Published online Dec 28, 2023. doi: 10.3748/wjg.v29.i48.6198
Peer-review started: September 3, 2023
First decision: November 1, 2023
Revised: November 13, 2023
Accepted: December 12, 2023
Article in press: December 12, 2023
Published online: December 28, 2023
Processing time: 114 Days and 12.8 Hours
Barrett’s esophagus (BE), which has increased in prevalence worldwide, is a precursor for esophageal adenocarcinoma. Although there is a gap in the detection rates between endoscopic BE and histological BE in current research, we trained our artificial intelligence (AI) system with images of endoscopic BE and tested the system with images of histological BE.
To assess whether an AI system can aid in the detection of BE in our setting.
Endoscopic narrow-band imaging (NBI) was collected from Chung Shan Medical University Hospital and Changhua Christian Hospital, resulting in 724 cases, with 86 patients having pathological results. Three senior endoscopists, who were instructing physicians of the Digestive Endoscopy Society of Taiwan, independently annotated the images in the development set to determine whether each image was classified as an endoscopic BE. The test set consisted of 160 endoscopic images of 86 cases with histological results.
Six pre-trained models were compared, and EfficientNetV2B2 (accuracy [ACC]: 0.8) was selected as the backbone architecture for further evaluation due to better ACC results. In the final test, the AI system correctly identified 66 of 70 cases of BE and 85 of 90 cases without BE, resulting in an ACC of 94.37%.
Our AI system, which was trained by NBI of endoscopic BE, can adequately predict endoscopic images of histological BE. The ACC, sensitivity, and specificity are 94.37%, 94.29%, and 94.44%, respectively.
Core Tip: The prevalence of Barrett’s esophagus (BE) diagnosed by endoscopy significantly differs from BE diagnosed by histology (7.8% vs 1.3%). Current research showed that image-enhanced endoscopy can only increase the detection ability for dysplasia lesions in BE. Our artificial intelligence prediction system, which was trained by endoscopic BE images with the Olympus narrow-band imaging system, still provided good prediction results for images of histological BE. The accuracy, sensitivity, and specificity are 94.37%, 94.29%, and 94.44%, respectively, in the final test, which indicates that endoscopic BE images have characteristics similar to images of histological BE.
- Citation: Tsai MC, Yen HH, Tsai HY, Huang YK, Luo YS, Kornelius E, Sung WW, Lin CC, Tseng MH, Wang CC. Artificial intelligence system for the detection of Barrett’s esophagus. World J Gastroenterol 2023; 29(48): 6198-6207
- URL: https://www.wjgnet.com/1007-9327/full/v29/i48/6198.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i48.6198
Barrett’s esophagus (BE), which is characterized by a columnar lined esophagus with documented intestinal metaplasia[1,2], is a precursor for esophageal adenocarcinoma (EAC)[3,4]. The prevalence of BE has been increasing in western countries[5,6] for decades, and this trend has also recently been observed in Asian countries[7]. The annual incidence of EAC is approximately 0.3% in non-dysplasia BE[8], while the annual incidence of EAC increases to 0.76% in low-grade dysplasia BE[9] and to 6% in high-grade dysplasia BE[10]. Although the importance of the detection of BE in patients is recognized by endoscopists worldwide, the prevalence of BE diagnosed by endoscopy significantly differs from BE diagnosed by histology[7] (7.8% vs 1.3%).
The methods of biopsy in Seattle protocol[11] have been under debate, as random four-quadrant biopsies at 2-cm intervals are time-consuming and intolerable under some situations. A recent study showed that targeted biopsy using image-enhanced endoscopy and molecular biomarkers can increase the detection ability for dysplasia lesions in BE[12]. The above issues indicate that there is a large degree of interobserver disagreement in the recognition and biopsy methods of BE. One previous study showed that the prevalence of histological BE is 2.6% in a health examination center data[13] but that the utilization of biopsy during endoscopy is far less in daily practice in Taiwan. We propose that an artificial intelligence (AI) system should be helpful for promoting awareness of BE in daily endoscopic practice.
AI systems are already applied in many fields of modern medicine, which started with diabetic retina observation[14,15], diabetes mellitus care[16], and the detection and classification of Alzheimer's disease[17] and later extended to the fields of gastroenterology and endoscopy. AI systems can help detect and differentiate the classification of colon polyps[18,19], gastric cancer[20], neoplastic and non-neoplastic small bowel lesions[21], pancreatic lesions[22,23], BE[24-26], and EAC[24,26,27]. Despite recent improvements in the AI prediction of BE and EAC, the accuracy (ACC) of such models has remained approximately 90%; one of the insurmountable problems is that there are insufficient images of histologically proven BE to train AI models. Our healthcare system faced the same dilemma, and we tried to conquer this problem by establishing a relationship between images of endoscopic BE and images of histological BE.
We trained our AI model with images of endoscopic BE and endoscopic images from patients without BE and then tested our AI prediction system using cases both with and without histologically proven BE. We also compared the outcomes of our prediction system with data from similar recent studies.
The endoscopic images of patients with typical gastroesophageal reflux symptoms were collected from Chung Shan Medical University Hospital (Taichung City, Taiwan) and Changhua Christian Hospital (Changhua County, Taiwan) retrospectively from January 1, 2020 to June 30, 2020, resulting in 724 cases, with 86 patients having complete histological results. The collection of clinical data was de-identification and sent for further image annotation. This study reviewed and approved by the institutional review board (IRB) on May 28, 2020 with IRB number CS1–20075 (the application of artificial intelligence in advanced endocopy) and conducted under IRB regulations to ensure the rights and welfare of the participants.
The images utilized in this study were obtained from the hospitals’ endoscopy systems, which were operated by professional physicians. The narrow-band imaging (NBI) mode in the Olympus endoscopy system was selected for capturing images. We collected both histological BE images and other image data from patients with typical symptoms of gastroesophageal reflux disease (GERD) to establish a study dataset.
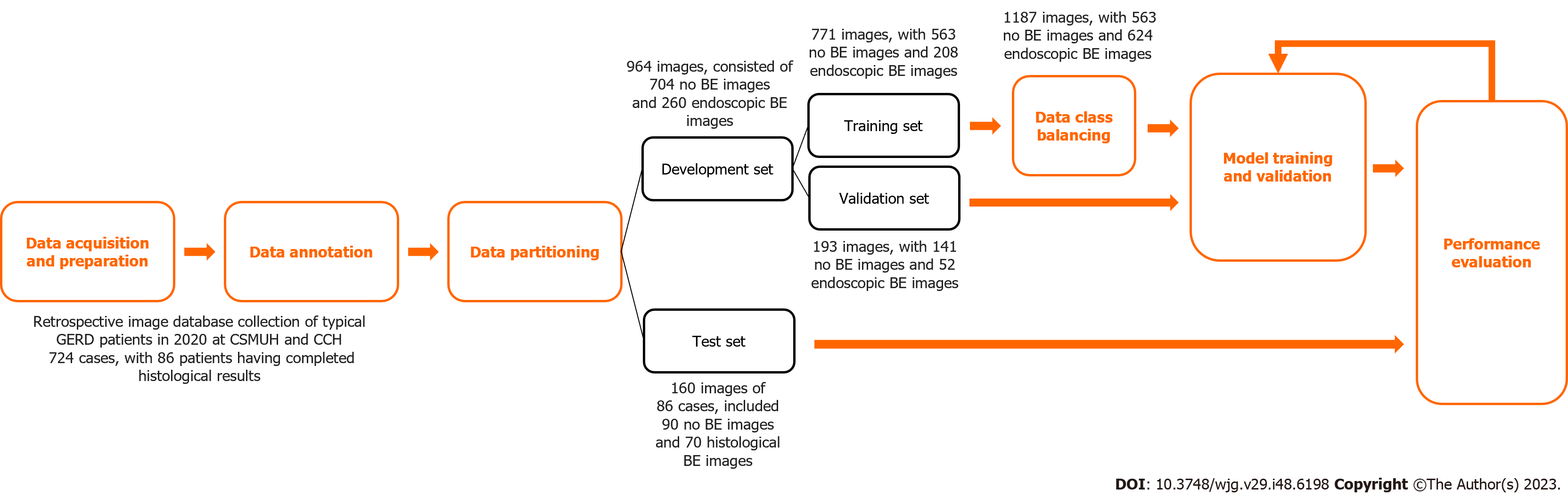
The development set of this study consisted of 964 images. Three senior endoscopists who were instructing physicians of the Digestive Endoscopy Society of Taiwan independently annotated the images in the development set to determine whether each image was classified as endoscopic BE. After a majority vote among the three senior endoscopists, the final classification was determined. The test set consisted of 160 images of 86 cases, and the categorization was based on the histological results.
The development set consisted of 704 images without BE and 260 endoscopic BE images. To avoid overfitting, the image dataset was divided using the holdout method, with 80% of the data employed as the training set and 20% of the data employed as the validation set for model training and development. The test set, which consists of complete histological result images, was used to evaluate the predictive performance of the model. The test set included 90 images without BE and 70 histological BE images. These details are shown in Table 1.
| NBI image number | Training set, n = 771 | Label balance, n = 1187 | Valid set, n = 193 | Test set, n = 160 | ||||
| n | % | n | % | n | % | n | % | |
| No Barrett’s esophagus | 563 | 73.02 | 563 | 47.43 | 141 | 73.06 | 90 | 56.25 |
| Barrett’s esophagus | 208 | 26.98 | 624 | 52.57 | 52 | 26.94 | 70 | 43.75 |
This section describes the deep learning model architecture utilized in the study to accomplish the binary classification task for BE. First, the image size was resized to 224 pixel × 224 pixel for inputting into the model. To address the issue of class imbalance in the dataset, this study employed a resampling technique to augment the data from the minority class. Data class balancing was performed only on the training set. The original training dataset consisted of 771 images, of which 563 images did not have BE, and 208 images showed endoscopic BE. After data augmentation, the training dataset expanded to 1,187 images, of which 563 did not show BE and 624 images showed endoscopic BE. The effects of data augmentation are listed in the Supplementary Table 1.
In this study, a transfer learning technique was applied for model training. Six pretrained models were compared, including EfficientNetV2B1, EfficientNetV2B2, EfficientNetV2B3, DenseNet201, ResNet50, and VGG16. Ultimately, EfficientNetV2B2 was selected as the backbone architecture for further evaluation due to its better prediction ACC.
The deep neural network in this study employed the sigmoid function as the activation function in the output layer. An early stopping technique was utilized to monitor the maximization of validation ACC, which allowed the model to stop training at a suitable convergence point. After completion of the training, the best model weights were saved. To further enhance the model’s generalization ability, the development set was repeatedly sampled to create multiple training sets for model training and validation, with the aim of developing an optimal AI model. A flow chart of the study design and data acquisition is shown in Figure 1.
The evaluation metrics applied in this study include ACC, sensitivity (SEN), and specificity (SPE), which determine the classification performance of the model. ACC indicates the classification ACC of the model, SEN reveals the proportion of actual positive samples correctly identified as positive, and SPE indicates the proportion of actual negative samples correctly identified as negative.
We collected endoscopic images of the gastroesophageal junction in a total of 724 cases, with 86 patients having complete histological results. There were 771 original images in the training set, including 563 images that did not show BE and 208 images that showed endoscopic BE (26.98% of the total number of images). A total of 193 original images were included in the validation set, including 141 images without BE and 52 images with endoscopic BE (26.94% of the validation images). Due to the imbalanced distribution in these two categories and the limited number of endoscopic BE images, we augmented the BE images in the training set to obtain a total of 624 images of endoscopic BE, which provided 52.57% of the images in the training set. These baseline characteristics are shown in Table 1.
We compared the prediction outcomes in different pre-trained models, which included EfficientNetV2B1, Effi-cientNetV2B2, EfficientNetV2B3, ResNet50, DenseNet201, and VGG16. ACC was comparable in the training process of most of the pre-trained models, with the exception of VGG16, which had an ACC of only 26.98%. The same trends were observed in the validation set, with an ACC of 78.76%, 84.97%, 78.24%, 82.90%, 77.72%, and 26.94% for EfficientNetV2B1, EfficientNetV2B2, EfficientNetV2B3, ResNet50, DenseNet201, and VGG16, respectively. For final testing, EfficientNetV2B2 achieved the best ACC of 85%, which was superior to the other pre-trained models; hence, we chose EfficientNetV2B2 as the training model for our BE prediction system. Detailed data on SEN, SPE, and ACC for the pre-trained model comparisons are shown in Table 2.
| Pre-trained model | Training | Validation | Test | ||||||
| ACC | SEN | SPE | ACC | SEN | SPE | ACC | SEN | SPE | |
| EfficientNetV2B1 | 0.9196 | 0.8702 | 0.9378 | 0.7876 | 0.6346 | 0.8440 | 0.7625 | 0.6857 | 0.8222 |
| EfficientNetV2B2 | 0.9702 | 0.9663 | 0.9716 | 0.8497 | 0.7500 | 0.8865 | 0.8500 | 0.8286 | 0.8667 |
| EfficientNetV2B3 | 0.9170 | 0.8798 | 0.9307 | 0.7824 | 0.6154 | 0.8440 | 0.8125 | 0.7429 | 0.8667 |
| ResNet50 | 0.8962 | 0.7885 | 0.9361 | 0.8290 | 0.5577 | 0.9291 | 0.7063 | 0.5429 | 0.8333 |
| DenseNet201 | 0.8755 | 0.8173 | 0.8970 | 0.7772 | 0.5577 | 0.8582 | 0.7312 | 0.6143 | 0.8222 |
| VGG16 | 0.2698 | 1.0000 | 0.0000 | 0.2694 | 1.0000 | 0.0000 | 0.4375 | 1.0000 | 0.0000 |
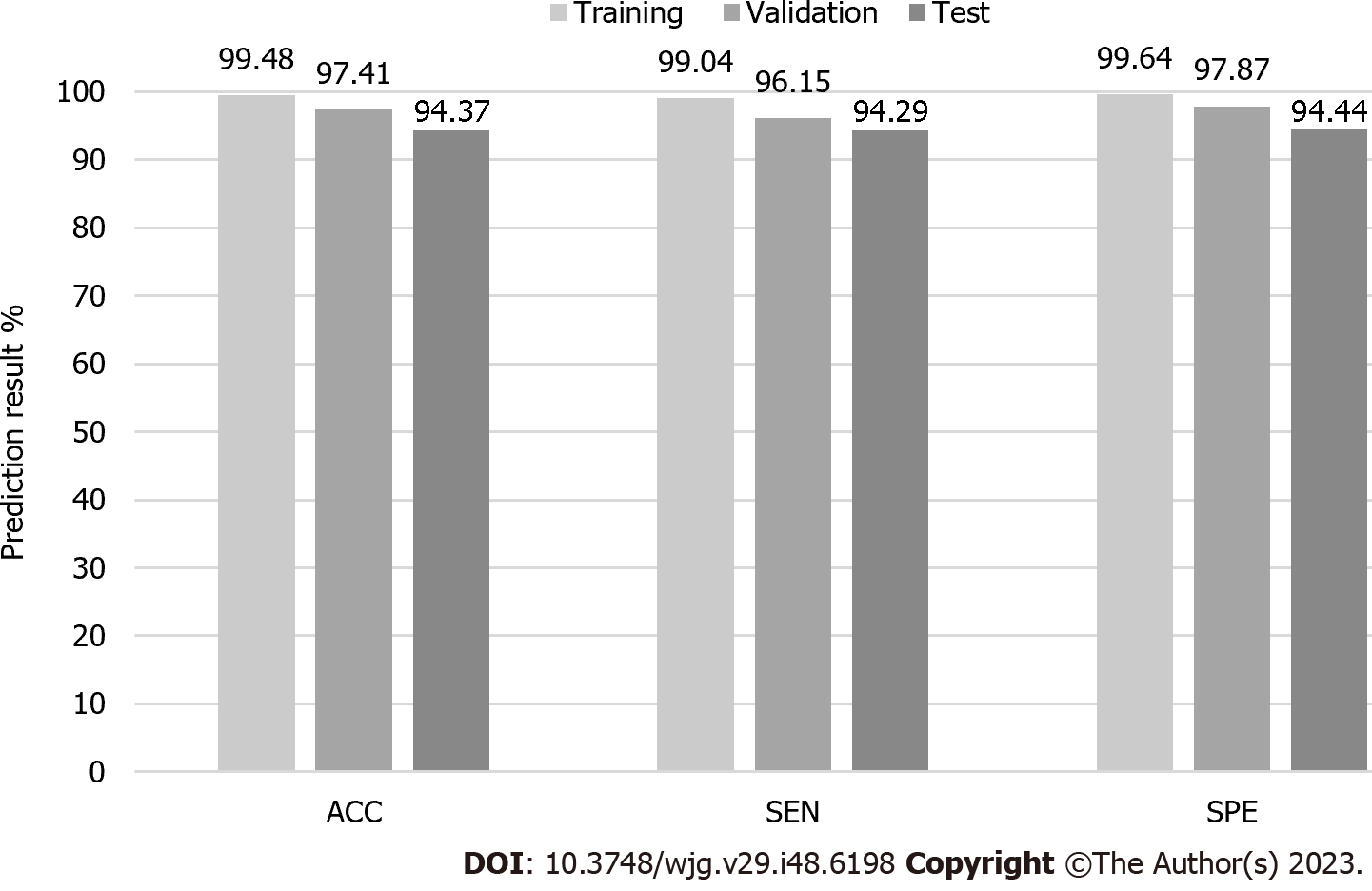
For the training set, two images were incorrectly classified as having no BE among 208 images of endoscopic BE. For images that showed no BE, two images were incorrectly classified as having BE, while the other 561 images were correctly classified as having BE. Overall, ACC, SEN, and SPE were 99.48%, 99.04%, and 99.64%, respectively.
For the validation set, two images were incorrectly classified as having no BE among 52 images of endoscopic BE, while three images were incorrectly classified as having BE among 141 non-BE images. Overall, ACC, SEN, and SPE were 97.41%, 96.15%, and 97.87%, respectively.
For the test set, four images were incorrectly classified as having no BE among 70 images of histological BE, and five images were incorrectly classified as having BE among 90 non-BE images (as proven by histology) in the final test. The AI BE prediction system had an ACC of 94.37%, a SEN of 94.29%, and a SPE of 94.44%. Detailed information is provided in Table 3 and Figure 2.
| Confusion matrices | Predicted class | ||||||
| Training, n = 771 | Validation, n = 193 | Test, n = 160 | |||||
| BE | No BE | BE | No BE | BE | No BE | ||
| Actual class | BE | 206 | 2 | 50 | 2 | 66 | 4 |
| No BE | 2 | 561 | 3 | 138 | 5 | 85 | |
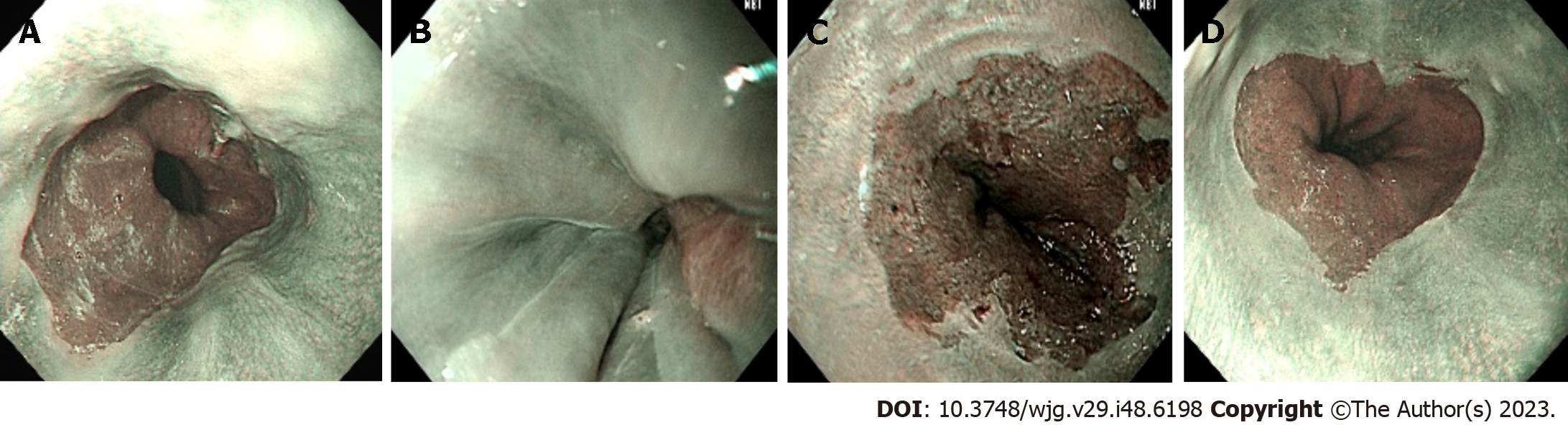
Representative images of BE with successful detection (panel A), no BE with successful detection (panel B), BE with false detection (panel C), and no BE with false detection (panel D) are demonstrated in Figure 3.
We used only NBI images from the Olympus endoscopy system in our study instead of white light endoscopy images because our previous data showed the superiority of NBI images over white light endoscopy images for the differentiation of GERD classification[28]. However, some studies have shown that NBI images do not display differences in detecting BE esophagus compared to white light endoscopy images[29,30], while another recent convincing study showed benefits in neoplasia detection with an NBI system[12]. The concept that targeted biopsies using NBI enable the detection of more dysplastic areas and thus reduce the number of biopsies required is widely accepted by the Japan Esophageal Society[12,31]. As a result, we exclusively utilized NBI still images in our AI prediction system training.
Our study is unique in that training of the pretrained model and selected AI prediction model was performed with images of endoscopic BE rather than images of histological BE. The AI prediction system was still able to maintain a high ACC in the final prediction test that used images of histological BE. This finding indicates that the images of endoscopic BE have characteristics similar to those of histological BE in our study. The final prediction results using the Effi-cientNetV2B2 training model showed that SEN, SPE, and ACC were 94.29%, 94.44%, and 94.37%, respectively. Previous data have highlighted that ACC is currently unsatisfied in AI systems of BE endoscopic interpretation[32], but the AI prediction of early cancer or dysplasia were impressive. Our database employed larger real-case numbers and more original images than previous studies and showed improved SEN, SPE, and ACC. We believe that the data augmentation technique limited the ACC in the final test results due to the large number of training images with similar detailed characteristics.
The most similar recent studies focused on BE neoplasia detection. Ebigbo et al[33] demonstrated an ACC of 89.9% using only 14 test patients in 2020, while Hussein et al[26] improved the ACC to 93% with 118 patients in 2022. Another study by Abdelrahim et al[25] in 2023, which utilized real-time video for the identification of BE dysplasia, showed a similar ACC of 92% with 75 cases.
Our study selected 771 original images from 579 patients as the training set, 193 original images from 145 patients as the validation set, and 160 images from 86 patients as the test set; the final ACC was 94.37%. Detailed information on these results is provided in Table 4. We focused on building an AI reminder system that helps endoscopists decide whether to perform a biopsy when BE is suspected instead of a system that reminds endoscopists which BE lesions may already have dysplastic changes.
| Ref. | Validation (train:valid:test) | Valid | Test |
| Ebigbo et al[33], 2020 | Holdout; Image (129:N/A:62); Test patients:14 | ACC: N/A | ACC: 89.9% |
| AUC: N/A | AUC: N/A | ||
| SEN: N/A | SEN: 83.7% | ||
| SPE: N/A | SPE: 100.0% | ||
| PPV: N/A | PPV: N/A | ||
| NPV: N/A | NPV: N/A | ||
| Hussein et al[26], 2022 | Holdout; Video (64:11:44); Patients: 118 | ACC: N/A | ACC: N/A |
| AUC: N/A | AUC: 93% | ||
| SEN: N/A | SEN: 91% | ||
| SPE: N/A | SPE: 79% | ||
| PPV: N/A | PPV: N/A | ||
| NPV: N/A | NPV: N/A | ||
| Abdelrahim et al[25], 2023 | Holdout; Image (816:471:N/A); Video (161:N/A:75); Case (161:34:75) | ACC: 94.7% | ACC: 92.0% |
| AUC: 0.898 | AUC: 0.964 | ||
| SEN: 95.3% | SEN: 93.8% | ||
| SPE: 94.5% | SPE: 90.7% | ||
| PPV: 83.6% | PPV: 88.2% | ||
| NPV: 98.6% | NPV: 95.1% | ||
| Our approach, 2023 | Holdout image; Image (771:193:160); Case (579:145:86) | ACC: 97.41% | ACC: 94.37% |
| AUC: 97.01% | AUC: 94.37% | ||
| SEN: 96.15% | SEN: 94.29% | ||
| SPE: 97.87% | SPE: 94.44% | ||
| PPV: 94.00% | PPV: 92.96% | ||
| NPV: 98.57% | NPV: 95.51% |
Due to the lack of awareness regarding the diagnosis of BE, cases of histological BE are relatively rare in central Taiwan. First, we initiated this study and collected endoscopic images of the lower esophagus and esophagogastric junction in two medical centers in central Taiwan. The endoscopic BE dataset was developed using the Delphi method, which involved blind voting on endoscopic images by three endoscopists, instead of histological BE images, which can lead to selection bias due to the previously known difference in prevalence between endoscopic BE and histologic BE[7] (7.8% vs. 1.3%). Second, the method of biopsy was not recorded in all cases, which meant that some of the cases involved random biopsies, while other cases were NBI-targeted biopsies. Third, the images in our database were obtained from an Olympus endoscopic system with lower esophageal still NBI pictures. It is not known whether our system can adequately perform for white light or other endoscopic systems. The main limitations were attributed to the limited cases and limited original images in our study and the limited number of cases of histological BE. However, our AI prediction system, which was trained by endoscopic BE images, still provided good prediction results for images of histological BE.
Our AI prediction system can provide good prediction results for images of histological BE obtained with Olympus NBI after training with images of endoscopic BE.
The prevalence of endoscopic Barrett’s esophagus (BE) differs significantly from histological BE. We believe the endoscopic characteristics are similar with endoscopic BE and histological BE.
We want to train an artificial intelligence (AI) system to identified images of BE under endoscopic environments.
To construct an AI system for the detection of endoscopic images of histological BE.
Endoscopic narrow-band images of 724 cases, were collected from two medical centers at central Taiwan, with 86 patients having pathological results. Images of endoscopic BE was classified using independent annotation by three senior endoscopists, who were instructing physicians of the Digestive Endoscopy Society of Taiwan. The test set consisted of 160 endoscopic images in 86 histological BE cases.
EfficientNetV2B2 [accuracy (ACC): 0.85] was selected as the backbone architecture from six training model due to better ACC result. In the final test, the AI system obtained 94.37%, 94.29%, and 94.44%, in ACC, sensitivity, and specificity respectively.
Our AI prediction system can provide good prediction results after training with images of endoscopic BE.
Our result implies that images of endoscopic BE share similar characteristics with images of histological BE even in the perspectives of AI system. The gap from endoscopic BE to histological BE maybe comes from biopsy or sampling bias. This opinion needs further prospective studies to confirm. Meanwhile, a better AI prediction system for endoscopic video BE detection is an ongoing task in the near future.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Taiwan
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C, C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Ma L, China; Mijwil MM, Iraq; Poddighe D, Kazakhstan; Shah OJ, India S-Editor: Lin C L-Editor: Filipodia P-Editor: Chen YX
| 1. | BARRETT NR. Chronic peptic ulcer of the oesophagus and 'oesophagitis'. Br J Surg. 1950;38:175-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 655] [Cited by in RCA: 546] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 2. | Balasubramanian G, Singh M, Gupta N, Gaddam S, Giacchino M, Wani SB, Moloney B, Higbee AD, Rastogi A, Bansal A, Sharma P. Prevalence and predictors of columnar lined esophagus in gastroesophageal reflux disease (GERD) patients undergoing upper endoscopy. Am J Gastroenterol. 2012;107:1655-1661. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 4. | Chang K, Jackson CS, Vega KJ. Barrett's Esophagus: Diagnosis, Management, and Key Updates. Gastroenterol Clin North Am. 2021;50:751-768. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 5. | DeMeester SR. Adenocarcinoma of the esophagus and cardia: a review of the disease and its treatment. Ann Surg Oncol. 2006;13:12-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 154] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 6. | Hayeck TJ, Kong CY, Spechler SJ, Gazelle GS, Hur C. The prevalence of Barrett's esophagus in the US: estimates from a simulation model confirmed by SEER data. Dis Esophagus. 2010;23:451-457. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 146] [Cited by in RCA: 129] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 7. | Shiota S, Singh S, Anshasi A, El-Serag HB. Prevalence of Barrett's Esophagus in Asian Countries: A Systematic Review and Meta-analysis. Clin Gastroenterol Hepatol. 2015;13:1907-1918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 66] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 8. | Desai TK, Krishnan K, Samala N, Singh J, Cluley J, Perla S, Howden CW. The incidence of oesophageal adenocarcinoma in non-dysplastic Barrett's oesophagus: a meta-analysis. Gut. 2012;61:970-976. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 423] [Cited by in RCA: 414] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 9. | Singh S, Manickam P, Amin AV, Samala N, Schouten LJ, Iyer PG, Desai TK. Incidence of esophageal adenocarcinoma in Barrett's esophagus with low-grade dysplasia: a systematic review and meta-analysis. Gastrointest Endosc. 2014;79:897-909.e4; quiz 983.e1, 983.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 160] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 10. | Rastogi A, Puli S, El-Serag HB, Bansal A, Wani S, Sharma P. Incidence of esophageal adenocarcinoma in patients with Barrett's esophagus and high-grade dysplasia: a meta-analysis. Gastrointest Endosc. 2008;67:394-398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 313] [Cited by in RCA: 292] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 11. | Wang KK, Sampliner RE; Practice Parameters Committee of the American College of Gastroenterology. Updated guidelines 2008 for the diagnosis, surveillance and therapy of Barrett's esophagus. Am J Gastroenterol. 2008;103:788-797. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 850] [Cited by in RCA: 786] [Article Influence: 46.2] [Reference Citation Analysis (1)] |
| 12. | Vithayathil M, Modolell I, Ortiz-Fernandez-Sordo J, Oukrif D, Pappas A, Januszewicz W, O'Donovan M, Hadjinicolaou A, Bianchi M, Blasko A, White J, Kaye P, Novelli M, Wernisch L, Ragunath K, di Pietro M. Image-Enhanced Endoscopy and Molecular Biomarkers Vs Seattle Protocol to Diagnose Dysplasia in Barrett's Esophagus. Clin Gastroenterol Hepatol. 2022;20:2514-2523.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 13. | Chen YH, Yu HC, Lin KH, Lin HS, Hsu PI. Prevalence and risk factors for Barrett's esophagus in Taiwan. World J Gastroenterol. 2019;25:3231-3241. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 14] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 14. | Dong L, He W, Zhang R, Ge Z, Wang YX, Zhou J, Xu J, Shao L, Wang Q, Yan Y, Xie Y, Fang L, Wang H, Wang Y, Zhu X, Wang J, Zhang C, Chen R, Wan Q, Yang J, Zhou W, Li H, Yao X, Yang Z, Xiong J, Wang X, Huang Y, Chen Y, Wang Z, Rong C, Gao J, Zhang H, Wu S, Jonas JB, Wei WB. Artificial Intelligence for Screening of Multiple Retinal and Optic Nerve Diseases. JAMA Netw Open. 2022;5:e229960. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 72] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 15. | Date RC, Jesudasen SJ, Weng CY. Applications of Deep Learning and Artificial Intelligence in Retina. Int Ophthalmol Clin. 2019;59:39-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 16. | Ellahham S. Artificial Intelligence: The Future for Diabetes Care. Am J Med. 2020;133:895-900. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 120] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 17. | Frizzell TO, Glashutter M, Liu CC, Zeng A, Pan D, Hajra SG, D'Arcy RCN, Song X. Artificial intelligence in brain MRI analysis of Alzheimer's disease over the past 12 years: A systematic review. Ageing Res Rev. 2022;77:101614. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 52] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 18. | Byrne MF, Chapados N, Soudan F, Oertel C, Linares Pérez M, Kelly R, Iqbal N, Chandelier F, Rex DK. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut. 2019;68:94-100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 363] [Cited by in RCA: 412] [Article Influence: 68.7] [Reference Citation Analysis (0)] |
| 19. | Le Berre C, Sandborn WJ, Aridhi S, Devignes MD, Fournier L, Smaïl-Tabbone M, Danese S, Peyrin-Biroulet L. Application of Artificial Intelligence to Gastroenterology and Hepatology. Gastroenterology. 2020;158:76-94.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 324] [Article Influence: 64.8] [Reference Citation Analysis (1)] |
| 20. | Lee JH, Kim YJ, Kim YW, Park S, Choi YI, Park DK, Kim KG, Chung JW. Spotting malignancies from gastric endoscopic images using deep learning. Surg Endosc. 2019;33:3790-3797. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 61] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 21. | Charisis VS, Hadjileontiadis LJ. Potential of hybrid adaptive filtering in inflammatory lesion detection from capsule endoscopy images. World J Gastroenterol. 2016;22:8641-8657. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 26] [Cited by in RCA: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 22. | Pereira SP, Oldfield L, Ney A, Hart PA, Keane MG, Pandol SJ, Li D, Greenhalf W, Jeon CY, Koay EJ, Almario CV, Halloran C, Lennon AM, Costello E. Early detection of pancreatic cancer. Lancet Gastroenterol Hepatol. 2020;5:698-710. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 303] [Article Influence: 60.6] [Reference Citation Analysis (1)] |
| 23. | Chang D, Chen PT, Wang P, Wu T, Yeh AY, Lee PC, Sung YH, Liu KL, Wu MS, Yang D, Roth H, Liao WC, Wang W. Detection of pancreatic cancer with two- and three-dimensional radiomic analysis in a nationwide population-based real-world dataset. BMC Cancer. 2023;23:58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 24. | Dumoulin FL, Rodriguez-Monaco FD, Ebigbo A, Steinbrück I. Artificial Intelligence in the Management of Barrett's Esophagus and Early Esophageal Adenocarcinoma. Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 25. | Abdelrahim M, Saiko M, Maeda N, Hossain E, Alkandari A, Subramaniam S, Parra-Blanco A, Sanchez-Yague A, Coron E, Repici A, Bhandari P. Development and validation of artificial neural networks model for detection of Barrett's neoplasia: a multicenter pragmatic nonrandomized trial (with video). Gastrointest Endosc. 2023;97:422-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 32] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 26. | Hussein M, González-Bueno Puyal J, Lines D, Sehgal V, Toth D, Ahmad OF, Kader R, Everson M, Lipman G, Fernandez-Sordo JO, Ragunath K, Esteban JM, Bisschops R, Banks M, Haefner M, Mountney P, Stoyanov D, Lovat LB, Haidry R. A new artificial intelligence system successfully detects and localises early neoplasia in Barrett's esophagus by using convolutional neural networks. United European Gastroenterol J. 2022;10:528-537. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 27] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 27. | Horie Y, Yoshio T, Aoyama K, Yoshimizu S, Horiuchi Y, Ishiyama A, Hirasawa T, Tsuchida T, Ozawa T, Ishihara S, Kumagai Y, Fujishiro M, Maetani I, Fujisaki J, Tada T. Diagnostic outcomes of esophageal cancer by artificial intelligence using convolutional neural networks. Gastrointest Endosc. 2019;89:25-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 272] [Article Influence: 45.3] [Reference Citation Analysis (0)] |
| 28. | Wang CC, Chiu YC, Chen WL, Yang TW, Tsai MC, Tseng MH. A Deep Learning Model for Classification of Endoscopic Gastroesophageal Reflux Disease. Int J Environ Res Public Health. 2021;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 29. | Maes S, Sharma P, Bisschops R. Review: Surveillance of patients with Barrett oesophagus. Best Pract Res Clin Gastroenterol. 2016;30:901-912. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 30. | Naveed M, Dunbar KB. Endoscopic imaging of Barrett's esophagus. World J Gastrointest Endosc. 2016;8:259-266. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 6] [Cited by in RCA: 10] [Article Influence: 1.1] [Reference Citation Analysis (1)] |
| 31. | Ishihara R, Goda K, Oyama T. Endoscopic diagnosis and treatment of esophageal adenocarcinoma: introduction of Japan Esophageal Society classification of Barrett's esophagus. J Gastroenterol. 2019;54:1-9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 32. | Madabhushi A, Toro P, Willis JE. Artificial Intelligence in Surveillance of Barrett's Esophagus. Cancer Res. 2021;81:3446-3448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Reference Citation Analysis (0)] |
| 33. | Ebigbo A, Mendel R, Probst A, Manzeneder J, Prinz F, de Souza LA Jr, Papa J, Palm C, Messmann H. Real-time use of artificial intelligence in the evaluation of cancer in Barrett's oesophagus. Gut. 2020;69:615-616. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 124] [Article Influence: 24.8] [Reference Citation Analysis (0)] |