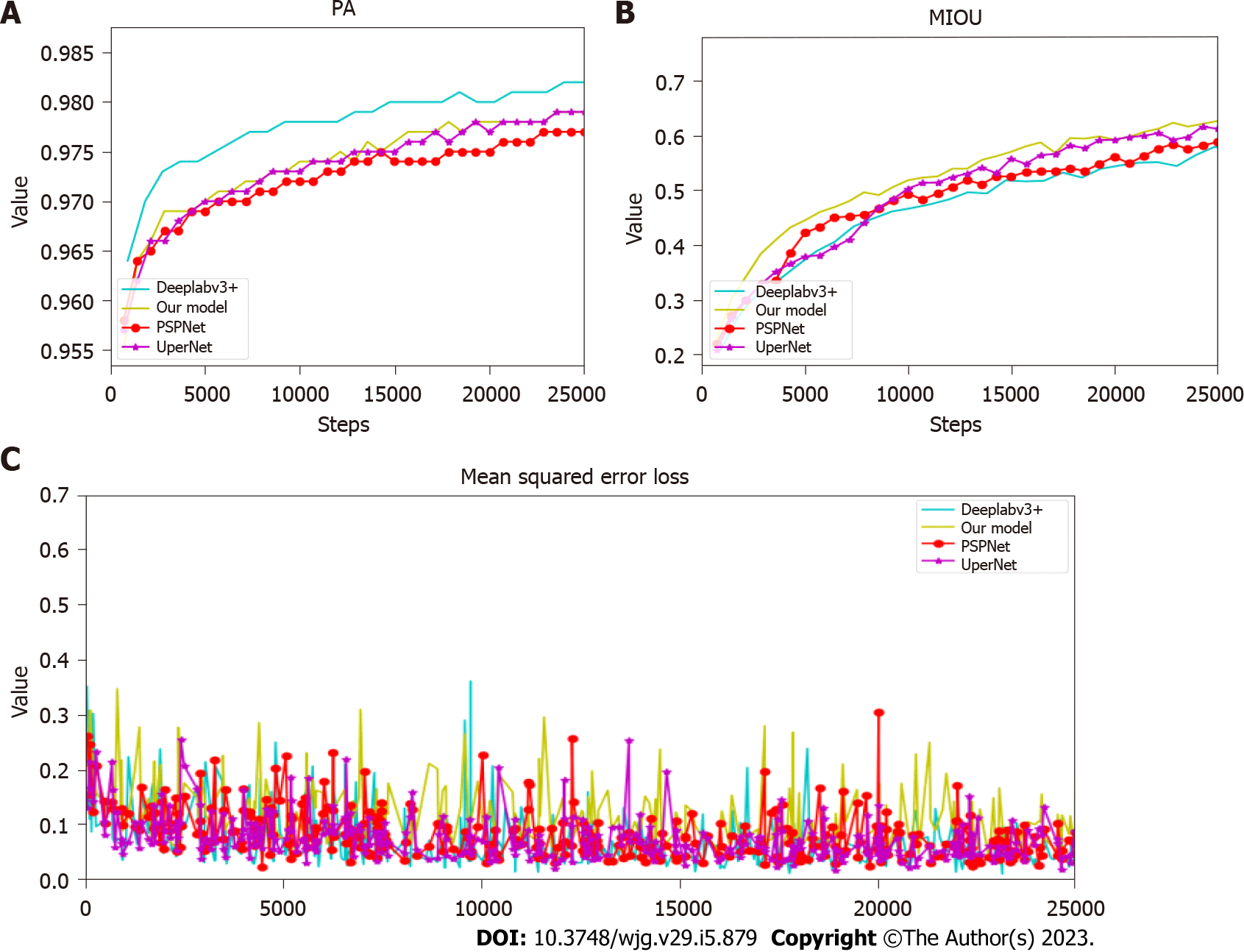
Python 3 is a good deep-learning programming language that supports multiple deep-learning frameworks. The model was implemented using Python 3 and Torch framework. The training server has a graphics processing unit. All images were first passed to the image data generation class in Pytorch, and the preprocessing operations were performed, including enhancement, resize, and normalization operations. Then, the generated images were sent to the model to start the training. The layers in the backbone network ResNet-50 used pre-trained weights on ImageNet. An optimizer (SGD) was used to train the model, after which a weight decay of 0.0001 and a momentum of 0.9 were applied. Each model ran approximately 25000 Epochs; each Epoch iterated eight times, and the batch size was 8. In the model training process, the loss change, PA index, and mIOU changes were detected (Figure 3).
On a test set consisting of 3000 image data, the following test indicators were compared on the four models: PPV, NPV, mean IOU, PA, parameter quantity, float calculation quantity, and duration. The results are shown in Table 2.
| Network type | PPV (%) | NPV (%) | mIOU | PA (%) | Parameter (M) | Float calculation amount (G) | Time (s) |
| PSPNet | 85.14 | 98.62 | 0.64 | 98 | 51.43 | 829.10 | 0.9 |
| DeeplabV3+ | 45.07 | 99.75 | 0.59 | 89 | 59.34 | 397.00 | 0.95 |
| UperNet | 92.55 | 95.69 | 0.69 | 98 | 126.08 | 34.94 | 0.9 |
| Our model | 94.27 | 98.74 | 0.69 | 99 | 46.38 | 467.2 | 0.6 |
Based on the method of fusion of shallow and deep features, the CNN segmentation network model was improved and optimized, and the segmentation and recognition of five types of angiodysplasias lesions, i.e., blue branch, blue cluster, red branch, red cluster, and red spider nevus, were realized. This method fully uses the shallow and deep features extracted from the skeleton network to perceive the global information and lesion texture information of the small intestine capsule image data as a whole. Thus, it significantly improves the PPV and NPV of the segmentation model in the angiodysplasias lesion image. In order to obtain the highest PPV, the NPV has to be the highest. The unified perception of the global and local information of the small intestine capsule data was completed through a CNN, which reduced the number of network model parameters, the number of float calculations, and the inference time of the deep learning model. Furthermore, a comparative experiment was designed and compared to the current advanced segmentation network models: PSPNet, DeeplabV3+, and UperNet. Our model showed that the NPV reached the highest 98.74% when the PPV was the highest.
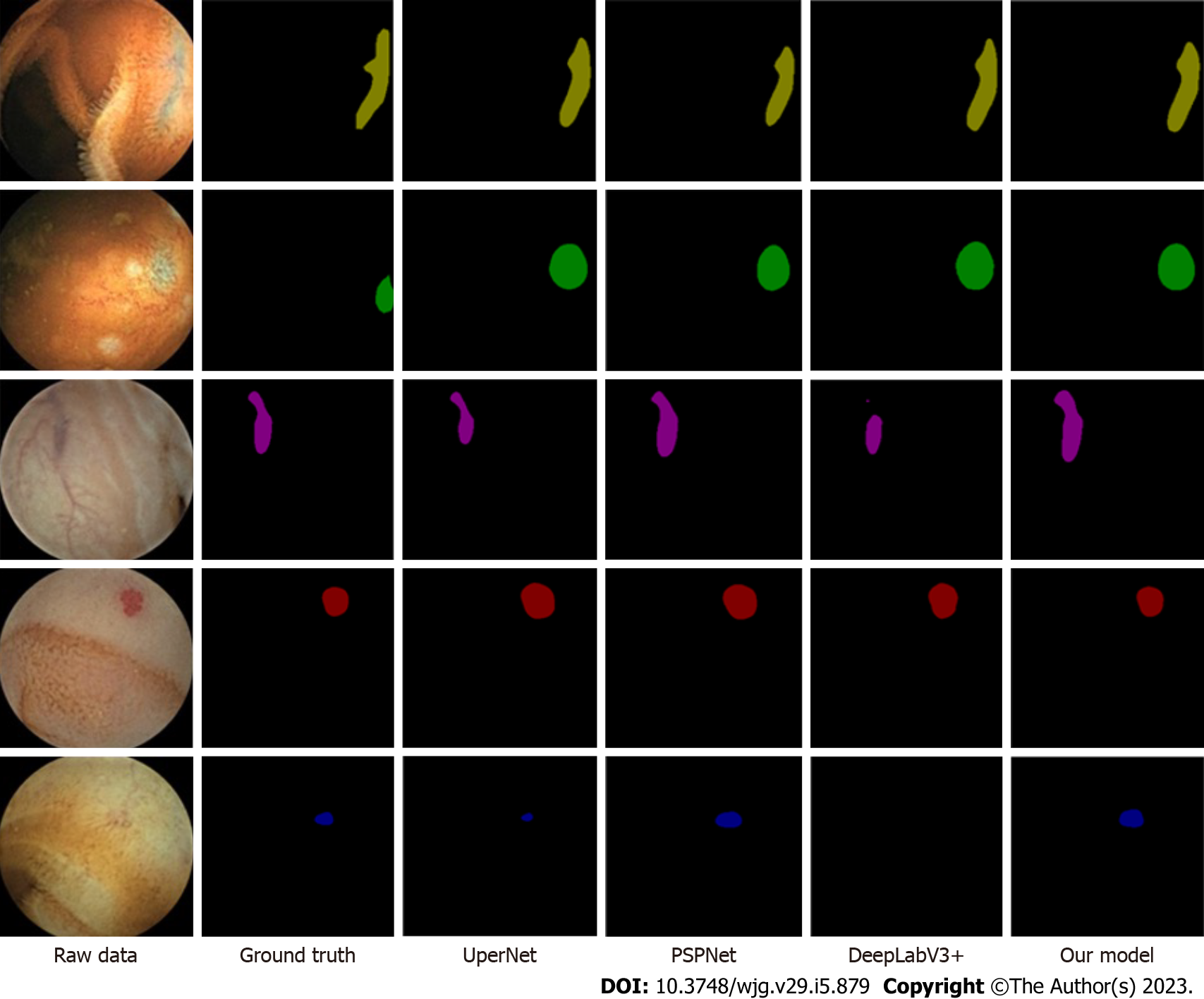
The comparison of the segmentation and recognition effects of the four models on the vascular aberration lesions of the CE small intestine data is shown in Figure 4. The model proposed in the study was similar to that of the expert’s annotation results.
Compared with relevant literature, Leenhardt et al[19] applied technology for segmentation, achieving the highest level of lesion detection, with an NPV value of 96%. However, the algorithm presented in this paper had some advantages in the test set. Also, our NPV value was 98%.
The classification network and the target detection network are the mainstream network structure that combines the deep learning model and the CE diagnosis method. In the present study, we introduced the segmentation network in deep learning, segmented and identified the angiodysplasias lesions, and completed the pixel-level segmentation task of the angiodysplasias lesions. The semantic segmentation network model had clinical practicality application as assessed using the training and test sets in comparative experiments.
The segmentation networks have been obviously developed in the field of deep learning. PSPNet uses the prior knowledge of the global feature layer to understand the semantics of various scenes, combined with the deep supervision loss to develop an effective optimization strategy on ResNet and embed difficult-to-analyze scene information features into the functional connectivity networks prediction framework to establish a pyramid. The pooling module aggregates the contextual information in different regions and improves the ability to obtain global information. This system was used for scene analysis and semantic segmentation and was 83% accurate on the COCO data set. The DeepLabV3+ model was based on an encoder-decoder structure, which improved the accuracy and saved the inference time; an accuracy rate of 89% was obtained in the COCO dataset. UperNet used unified perception analysis to build a network with a hierarchical structure to ensure that multiple levels were resolved at visual concepts, learn the differentiated data in various image datasets, achieve joint reasoning, and explore the rich visual knowledge in the images. Finally, 79.98%-PA was obtained on the ADE20K data set. UperNet used a unified perception analysis module from scenes, objects, parts, materials, and textures to simultaneously analyze the multilevel visual concepts of images, such that many objects could be segmented and recognized, and the rate of missed objects could be reduced. The CE small intestine image data has a simple scene and fewer semantic levels. The use of large segmentation network models would cause over-fitting in training and high computational complexity. This study was inspired by UperNet and optimized basic CNN segmentation network, which led to the creation of a network model suitable for the segmentation and recognition of angiodysplasias with CE.
On the other hand, a case-based dataset encompassing typical vascular malformation images, atypical angiodysplasias images, and normal images was constructed, including pictures with poor intestinal cleanliness. According to the color and morphology of the angiodysplasias lesions in the cases, the five types of angiodysplasias lesions were summarized as blue branched, blue cluster, red branched, red cluster, and red spider nevus. The dataset constructed in this study verified the clinical applicability of the semantic segmentation model. Thus, the dataset was essential in diagnosing CE small bowel vascular malformation based on the deep learning model.
The deep learning model constructed in this study showed high PPV and NPV for the segmentation and recognition of angiodysplasias lesions. In the future, it could be used to assist capsule endoscopists in the real-time diagnosis of angiodysplasias lesions. Deep learning does not require prior knowledge, as it can directly learn the most predictive features from image data, as well as segment and recognize the image. The larger the amount of data, the higher the advantages of deep learning and the higher the recognition accuracy. AI facilitates grassroots’ CE to obtain the same diagnosis effect as senior experts. However, the current uneven distribution of medical resources and the technical level of grassroots CE are the driving forces for the development of AI. In conclusion, the segmentation model based on deep learning can assist doctors in identifying the lesions of small intestinal vascular malformations.
Small intestinal vascular malformations (angiodysplasias) commonly cause small intestinal bleeding. Therefore, capsule endoscopy has become the primary diagnostic method for angiodysplasias. Nevertheless, manual reading of the entire gastrointestinal tract is a time-consuming heavy workload, which affects the accuracy of diagnosis.
The doctor’s manual reading of the entire gastrointestinal tract is time-consuming, and the heavy workload affects the accuracy of the diagnosis. Also, significant progress has been made in semantic segmentation in the field of deep learning.
This study aimed to assist in the diagnosis and increase the detection rate of angiodysplasias in the small intestine, achieve automatic disease detection, and shorten the capsule endoscopy (CE) reading time.
A convolutional neural network semantic segmentation model with feature fusion automatically recognizes the category of vascular dysplasia under CE and draws the lesion contour, thus improving the efficiency and accuracy of identifying small intestinal vascular malformation lesions, was proposed.
The test set constructed in the study achieved satisfactory results: pixel accuracy was 99%, mean intersection over union was 0.69, negative predictive value was 98.74%, and positive predictive value was 94.27%. The model parameter was 46.38 M, the float calculation was 467.2 G, and the time needed to segment and recognize a picture was 0.6 s.
Constructing a segmentation network based on deep learning to segment and recognize angiodysplasias lesions is an effective and feasible method for diagnosing angiodysplasias lesions.
The model detects the small intestinal malformation lesions in the capsule endoscopy image data and draws the lesion area through segmentation.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Garcia-Pola M, Spain; Morya AK, India S-Editor: Zhang H L-Editor: A P-Editor: Zhang H
| 1. | Leighton JA, Triester SL, Sharma VK. Capsule endoscopy: a meta-analysis for use with obscure gastrointestinal bleeding and Crohn's disease. Gastrointest Endosc Clin N Am. 2006;16:229-250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 75] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 2. | Sakai E, Endo H, Taniguchi L, Hata Y, Ezuka A, Nagase H, Yamada E, Ohkubo H, Higurashi T, Sekino Y, Koide T, Iida H, Hosono K, Nonaka T, Takahashi H, Inamori M, Maeda S, Nakajima A. Factors predicting the presence of small bowel lesions in patients with obscure gastrointestinal bleeding. Dig Endosc. 2013;25:412-420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 3. | Yano T, Yamamoto H, Sunada K, Miyata T, Iwamoto M, Hayashi Y, Arashiro M, Sugano K. Endoscopic classification of vascular lesions of the small intestine (with videos). Gastrointest Endosc. 2008;67:169-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 130] [Article Influence: 7.6] [Reference Citation Analysis (1)] |
| 4. | Nguyen DC, Jackson CS. The Dieulafoy's Lesion: An Update on Evaluation, Diagnosis, and Management. J Clin Gastroenterol. 2015;49:541-549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 5. | Chung CS, Chen KC, Chou YH, Chen KH. Emergent single-balloon enteroscopy for overt bleeding of small intestinal vascular malformation. World J Gastroenterol. 2018;24:157-160. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 8] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 6. | Molina AL, Jester T, Nogueira J, CaJacob N. Small intestine polypoid arteriovenous malformation: a stepwise approach to diagnosis in a paediatric case. BMJ Case Rep. 2018;2018:bcr2018224536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 7. | Iddan G, Meron G, Glukhovsky A, Swain P. Wireless capsule endoscopy. Nature. 2000;405:417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1994] [Cited by in RCA: 1384] [Article Influence: 55.4] [Reference Citation Analysis (1)] |
| 8. | Rondonotti E, Spada C, Adler S, May A, Despott EJ, Koulaouzidis A, Panter S, Domagk D, Fernandez-Urien I, Rahmi G, Riccioni ME, van Hooft JE, Hassan C, Pennazio M. Small-bowel capsule endoscopy and device-assisted enteroscopy for diagnosis and treatment of small-bowel disorders: European Society of Gastrointestinal Endoscopy (ESGE) Technical Review. Endoscopy. 2018;50:423-446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 333] [Cited by in RCA: 288] [Article Influence: 41.1] [Reference Citation Analysis (0)] |
| 9. | Liao Z, Gao R, Xu C, Li ZS. Indications and detection, completion, and retention rates of small-bowel capsule endoscopy: a systematic review. Gastrointest Endosc. 2010;71:280-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 561] [Cited by in RCA: 476] [Article Influence: 31.7] [Reference Citation Analysis (0)] |
| 10. | Lecleire S, Iwanicki-Caron I, Di-Fiore A, Elie C, Alhameedi R, Ramirez S, Hervé S, Ben-Soussan E, Ducrotté P, Antonietti M. Yield and impact of emergency capsule enteroscopy in severe obscure-overt gastrointestinal bleeding. Endoscopy. 2012;44:337-342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 74] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 11. | ASGE Technology Committee; Wang A, Banerjee S, Barth BA, Bhat YM, Chauhan S, Gottlieb KT, Konda V, Maple JT, Murad F, Pfau PR, Pleskow DK, Siddiqui UD, Tokar JL, Rodriguez SA. Wireless capsule endoscopy. Gastrointest Endosc. 2013;78:805-815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 194] [Article Influence: 16.2] [Reference Citation Analysis (2)] |
| 12. | Pennazio M, Spada C, Eliakim R, Keuchel M, May A, Mulder CJ, Rondonotti E, Adler SN, Albert J, Baltes P, Barbaro F, Cellier C, Charton JP, Delvaux M, Despott EJ, Domagk D, Klein A, McAlindon M, Rosa B, Rowse G, Sanders DS, Saurin JC, Sidhu R, Dumonceau JM, Hassan C, Gralnek IM. Small-bowel capsule endoscopy and device-assisted enteroscopy for diagnosis and treatment of small-bowel disorders: European Society of Gastrointestinal Endoscopy (ESGE) Clinical Guideline. Endoscopy. 2015;47:352-376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 482] [Cited by in RCA: 560] [Article Influence: 56.0] [Reference Citation Analysis (1)] |
| 13. | Zheng Y, Hawkins L, Wolff J, Goloubeva O, Goldberg E. Detection of lesions during capsule endoscopy: physician performance is disappointing. Am J Gastroenterol. 2012;107:554-560. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 61] [Article Influence: 4.7] [Reference Citation Analysis (1)] |
| 14. | Leenhardt R, Vasseur P, Li C, Saurin JC, Rahmi G, Cholet F, Becq A, Marteau P, Histace A, Dray X; CAD-CAP Database Working Group. A neural network algorithm for detection of GI angiectasia during small-bowel capsule endoscopy. Gastrointest Endosc. 2019;89:189-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 148] [Article Influence: 24.7] [Reference Citation Analysis (1)] |
| 15. | Wang S, Xing Y, Zhang L, Gao H, Zhang H. Deep Convolutional Neural Network for Ulcer Recognition in Wireless Capsule Endoscopy: Experimental Feasibility and Optimization. Comput Math Methods Med. 2019;2019:7546215. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 16. | Korman LY, Delvaux M, Gay G, Hagenmuller F, Keuchel M, Friedman S, Weinstein M, Shetzline M, Cave D, de Franchis R. Capsule endoscopy structured terminology (CEST): proposal of a standardized and structured terminology for reporting capsule endoscopy procedures. Endoscopy. 2005;37:951-959. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 99] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 17. | Gulati S, Emmanuel A, Patel M, Williams S, Haji A, Hayee B, Neumann H. Artificial intelligence in luminal endoscopy. Ther Adv Gastrointest Endosc. 2020;13:2631774520935220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 18. | Molder A, Balaban DV, Jinga M, Molder CC. Current Evidence on Computer-Aided Diagnosis of Celiac Disease: Systematic Review. Front Pharmacol. 2020;11:341. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 19. | Leenhardt R, Li C, Le Mouel JP, Rahmi G, Saurin JC, Cholet F, Boureille A, Amiot X, Delvaux M, Duburque C, Leandri C, Gérard R, Lecleire S, Mesli F, Nion-Larmurier I, Romain O, Sacher-Huvelin S, Simon-Shane C, Vanbiervliet G, Marteau P, Histace A, Dray X. CAD-CAP: a 25,000-image database serving the development of artificial intelligence for capsule endoscopy. Endosc Int Open. 2020;8:E415-E420. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 20. | Bianchi F, Masaracchia A, Shojaei Barjuei E, Menciassi A, Arezzo A, Koulaouzidis A, Stoyanov D, Dario P, Ciuti G. Localization strategies for robotic endoscopic capsules: a review. Expert Rev Med Devices. 2019;16:381-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 21. | Min JK, Kwak MS, Cha JM. Overview of Deep Learning in Gastrointestinal Endoscopy. Gut Liver. 2019;13:388-393. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 116] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 22. | Hwang Y, Park J, Lim YJ, Chun HJ. Application of Artificial Intelligence in Capsule Endoscopy: Where Are We Now? Clin Endosc. 2018;51:547-551. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 23. | Alaskar H, Hussain A, Al-Aseem N, Liatsis P, Al-Jumeily D. Application of Convolutional Neural Networks for Automated Ulcer Detection in Wireless Capsule Endoscopy Images. Sensors (Basel). 2019;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 113] [Cited by in RCA: 81] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 24. | Wang S, Xing Y, Zhang L, Gao H, Zhang H. A systematic evaluation and optimization of automatic detection of ulcers in wireless capsule endoscopy on a large dataset using deep convolutional neural networks. Phys Med Biol. 2019;64:235014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 25. | Fan S, Xu L, Fan Y, Wei K, Li L. Computer-aided detection of small intestinal ulcer and erosion in wireless capsule endoscopy images. Phys Med Biol. 2018;63:165001. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 90] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 26. | Aoki T, Yamada A, Aoyama K, Saito H, Tsuboi A, Nakada A, Niikura R, Fujishiro M, Oka S, Ishihara S, Matsuda T, Tanaka S, Koike K, Tada T. Automatic detection of erosions and ulcerations in wireless capsule endoscopy images based on a deep convolutional neural network. Gastrointest Endosc. 2019;89:357-363.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 175] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 27. | Pogorelov K, Suman S, Azmadi Hussin F, Saeed Malik A, Ostroukhova O, Riegler M, Halvorsen P, Hooi Ho S, Goh KL. Bleeding detection in wireless capsule endoscopy videos - Color versus texture features. J Appl Clin Med Phys. 2019;20:141-154. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 28. | Blanes-Vidal V, Baatrup G, Nadimi ES. Addressing priority challenges in the detection and assessment of colorectal polyps from capsule endoscopy and colonoscopy in colorectal cancer screening using machine learning. Acta Oncol. 2019;58:S29-S36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 54] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 29. | Kundu AK, Fattah SA, Rizve MN. An Automatic Bleeding Frame and Region Detection Scheme for Wireless Capsule Endoscopy Videos Based on Interplane Intensity Variation Profile in Normalized RGB Color Space. J Healthc Eng. 2018;2018:9423062. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 30. | Hajabdollahi M, Esfandiarpoor R, Najarian K, Karimi N, Samavi S, Reza Soroushmehr SM. Low Complexity CNN Structure for Automatic Bleeding Zone Detection in Wireless Capsule Endoscopy Imaging. Annu Int Conf IEEE Eng Med Biol Soc. 2019;2019:7227-7230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 31. | Zhao H, Shi J, Qi X, Wang X, Jia J. Pyramid Scene Parsing Network. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). Honolulu, HI, USA, 2017: 6230-6239. [DOI] [Full Text] |
| 32. | Chen LC, Zhu Y, Papandreou G, Schroff F, Adam H. Encoder-Decoder with Atrous Separable Convolution for Semantic Image Segmentation. In: Ferrari V, Hebert M, Sminchisescu C, Weiss Y. Computer Vision – ECCV 2018. 2018: 833-851, Springer, Cham. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2522] [Cited by in RCA: 1600] [Article Influence: 228.6] [Reference Citation Analysis (0)] |
| 33. | Xiao T, Liu Y, Zhou B, Jiang Y, Sun J. Unified Perceptual Parsing for Scene Understanding. In: Ferrari V, Hebert M, Sminchisescu C, Weiss Y. Computer Vision – ECCV 2018. 2018: 432–448, Springer, Cham. [RCA] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 152] [Article Influence: 21.7] [Reference Citation Analysis (0)] |